Abstract
The sterilising cytoplasm from Triticum timopheevii is presently considered to be the most promising as regards to the seed production of triticale hybrid cultivars. This study was aimed at the utilisation of Diversity Arrays Technology (DArT) for the preliminary identification of genomic regions with loci controlling male sterility/fertility in the field-grown F2 generation of the interline hybrid between male sterile line CMS-Salvo 15/1 and restorer line Stan I. The fertility of plants was examined by visual scoring as well as by the assessment of seed setting within bagged spikes. For DNA analyses, 92 individuals representing opposite phenotypes (male sterile vs. fully male fertile) were chosen from the whole F2 population, which consisted of 414 plants. The constructed genetic map consists of 759 DArT markers distributed in 24 linkage groups that cover a distance of 974.4 cM. Application of the interval mapping method and the Kruskal–Wallis test enabled the identification of six genomic regions engaged in the restoration of male fertility within the mapping population. The most effective restorer genes were found on chromosomes of the sixth homeologic group, i.e. on 6R (the most efficient), 6A and 6B. Additionally, linkage groups assigned to chromosomes 1BS, 3A and 3A/3B were important for the determination of male fertility.
Keywords: Cytoplasmic male sterility, Triticale, Triticum timopheevii cytoplasm
Triticale (× Triticosecale Wittm.) is an artificial allo-hexaploid species obtained by breeders as a result of intergeneric crosses between wheat and rye. The first cultivars of this crop were registered over 50 years ago. Recently, this cereal has become successively more common year on year. Over the last two decades, breeders of triticale have become increasingly more interested in the exploration of heterosis (Oettler et al. 2003, 2005; Góral et al. 2005; Tams et al. 2005). Practicable organisation of seed production for hybrid cultivars could be achieved via application of the cytoplasmic male sterility (CMS) system in triticale. Among the few sources of sterilising cytoplasm available for this crop, the cytoplasm of Triticum timopheevii Zhuk. is considered to be one of the most promising, due to its small number of deleterious effects (Cauderon et al. 1985; Nalepa 1990; Spiss and Góral 1994). Utilisation of the T. timopheevii CMS system in the breeding of hybrid cultivars may be strongly facilitated by a better understanding of the genetic mechanisms leading to microsporogenesis disorders in triticale plants carrying this cytoplasmic system. Unfortunately, current knowledge about the genetic background of male sterility in this cereal is very limited. The majority of research concerning genes restoring male fertility in plants with T. timopheevii cytoplasm is focused on wheat. The cytoplasm originating from Triticum timopheevii is also important in wheat hybrid cultivar breeding programmes, as it leads to male sterility symptoms in this cereal. Curtis and Lukaszewski (1993) reported on the capability of a rye gene located on the long arm of the 6R chromosome to restore male fertility in wheat with the T. timopheevii cytoplasm. The same authors indicated that the next less effective restorer gene is located on the 4RL chromosome. Several genes controlling the production of pollen in the CMS–T. timopheevii system have been reported on six chromosomes of Triticum aestivum L. (Rf1-1A; Rf2-7D; Rf3-1B; Rf4-6B; Rf5-6D; Rf7-7B) and the effective Rf3 locus located on the 1BS chromosome has been precisely mapped (Ma and Sorrells 1995; Kojima et al. 1997). Quantitative trait loci (QTL) analysis allowed for the localisation of additional less efficient restorers on chromosomes 2A, 4B and 6A (Ahmed et al. 2001). In rye (the second ancestor of the triticale), QTL analyses performed on F2 mapping populations resulted in the detection of genes restoring male fertility in CMS systems on chromosomes 1R, 3R, 4R, 5R and 6R (Miedaner et al. 2000; Stojałowski et al. 2004).
Different molecular markers, like RAPD, ISSR, SSR, AFLP and SCAR, were applied for genetic research on triticale (Masojć 2000; Stojałowski and Góral 2002; Góral et al. 2005; Tams et al. 2005; Stojałowski et al. 2006) but none of these techniques resulted in the construction of a genetic map. The development of Diversity Arrays Technology (DArT, Jaccoud et al. 2001) opened up new possibilities in the exploration of triticale genome, because it allowed for the construction of the first genetic maps covering the entire genome of this artificial species (Tyrka et al. 2011; Alheit et al. 2011). This study was aimed at the application of DArT markers for mapping the triticale genome, as well as the utilisation of a developed linkage map for the localisation of genes restoring male fertility in plants carrying the T. timopheevii cytoplasm.
The F2 population used in this study consisted of 414 individuals and was developed by the pollination of male sterile inbred line CMS-Salvo 15/1 by restorer line Stan I. The male sterile maternal component of the cross was kindly provided by H. Góral (The Agricultural University in Krakow, Poland) and the pollinator line was obtained from A. Łukaszewski (University of California, Riverside, CA, USA). All individuals of F1 progeny were fully male fertile. Seeds of the F2 population were harvested after the self-fertilisation of one F1 plant.
Individual plants of the F2 progeny were grown with a 25 × 25-cm spacing during the 2010/11 vegetation season in a field belonging to the West Pomeranian University of Technology in Szczecin, Poland. In two subsequent days with stable weather conditions, the evaluation of male fertility was conducted via visual scoring of two to three spikes per plant at the flowering stage, according to the 5-step scale proposed by Góral et al. (2006), where 1 corresponds to absolute sterility and 5 to full fertility of anthers. In addition, three to five spikes of each plant were bagged before flowering and visual observations of fertility were verified after harvest, by the evaluation of seed setting under isolation.
DNA was isolated from young leaves harvested in early spring and kept in a deep freezer at −70 °C. The GenElute Plant Mini Kit from Sigma-Aldrich was employed for this purpose. Marker analyses were performed on parental lines, as well as on 92 individuals taken from the segregating F2 population. The group of segregants consisted of plants representative of two opposite phenotypes: male sterile (MS) and male fertile (MF). As only 12 fully sterile individuals were identified in the whole population, the MS group was supplemented by 26 partially male sterile plants that produced few (1–19) grains per spike. The MF group consisted of 54 completely fertile individuals (producing not less than 54 grains per spike and visually scored as 5 on the valuation scale). Only plants revealing a consistent assessment by visual scoring and observation of seed setting under bags were considered for the marker analyses. These were performed with the use of DArT (Jaccaud et al. 2001).
The genetic map of triticale was constructed with the use of JoinMap® 3.0 software (Van Ooijen and Voorrips 2001). The localisation of DArT markers on triticale and rye genetic maps reported by Alheit et al. (2011), Milczarski et al. (2011) and Tyrka et al. (2011) were applied for the assignment of linkage groups to entire chromosomes. For grouping analyses, the LOD = 4.0 level was initially established, but in some cases, when markers formerly mapped on different chromosomes were included in the same linkage group, the LOD level was increased to 5.0 or 6.0.
Agreement of the distribution of male sterility within the studied population (data obtained by visual scoring and analysis of seed setting under bags) with the normal population was tested using the Lilliefors test. Interval mapping, the permutation test and the Kruskal–Wallis test (K-W test) were performed with the use of MapQTL® 5.0 software, revealing relationships between segregations of molecular markers and male fertility restoration (Van Ooijen 2004). Putative localisation of the gene(s) controlling male fertility restoration was considered when the LOD value exceeded 3.0, and additional thresholds of significance for declaring the presence of a given QTL were estimated from 1,000 permutations of the data (Doerge and Churchill 1996). As suggested by the author of the software (Van Ooijen 2004), during analyses using the K-W test, molecular markers were considered to be significantly associated with the studied trait if P < 0.01.
Within the studied F2 population containing over 400 individuals, high phenotypic variation was observed. The number of kernels produced within bagged spikes ranged from 0 to 103. Twelve plants were fully male sterile. Partially male sterile plants were also not numerous, leading to a limited number of MS genotypes available for marker analyses. About 20 % of the population consisted of fully fertile individuals, but the most numerous group contained plants classified as partially male fertile (over half of the entire studied population). The results of visual assessment and analyses of seed setting in isolated spikes were consistent in the majority of cases, but some contradictories were also noticed (such individuals were not considered for mapping analyses). Generally, the correlation coefficient between results obtained by two applied methods of sterility/fertility estimation was 0.55. The distribution of male sterility/fertility (independent of the method of assessment—visual vs. seed setting on bagged ears) deviated significantly from the normal distribution, but these deviations were relatively small in the case of seed setting data (data not shown). For this reason, the results of seed setting under bags were chosen for the performance of an interval mapping analysis of loci responsible for the restoration of male fertility.
DArT resulted in 822 polymorphic markers. The segregation of 57 DArT markers deviated significantly from the expected 3:1 ratio, but due to the selection of individuals for mapping from groups of opposite phenotypes, these markers were not excluded from the linkage analyses. Grouping analysis allowed for the construction of 24 linkage maps (Fig. 1). The number of markers within particular linkage groups varied widely: chromosome 1A was represented by only four linked markers but 99 loci of DArT markers were located on the 6R chromosome. No group assigned to the 4A chromosome was obtained. Due to the homeology of chromosomes within the triticale genome, some linkage maps could not be clearly associated with a given chromosome. The coverage of rye chromosomes by DArT markers was significantly more efficient than chromosomes belonging to the wheat A and B genomes. In total, the constructed genetic map of the triticale genome contains 759 DArT markers and cumulatively spans a distance of 974.4 cM (Fig. 1).
Fig. 1.
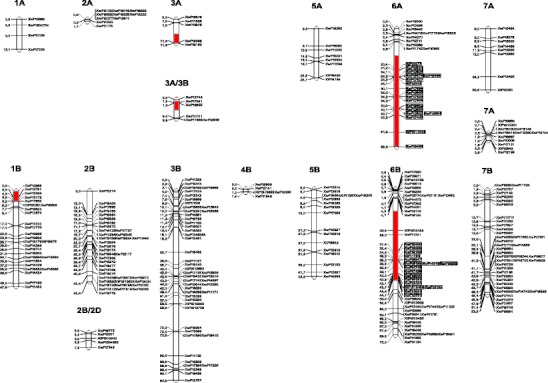
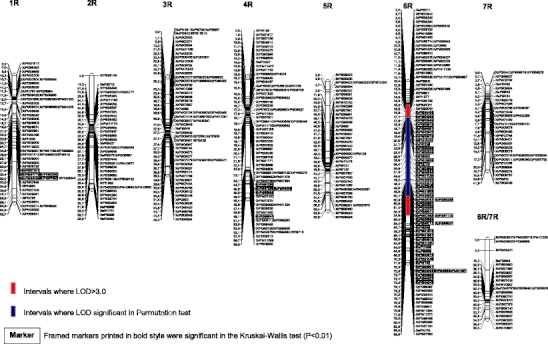
Linkage groups of Diversity Arrays Technology (DArT) markers and localisation of genomic regions determining male fertility restoration in the F2 generation of the cross [CMS-Salvo 15/1 × Stan I] of winter triticale with the Triticum timopheevii cytoplasm
Interval mapping analysis revealed six genomic regions associated with the determination of male fertility of triticale with T. timopheevii cytoplasm (Table 1, Fig. 1). The most effective genes were identified on chromosomes 6A, 6B and 6R. Their estimated additive effects ranged from approximately 20 to 30 grains per spike (differences in kernel number produced within a bagged spike) and restorer alleles originating from paternal component (line Stan I) of the studied hybrid. All these QTL detected using the interval mapping method were confirmed when the non-parametric K-W test was applied (Table 1). The most efficient as regards to the restoration of fertility seems to be a gene located on the 6R chromosome-the only gene that was statistically significant when the permutation test was utilised. The next two loci detected by means of the interval mapping method were located on chromosome 1B and the linkage group ambiguously assigned to chromosomes 3A or 3B. Both genes revealed an additive effect at approximately 10 kernels per self-pollinated ear, but alleles increasing male fertility seemed to be introduced by the maternal line CMS-Salvo 15/1. Localisations of these genes were not confirmed by the K-W test. The less effective QTL was detected by the interval mapping method (but not by the K-W test) on the 3A linkage group. Calculated variance explained by all of the identified QTL exceeded 20 %, but these values are probably overestimated for two reasons: the lack of co-dominant markers on the linkage maps and the selection of opposite phenotypes for map construction and interval mapping analyses. Some additional markers not indicated by the interval mapping method were considered as being significantly associated with male fertility by the K-W test. These markers were located on chromosomes 5B, 1R and 4R (Fig. 1).
Table 1.
Characteristics of quantitative trait loci (QTL) controlling the restoration of fertility in the [CMS-Salvo 15/1 × Stan I] F2 intercross (results based on the seed setting data)
| Chromosome | Interval mapping | Kruskal–Wallis test | ||||||
|---|---|---|---|---|---|---|---|---|
| Interval (cM)a | Maximum value of LOD (position in cM)b | Additive effect of maternal allele | Variance explained (%) | Kmax c | Marker revealing the max. value of the K statistic | Mean value of the genotype CMS-Salvo 15/1 | Mean value of genotype Stan I | |
| 3A | 7.91–11.86 | 13.08 (7.91 cM) | 1.59 | 93.3 | 1.10NS | XwPt0398 | 33.5 | 44.3 |
| 3A/3B | 1.00–4.89 | 3.97 (3.89 cM) | 13.84 | 77.3 | 0.50NS | XwPt8855 | 42.5 | 37.4 |
| 6A | 14.8–59.2 | 6.03 (44.2 cM) | −23.51 | 28.8 | 18.86** | XwPt7445 | 16.5 | 50.3 |
| 1B | 0.00–5.30 | 6.72 (1.5 cM) | 10.77 | 79.8 | 5.08NS | XwPt3172 | 28.6 | 45.3 |
| 6B | 11.7–44.7 | 4.64 (27.0 cM) | −20.09 | 25.6 | 11.85* | XwPt1241 | 22.5 | 52.8 |
| 6R | 11.4–64.8 | 21.08 (29.2 cM) | −30.00 | 93.6 | 33.18** | XtPt3774 | 15.5 | 61.5 |
aMapping interval where LOD > 3.0
bEntry in bold indicates significance in the permutation test
c K max maximum value of the K statistic within the interval; NS not significant; *Significant at P < 0.001; **Significant at P < 0.0001
The restoration of male fertility in triticale with T. timopheevii cytoplasm remains under the control of several Rf genes (Góral et al. 2010), but information about their characterisation and localisation is still very limited. The present study attempted to broaden knowledge concerning this problem. It should be noted, however, that the above results were obtained on the basis of the phenotyping of individual plants in only one environment. The significance of genotype by environment interactions for the detection of QTL controlling different traits is usually investigated by the assessment of “immortal” mapping populations (recombinant inbred lines, DH lines) or by the verification of results from F2 in the F3 generation. Unfortunately, research on male sterility restoration is frequently limited to the one generation (Miedaner et al. 2000; Stojałowski et al. 2004) because the reproduction of male sterile genotypes by self-pollination is impossible. Application of the partial F3 generation obtained from only fertile plants can be recommended for the identification of heterozygous F2 plants, which is reasonable for investigations on one effective restorer gene (Börner et al. 1998; Hackauf et al. 2012). The assessment of male fertility in different localisations but in the same year of study is sometimes performed by vegetative cloning of individual plants (Miedaner et al. 2000; Hackauf et al. 2012). The sensitivity of pollen production to variable environmental conditions (Góral et al. 2006) leads to the recommendation that the male sterility/fertility of triticale plants carrying a sterilising cytoplasm should be assessed in different environments. On the other hand, it was stated also that donors of the most effective non-restoring alleles are environmentally insensitive genotypes (Góral et al. 2006). Geiger et al. (1995) reported that the influence of environment on male sterility/fertility in rye is the most significant for partially fertile individuals. This rule probably also applies to triticale. Therefore, with the aim to increase the proportion of variation affected only by plant genotype, we decided to ignore all intermediates and to analyse, via molecular markers, only extreme (male sterile and male fertile) phenotypes selected within the studied mapping population. Additionally, Carey and Williamson (1991) as well as Xu et al. (2005) indicated that the selection of individuals representing opposite phenotypes from the whole F2 population should significantly increase the powerful detection of QTL controlling the studied trait.
Obviously, the results of the present study should be considered as preliminary; however, their close coincidence with previous reports investigating the localisation of restorer genes in T. timopheevii cytoplasm is intriguing. To the present day, the only research focusing on the localisation of rye genome genes acting as restorers in T. timopheevii cytoplasm indicated the importance of 6RL and 4RL chromosomes (Curtis and Lukaszewski 1993). In wheat with the same type of sterilising cytoplasm, chromosomes 1BS, 2A, 4B and 6A were reported as containing Rf loci (Ma and Sorrells 1995; Kojima et al. 1997; Ahmed et al. 2001). The results of our study on the cross [CMS-Salvo 15/1 × Stan I] confirm that the 6R chromosome is the most significant as regards to male sterility restoration in triticale with the T. timopheevii cytoplasm. Within the studied population, the remaining chromosomes of the sixth homeologic group (6A and 6B) were also found to be important for male fertility. The lowest effect on pollen fertility was identified for loci located on the 1B and 3A (optionally 3B) chromosomes. The short arm of chromosome 1B might carry a restorer gene Rf3, but this effective allele introduced into wheat from Triticum spelta (Kojima et al. 1997; Ahmed et al. 2001) was not detected among the hybrid F2 generation investigated in the present study.
The presented research is of preliminary character and its data need to be verified in further studies on a broader plant material (e.g. vegetative clones of F2 plants or DH lines carrying normal cytoplasm analysed by test-crosses with CMS sources) examined under different environmental habitats. Nevertheless, the initial results obtained appear to confirm a complex genetic control of male fertility restoration in triticale with T. timopheevii cytoplasm previously reported by Góral et al. (2010). It is noticeable also that genes located on all chromosomes of the sixth homeologic group are important for the abundance of pollen production and the 6R chromosome is especially significant for this phenomenon.
References
- Ahmed TA, Tsujimoto H, Sasakuma T. QTL analysis of fertility-restoration against cytoplasmic male sterility in wheat. Genes Genet Syst. 2001;76:33–38. doi: 10.1266/ggs.76.33. [DOI] [PubMed] [Google Scholar]
- Alheit KV, Reif JC, Maurer HP, Hahn V, Weissmann EA, Miedaner T, Würschum T. Detection of segregation distortion loci in triticale (× Triticosecale Wittmack) based on a high-density DArT marker consensus genetic linkage map. BMC Genomics. 2011;12:380. doi: 10.1186/1471-2164-12-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner A, Korzun V, Polley A, Malyshev S, Melz G. Genetics and molecular mapping of a male fertility restoration locus (Rfg1) in rye (Secale cereale L.) Theor Appl Genet. 1998;97:99–102. doi: 10.1007/s001220050871. [DOI] [Google Scholar]
- Carey G, Williamson J. Linkage analysis of quantitative traits: increased power by using selected samples. Am J Hum Genet. 1991;49:786–796. [PMC free article] [PubMed] [Google Scholar]
- Cauderon Y, Cauderon A, Gay G, Roussel J (1985) Alloplasmic lines and nucleo-cytoplasmic interactions in triticale. In: Genetics and breeding of triticale. EUCARPIA Meeting, Clermont-Ferrand, France, July 2–5 1984. INRA, Paris, pp 177–191
- Curtis CA, Lukaszewski AJ. Localization of genes in rye that restore male fertility to hexaploid wheat with timopheevi cytoplasm. Plant Breeding. 1993;111:106–112. doi: 10.1111/j.1439-0523.1993.tb00615.x. [DOI] [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger HH, Yuan Y, Miedaner T, Wilde P. Environmental sensitivity of cytoplasmic genic male sterility (CMS) in Secale cereale L. In: Kück U, Wricke G, editors. Genetic mechanisms for hybrid breeding. Advances in plant breeding 18. Berlin: Blackwell Wissenschafts-Verlag; 1995. pp. 7–17. [Google Scholar]
- Góral H, Tyrka M, Spiss L. Assessing genetic variation to predict the breeding value of winter triticale cultivars and lines. J Appl Genet. 2005;46:125–131. [PubMed] [Google Scholar]
- Góral H, Warzecha T, Stojałowski S, Pojmaj M, Kurleto D, Trąbka A, Spiss L. Stability of male sterility and fertility restoration in the CMS T. timopheevi system in triticale. Folia Univ Agric Stetin. 2006;247:55–62. [Google Scholar]
- Góral H, Stojałowski S, Tyrka M, Wędzony M. Inheritance of fertility restoration in winter triticale with cytoplasm of Triticum timopheevi. Folia Pomer Univ Technol Stetin. 2010;276:11–18. [Google Scholar]
- Hackauf B, Korzun V, Wortmann H, Wilde P, Wehling P. Development of conserved ortholog set markers linked to the restorer gene Rfp1 in rye. Mol Breed. 2012;30:1507–1518. doi: 10.1007/s11032-012-9736-5. [DOI] [Google Scholar]
- Jaccoud D, Peng K, Feinstein D, Kilian A. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001;29:E25. doi: 10.1093/nar/29.4.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T, Tsujimoto H, Ogihara Y. High-resolution RFLP mapping of the fertility restoration (Rf3) gene against Triticum timopheevi cytoplasm located on chromosome 1BS of common wheat. Genes Genet Syst. 1997;72:353–359. doi: 10.1266/ggs.72.353. [DOI] [Google Scholar]
- Ma Z-Q, Sorrells ME. Genetic analysis of fertility restoration in wheat using restriction fragment length polymorphisms. Crop Sci. 1995;35:1137–1143. doi: 10.2135/cropsci1995.0011183X003500040037x. [DOI] [Google Scholar]
- Masojć P. Identification of triticale cultivars by means of RAPD markers. Folia Univ Agric Stetin. 2000;206:179–183. [Google Scholar]
- Miedaner T, Glass C, Dreyer F, Wilde P, Wortmann H, Geiger HH. Mapping of genes for male-fertility restoration in ‘Pampa’ CMS winter rye (Secale cereale L.) Theor Appl Genet. 2000;101:1226–1233. doi: 10.1007/s001220051601. [DOI] [Google Scholar]
- Milczarski P, Bolibok-Brągoszewska H, Myśków B, Stojałowski S, Heller-Uszyńska K, Góralska M, Brągoszewski P, Uszyński G, Kilian A, Rakoczy-Trojanowska M. A high density consensus map of rye (Secale cereale L.) based on DArT markers. PLoS One. 2011;6(12):e28495. doi: 10.1371/journal.pone.0028495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalepa S (1990) Hybrid triticale: present and future. In: Proceedings of the 2nd International Triticale Symposium, Passo Fundo, Rio Grande do Sul, Brazil, October 1–5 1990. CIMMYT, Mexico, pp 402–407
- Oettler G, Burger H, Melchinger AE. Heterosis and combining ability for grain yield and other agronomic traits in winter triticale. Plant Breed. 2003;122:318–321. doi: 10.1046/j.1439-0523.2003.00877.x. [DOI] [Google Scholar]
- Oettler G, Tams SH, Utz HF, Bauer E, Melchinger AE. Prospects for hybrid breeding in winter triticale: I. Heterosis and combining ability for agronomic traits in European elite germplasm. Crop Sci. 2005;45:1476–1482. doi: 10.2135/cropsci2004.0462. [DOI] [Google Scholar]
- Spiss L, Góral H. Breeding male sterile and fertility restoring forms in triticale. Zesz Nauk AR Szczecin. 1994;162:243–246. [Google Scholar]
- Stojałowski S, Góral H. The use of RAPD and ISSR markers for differentiation of CMS-lines of winter triticale with T. timopheevi cytoplasm. Folia Univ Agric Stetin. 2002;228:161–166. [Google Scholar]
- Stojałowski S, Łapiński M, Masojć P. RAPD markers linked with restorer genes for the C-source of cytoplasmic male sterility in rye (Secale cereale L.) Plant Breed. 2004;123:428–433. doi: 10.1111/j.1439-0523.2004.01009.x. [DOI] [Google Scholar]
- Stojałowski S, Drozd A, Jaciubek M, Masojć P. Polymorphism of SCAR and SSR markers from rye genome in cultivars and lines of triticale. Folia Univ Agric Stetin. 2006;247:193–200. [Google Scholar]
- Tams SH, Melchinger AE, Bauer E. Genetic similarity among European winter triticale elite germplasms assessed with AFLP and comparisons with SSR and pedigree data. Plant Breed. 2005;124:154–160. doi: 10.1111/j.1439-0523.2004.01047.x. [DOI] [Google Scholar]
- Tyrka M, Bednarek PT, Kilian A, Wędzony M, Hura T, Bauer E. Genetic map of triticale compiling DArT, SSR, and AFLP markers. Genome. 2011;54:391–401. doi: 10.1139/g11-009. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW. MapQTL® 5. Software for the mapping of quantitative trait loci in experimental populations. Wageningen: Kyazma B.V.; 2004. [Google Scholar]
- Van Ooijen JW, Voorrips RE. JoinMap® 3.0. Software for the calculation of genetic linkage maps. Wageningen: Plant Research International; 2001. [Google Scholar]
- Xu Z, Zou F, Vision TJ. Improving quantitative trait loci mapping resolution in experimental crosses by the use of genotypically selected samples. Genetics. 2005;170:401–408. doi: 10.1534/genetics.104.033746. [DOI] [PMC free article] [PubMed] [Google Scholar]


