Abstract
Chromosome identification using fluorescence in situ hybridization (FISH) is widely used in cytogenetic research. It is a diagnostic tool helpful in chromosome identification. It can also be used to characterize alien introgressions, when exercised in a combination with genomic in situ hybridization (GISH). This work aims to find chromosome identification of Aegilops species and Aegilops × Secale amphiploids, which can be used in cereal breeding as a source of favourable agronomic traits. Four diploid and two tetraploid Aegilops species and three Aegilops × Secale hybrids were analysed using FISH with pSc119.2, pAs1, 5S rDNA and 25S rDNA clones to differentiate the U-, M-, Ssh- and D-subgenome chromosomes of Aegilops genus. Additionally, GISH for chromosome categorization was carried out. Differences in the hybridization patterns allowed to identify all U-, M-, Ssh- and D-subgenome chromosomes. Some differences in localization of the rDNA, pSc119.2 and pAs1 sequences between analogue subgenomes in diploid and tetraploid species and Aegilops × Secale hybrids were detected. The hybridization pattern of the M and S genome was more variable than that of the U and D genome. An importance of the cytogenetic markers in plant breeding and their possible role in chromosome structure, function and evolution is discussed.
Keywords: Aegilops species, Aegilops × Secale hybrids, Cytogenetic markers, Fluorescence and genomic in situ hybridization
Introduction
Interspecific hybridization is the most common method used to transfer genes of agronomic value from wild relatives into cultivated cereals (Benavente et al. 1996). This is of great importance for human nutrition, since cereal species are a staple of diet. One of the main domains in this field of research is resistance breeding. Aegilops (goatgrass) species are rich germplasm sources of resistance and tolerance to biotic and abiotic stresses. The genus Aegilops belongs to the tribe Triticeae, subtribe Triticinae, where we can also distinguish the genera Triticum, Secale and Brachypodium. Some of the species of Aegilops genus like: Ae. speltoides Tausch., Ae. sharonensis Eig., Ae. bicornis (Forsk.) Jaub. & Spach., Ae. longissima Schweinf. & Muschl., and Ae. searsii Feldman & Kislev ex K. Hammer (S-genome species), are proposed to be the donors of the B-genome of bread wheat (Triticum aestivum L.) (Feldman et al. 1995; Belyayev et al. 2001; Raskina et al. 2002), while Ae. tauschii Coss. is assumed to be a D-genome donor (Hedge et al. 2002). There are two proposals for B genome origin. The first, called the polyphyletic theory, states that permanent hybridizations and exchanges of genetic information between ancestral AASS amphiploids and related S-genome diploid progenitors singled out the B genome. The second theory assumes that the B-genome originated from a single S-genome ancestor, probably Ae. speltoides (Raskina et al. 2002; Belyayev and Raskina 2012). Taking into consideration the close relationship within Triticeae tribe, many genes have been transferred from Aegilops species to cultivated wheat, such as those for resistance to leaf rust, stem rust, yellow rust and powdery mildew (Schneider et al. 2008). In interspecific hybridization analysis, it is fundamental to be able to describe the genomic composition of plants carrying the desired traits (Sepsi et al. 2008). Molecular cytogenetics has revolutionized the genetic analysis of plant genomes and has also provided plant breeders with new tools to identify chromatin involved in resistance to stresses and quality traits. Genomic in situ hybridization (GISH) is successfully used to discriminate the genome composition of hybrids (Mahelka et al. 2011), whereas fluorescence in situ hybridization (FISH), coupled with observations of chromosome morphology (Castilho and Heslop-Harrison 1995), is a method for chromosome identification using specific probes, such as repetitive DNA sequences (Cuadrado et al. 2000). Also Giemsa C-banding and N-banding are successful methods in chromosome identification and detection of chromosome rearrangements (e.g. Gill and Kimber 1977; Łukaszewski and Gustafson 1983; Apolinarska et al. 2010). The most common repetitive DNA sequences used in physical mapping in the genus Triticum are: pSc119.2, pSc200 (derived from rye), pAs1 (Afa family, from wheat), spelt 1 and spelt 52 (from Ae. speltoides) (Salina et al. 2006). The vast variability of goatgrass species is expressed in a different location of repetitive sequences, used as probes in FISH analysis, as reported in many papers so far. For example, the chromosomes of Ae. biuncialis Vis. and its ancestors: Ae. umbellulata Zhuk. and Ae. comosa Sm., were examined by Schneider et al. (2005) in order to identify wheat addition lines. There are also several other papers discussing the distribution of repetitive sequences with regard to genome differentiation (e.g. Badaeva et al. 1996a, b; Cuadrado and Jouve 2002; Cuadrado et al. 2008; Raskina et al. 2008; Salina et al. 2006, 2009). From our research perspective, this indicated a need for creating physical maps of cytogenetic markers of Aegilops accessions used in a further widening of the genetic diversity of triticale. The present work is a continuation of a research cycle (Kwiatek et al. 2012), that deals with genetic diversification of cultivated triticale by introgressing alien Aegilops chromatin. Considering the progressive loss of effective resistance genes in triticale because of the appearance of new virulent disease pathotypes (Arseniuk 1996), the adaptation of methods used in gene transfer from alien species is a key issue. The main goal of this research was: (1) to generate physical maps using four repetitive DNA sequences (pSc119.2, pAs1, 5S rDNA, 25SrDNA) of U, M, S and D subgenomes; and (2) to categorize the amphiploid chromosomes with respect to the subgenomes. This paper reports FISH/GISH chromosome characterization and categorization of Aegilops kotschyi Boiss., Ae. ovata Roth., Ae. tauschii and amphiploids derived from Aegilops spp. × rye crossing. The ancestors of the introduced subgenomes, namely Ae. umbellulata, Ae. comosa and Ae. sharonensis, were analysed to help to identify the specific signal patterns. It is hoped that the development of these amphiploid forms will allow us to transfer agronomically useful traits (drought and salt tolerance, disease resistance) from goatgrasses into triticale.
Materials and methods
Plant material
Seeds of Aegilops umbellulata, Ae. comosa and Ae. sharonensis were kindly supplied for the study from the National Small Grains Germplasm Research Facility, National Small Grains Collection (Aberdeen, Idaho, USA). Seeds of Ae. kotschyi, Ae. ovata, Ae. tauschii were received by Wojciechowska (1996) from a collection of Professor M. Feldman (Weizmann Institute of Science, Rehovot, Israel). Seeds of Secale cereale were provided by a collection of the Institute of Plant Genetics, Polish Academy of Sciences in Poznań, Poland (Table 1). Amphiploids were generated from interspecific crosses between these Aegilops spp. and rye (Wojciechowska and Pudelska 1999, 2002a, b, 2005). Twenty seeds of each accession were germinated on moist filter paper in Petri dishes for 3–4 days. The root-tips were collected and ice-cooled for 26 h for metaphase accumulation. The fixation was made using ethanol and acetic acid (3:1, v/v). The chromosome preparations were made according to Pijnacker and Ferwerda (1984).
Table 1.
Plant material used for fluorescence in situ hybridization polymorphism analysis
| Accession/form | Number of plants | Genome structure | Source |
|---|---|---|---|
| Aegilops umbellulata | 20 | 2n = 2x = 14, UU | National Small Grains Germplasm Research Facility, National Small Grains Collection (Aberdeen, Idaho, USA) |
| Aegilops comosa | 20 | 2n = 2x = 14, MM | National Small Grains Germplasm Research Facility, National Small Grains Collection (Aberdeen, Idaho, USA) |
| Aegilops sharonensis | 20 | 2n = 2x = 14, SshSsh | National Small Grains Germplasm Research Facility, National Small Grains Collection (Aberdeen, Idaho, USA) |
| Aegilops tauschii | 20 | 2n = 2x = 14, DD | National Small Grains Germplasm Research Facility, National Small Grains Collection (Aberdeen, Idaho, USA) |
| Secale cereale | n/aa | 2n = 2x = 14, RR | Institute of Plant Genetics, Polish Academy of Sciences |
| Aegilops kotschyi | 20 | 2n = 4x = 28, UUSS | Weizmann Institute of Science, Rehovot, Israel |
| Aegilops ovata | 20 | 2n = 4x = 28, UUMM | Weizmann Institute of Science, Rehovot, Israel |
| Aegilops tauschii × Secale cereale | 20 | 2n = 4x = 28, DDRR | Institute of Plant Genetics, Polish Academy of Sciences |
| Aegilops kotschyi × Secale cereale | 20 | 2n = 6x = 42, UUSSRR | Institute of Plant Genetics, Polish Academy of Sciences |
| Aegilops ovata × Secale cereale | 20 | 2n = 6x = 42, UUMMRR | Institute of Plant Genetics, Polish Academy of Sciences |
aLeaf tissue for DNA isolation
DNA isolation
The DNA isolation from leaf tissue was made after Lombard and Delourme (2001). The pSc 119.2, 25S rDNA and 5S rDNA were amplified using M13 sequencing primers. The pAs1 (Afa family) was amplified according to Nagaki et al. (1995).
In situ hybridization
Three in situ hybridizations on the same chromosome preparations were carried out. First FISH was made according to Książczyk et al. (2010) with minor modifications, using 25S (used for detection of 35S rDNA loci) (Unfried and Gruendler 1990) and 5S rDNA (pTa794) (Gerlach and Dyer 1980) labelled by PCR with digoxygenin-11-dUTP and tetramethyl-rhodamine-5-dUTP, respectively (ROCHE). The hybridization mixture (40 μL per slide) contained 90 ng of each probe in the presence of salmon sperm DNA. After documentation of the FISH sites, the slides were washed (2 × 45 min in 4 × SSC Tween, 2 × 5 min in 2 × SSC, at room temperature).
During the second FISH, pSc119.2 and pAs1 (labelled with digoxygenin-11-dUTP and tetramethyl-rhodamine-5-dUTP, respectively) were used as probes.
After second reprobing, GISH was carried out according to Książczyk et al. (2010) with modifications. GISH on chromosomes of tetraploid Aegilops species was carried out using U probes (generated from Ae. umbellulata), which were labelled with tetramethyl-rhodamine-5-dUTP using nick translation kit (ROCHE) and M probes (Ae. comosa) and S probes (Ae. sharonensis) labelled with digoxigenin-11-dUTP. GISH on amphiploids (Aegilops ssp. × S. cereale) forms was carried out using properly probes, generated from Ae. kotschyi, Ae. ovata, Ae. tauschii genomic DNAs, labelled by nick translation with digoxigenin-11-dUTP. The blocking DNA was obtained by autoclaving the total DNA of rye. The GISH mixture (40 mL per slide), containing 50 % formamide, 2 × SSC, 10 % dextran sulphate, 90 ng each of the genome probes, and 4.5 mg blocking DNA, was denatured at 75 °C for 10 min and stored on ice for 10 min. Chromosomal DNA was denatured in the presence of the hybridization mixture at 75 °C for 6 min and allowed to hybridise overnight at 37 °C. For detection of the hybridization signals, anti-digoxigenin-rhodamine conjugated with FITC (ROCHE) was used. Mitotic cells were examined with an Olympus XM10 CCD camera attached to an Olympus BX 61 automatic epifluorescence microscope. Image processing was carried out using Olympus Cell-F imaging software and Micrographx Picture Publisher software.
Results
U chromosomes
In Ae. umbellulata, 5S rDNA and 35S rDNA were found in short arms of two pairs of chromosomes: 1U and 5U (Fig. 1a). The 5S rDNA landmarks were more distant from the centromere than 35S rDNA in 1U chromosomes, but in chromosome pair 5U their location was reversed. The same arrangement was observed in Ae. kotschyi (Fig. 1b), Ae. ovata (Fig. 1c), and amphiploids: Ae. kotschyi × S. cereale and Ae. ovata × S. cereale (Fig. 2). There was only one exception: in chromosome 5Uo of plant no. 3/4 of Ae. ovata × S. cereale a deletion of the 5S and 35S rDNA sites was detected (Fig. 2a, d). The pSc 119.2 labelling was detected at telomeric locations of each U chromosome of Ae. umbellulata except short arm of 6U chromosome and long arms of 1U and 2U chromosomes. Moreover, a subtelomeric site in 7U chromosomes was observed. In tetraploid accessions containing the U subgenome, the labelling pattern of pSc 119.2 was similar as in Ae. umbellulata but additionally in chromosome 6Uo of Ae. ovata and amphiploids in the middle of the long arm a considerable signal was detected. Furthermore, in 1Uo chromosomes in Ae. ovata (and amphiploid) only short-arm signals were found, when in 1Uk chromosome of Ae. kotschyi two sites were observed (telomeric and distal region of long arm). In the long arm of 7Uo chromosome of Ae. ovata and Ae. ovata × S. cereale, two subtelomeric sites were observed.
Fig. 1.
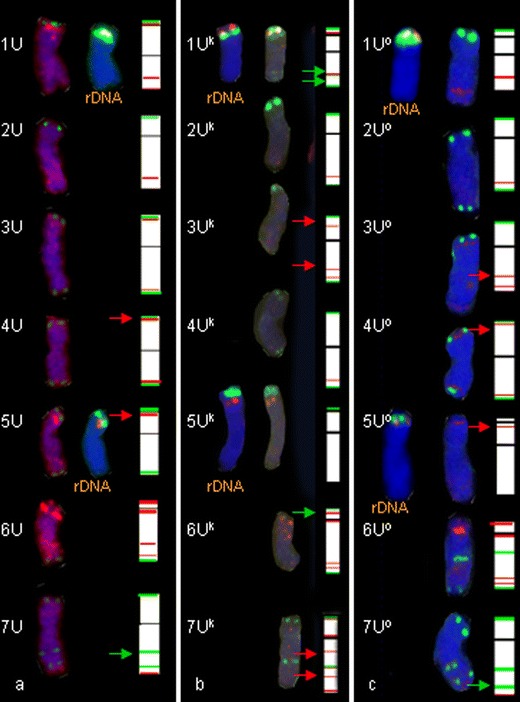
Fluorescence in situ hybridization (FISH) pattern and graphical presentation showing the location of 5S (red) and 35S (green) rDNA, pSc119.2 (green) and pAs1 (red) repetitive clones on the individual somatic chromosomes of Ae. umbellulata (a), Ae. kotschyi (b) and Ae. ovata (c)
Fig. 2.
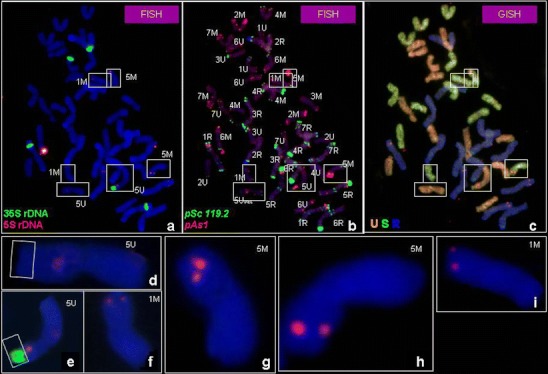
a–i Mitotic metaphase cell division of Ae. ovata × S. cereale (plant 3/4) analysed using: a FISH pattern showing the location of 5S rDNA (red) and 35S rDNA (green), followed by b FISH pattern showing the location of pSc119.2 (green) and pAs1 (red) repetitive clones and c genomic in situ hybridization (GISH) with total genomic DNA of Ae. umbellulata (red), Ae. comosa (green) used as probes and S. cereale (blue) used as blocking DNA. d Chromosome 5U with 35S rDNA deletion, e chromosome 5U, f, i chromosome 1M, g, h chromosome 5M
The pAs1 signals were observed in each chromosome of Ae. umbellulata. In 1U, 2U, 3U, 6U and 7U chromosomes, the pAs1 probe was detected in the interstitial region of the long arm. Faint signals were also observed in the distal part of the long arm of 7U chromosomes. The most significant pAs1 signal was detected in the pericentromeric region of 6U chromosomes. The pAs1 labelling patterns in tetraploids and amphiploids were similar to those in Ae. umbellulata with the exception of Ae. kotschyi and amphiploids, where short arms of 3U had faint signals of the Afa family. Furthermore, 7Uk chromosomes of Ae. kotschyi carried more signals of pAs1 than 7U chromosomes of other species taken into consideration.
M chromosomes
The FISH analysis of Ae. comosa using 5S and 35S rDNA probes showed 2 pairs of chromosomes carrying these landmarks (Fig. 3a). In 1M chromosome satellites, 5S rDNA was more distant than the 35S rDNA site, whereas in 5M the location of the 35S rDNA site was telomeric (on the satellite) and the 5S rDNA signal was situated close to nucleolus organizer region (NOR). However chromosome 5Mo in Ae. ovata and the amphiploids (containing M chromosomes inherited from the above-mentioned accessions), carried only the 5S signal.
Fig. 3.
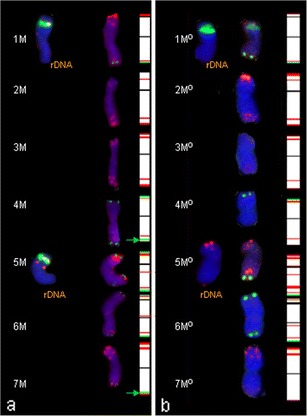
Fluorescence in situ hybridization (FISH) pattern and graphical presentation showing the location of 5S (red) and 35S (green) rDNA, pSc119.2 (green) and pAs1 (red) repetitive clones on the individual somatic chromosomes of Ae. comosa (a), Ae. ovata (b)
All Ae. comosa chromosomes with the exception of 2M and 3M were labelled by the pSc119.2 probe. The telomeric region of both arms was labelled in 4M, 5M and 6M chromosomes. The same labelling pattern appeared in Ae. ovata and Ae. ovata × S. cereale with the exception of 7Mo and the long arm of 4Mo chromosomes in Ae. ovata and its amphiploids, where no signals were observed. The hybridization of the pAs1 probe in Ae. comosa was present in each chromosome. The most recognizable were 2M chromosomes with two bold sites at telomeric locations of the short arm and dispersed signals in the distant region of the long arm. The pAs1 pattern of 7M chromosome was similar to the 2M one, but the signals in 7M chromosome in the long arm were located in the telomeric region. The most significant differences were in 5M and 6M chromosomes. These chromosomes in Ae. ovata carried more pAs1 landmarks than their analogues in Ae. comosa.
S chromosomes
The 5S rDNA sites were observed in the short arm of two 1S and 5S chromosomes of Ae. sharonensis (Fig. 4a). Two chromosomes (5Ssh and 6Ssh) carried 35S rDNA sites. However, in Ae. kotschyi, (Fig. 3a, b) and amphiploids, only 5S rDNA signals were observed in 1S and 5S chromosomes.
Fig. 4.
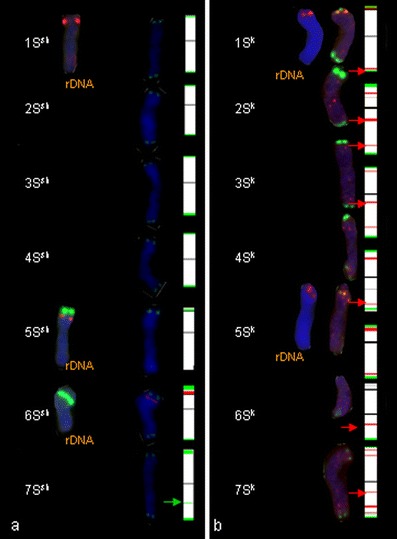
Fluorescence in situ hybridization (FISH) pattern and graphical presentation showing the location of 5S (red) and 35S (green) rDNA, pSc119.2 (green) and pAs1 (red) repetitive clones on the individual somatic chromosomes of Ae. sharonensis (a), Ae. kotschyi (b)
In Ae. sharonensis, Ae. kotschyi and amphiploids, chromosomes were labelled with the pSc119.2 probe at telomeric regions. The most significant site is located in the short arm of 6Ssh chromosome.
No pAs1 labelling was detected in Ae. sharonensis chromosomes. Very weak and dispersed but comparable signals were observed in distant regions of long arms of 2Sk, 3Sk, 6Sk and 7Sk in Ae. kotschyi and amphiploids: Ae. kotschyi × S. cereale. Significant signals were also observed in the short arm of 2Sk chromosome of Ae. kotschyi (Fig. 4).
D chromosomes
The 5S and 35S rDNA sites were observed in short arms of the 5D chromosome of Ae. tauschii (Fig. 5a). The 35S rDNA site was wide and more distal than 5S rDNA signals. On the short arm of 1D only 5S rDNA signals were found. Three chromosomes (2D, 3D and 4D) carried pSc119.2 signals in telomeric regions but only chromosomes 4D had signals in telomeric regions of both arms. The Afa family labelling in Ae. tauschii was very strong and characteristic. The most recognizable was chromosome 3D, with a large site at a distal location (including the telomere) of the short arm. Well-labelled sites were also observed in telomeric parts of both arms of chromosome 7D. Interstitial sites were found in long arms of chromosomes 2D, 4D, 5D and 6D. In chromosome 4D, significant pericentromeric signals were observed. The same labelling pattern of the pAs1 probe was observed in Ae. tauschii × Secale cereale amphiploids (Fig. 5b).
Fig. 5.
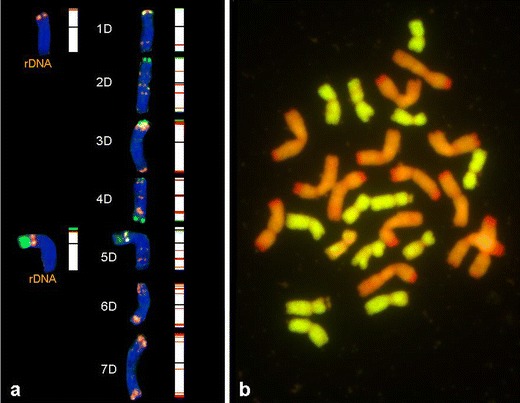
a Fluorescence in situ hybridization (FISH) pattern and graphical presentation showing the location of 5S (red) and 35S (green) rDNA, pSc119.2 (green) and pAs1 (red) repetitive clones on the individual somatic chromosomes of Ae. tauschii; b genomic in situ hybridization (GISH) of Ae. tauschii × S. cereale analysed using total genomic DNA of Ae. tauschii (green) used as probe and S. cereale (red) used as blocking DNA
Discussion
This study focused on U, M, S and D genomes, which are fundamental in the genus Aegilops (Van Slageren 1994). Diploid goatgrass species were compared with their polyploid evolutionary descendants to capture differences in the distribution of repetitive sequences. Creation of physical maps of cytogenetic markers could be a very useful tool helping to identify Aegilops chromosomes in Aegilops × Secale amphiploids and Aegilops × triticale crosses (in the future stages of this experimental program). Mukai et al. (1993) combined pAs1 and pSc119.2 for 2-colour FISH and were able to identify 17 of the 21 wheat chromosome pairs (Pedersen and Langridge 1997). In our study, pAs1 and pSc119.2 with addition of rDNA-FISH were used to identify Aegilops chromosomes and to track them in polyploid descendants and Aegilops × Secale amphiploids. Taking into consideration the subgenomic structure of the majority of tetraploid goatgrasses, it can be assumed that the U genome is pivotal in the genus Aegilops. According to the theory of pivotal-differential evolution, several polyploid species contain the U genome, which was derived from the diploid Ae. umbellulata (Zohary and Feldman 1962). This genome is considered to be unmodified in the diploids, based on chromosome pairing analysis (Talbert et al. 1993). It is suggested that the pivotal genome is a buffer to provide hybrid fertility, while genetic exchange can occur between the differential genomes (Chee et al. 1995). The present study corroborates this hypothesis, taking into account the small number of differences in the distribution of repetitive sequences of the U genome in Ae. umbellulata in comparison with Ae. ovata (UUMM), Ae. kotschyi (UUSS) and amphiploids (UUMMRR, UUSSRR). Insignificant dissimilarities appeared in 1U, 3U, 4U 6U and 7U, but have not resulted in chromosome identification. This tendency has also been reported in a similar study in which Ae. umbellulata and Ae. comosa were compared with Ae. biuncialis (Schneider et al. 2005). The hybridization patterns with repetitive DNA probes showed that the M and S genomes are much more diverse. Badaeva et al. (2004) showed an elimination of NOR and 35S rDNA site in chromosome 1M and the same arrangement of chromosome 5M (with 5S rDNA and 35S rDNA sites) of Ae. ovata (syn. Ae. geniculata) in comparison with Ae. comosa. 35S rDNA signals was also observed in chromosome 6M in both species, however chromosome 6M of Ae. comosa had a major NOR exclusively (Badaeva et al. 2004). On the other hand, Molnár et al. (2011) describe 1M and 6M as satellited chromosomes with strong pTa71 (18S–5.8S–26S rDNA) signals present in Ae. comosa, while Ae. ovata had no satellited M chromosome and no pTa71 signals were detected. Regarding the quoted studies, in this research we have additionally detected an elimination of 35S rDNA in chromosome 5M of Ae. ovata and Ae. ovata × S. cereale (Fig. 5a, b, g, h). It is possible that rDNA loci change their position by the same process, responsible for a dispersion of heterochromatic sequences through a genome (Dubcovsky and Dvoŕák 1995). Furthermore, rDNA elimination in S-genome chromosomes in the synthetic polyploid Ae. umbellulata × Ae. comosa appears to be similar to rDNA changes in natural allotetraploids (Shcherban et al. 2008). Such deletions of major rDNA sites and their replacements can lead to sudden fluctuations in the rDNA consensus sequence in an evolutionary lineage (Dubcovsky and Dvoŕák 1995). The differences in location of pSc 119.2 and pAs1 landmarks in 5M, 6M and 7M indicate a large variability in the M genome, which is comparable with molecular research made by Resta et al. (1996). An analysis reported by Kihara (1963) showed that Ae. comosa (MM) is less evolutionary stable than Ae. umbellulata, as revealed by the morphological variability and greater number of subspecies of Ae. comosa.
Ae. sharonensis, like Ae. longissima, Ae. searsii and Ae. bicornis, belong to the section Sitopsis of the genus Aegilops. This species is considered as a donor of the S-genome in Ae. kotschyi and Ae. variabilis. Cytogenetic analysis made in this study confirms the conclusions drawn by Badaeva et al. (1996a) that a similar labelling pattern of the pSc119.2 probe was present in Ae. sharonensis as well as in polyploid descendants. However, there were some differences in pAs1 labelling patterns between the ancestral S-genome and descendant ones, which suggests that this genome is evolutionally variable. That confirms the pivotal theory. Moreover, the lack of 35 rDNA landmarks in Ae. kotschyi was observed, which is novel according to the results reported by Badaeva et al. (1996a).
Another species in the genus Aegilops is Ae. tauschii (syn. Ae. squarossa). The distribution of repetitive sequences in the D-genome is quite conservative and similar to previous reports. However, the presence of pSc119.2 sites is variable. In previous papers, presence of small telomeric pSc119.2 sites in the short arms of 2D, 3D and 5D (inconsistent appearance) chromosomes of Ae. squarossa was reported by Badaeva et al. (1996a). Furthermore, the small telomeric pSc119.2 sites were also observed in both arms of 1D, 2D and 4D chromosomes (Schneider et al. 2003) and 7D chromosomes (Kwiatek, unpubl. data) of T. aestivum. Combining pAs1 and pSc119.2 with 5S and 35S rDNA probes is an effective cytogenetic tool for identification of complete Aegilops chromosomes and detection of their small rearrangements. However, some difficulties may appear during translocation analyses. The labelling landmarks were faint or localized only in the telomeric region in some chromosomes. In the case of a translocation, it might be impossible to distinguish and identify such a chromosome fragment. Taking this into consideration, it is advisable to develop other cytogenetic markers, such as repetitive sequences, synthetic oligonucleotides or bacterial artificial chromosomes (BACs) that can secure the mapping of each chromosome region. In accordance with Zhang et al. (2004), the use of genomic DNA cloned in large-insert vectors, such as BACs (Shizuya et al. 1992), in combination with FISH, called BAC-FISH, has been an effective approach for physical mapping of such specific DNA sequences and identifying individual chromosomes. On the other hand, our study shows that repetitive DNA probes could be considered as a subspecies discrimination factor, especially in variable species containing less stable genomes, such as S or M.
The distribution of cytogenetic landmarks reported in this paper may contribute to study the evolution in Triticeae and breeding research aiming at widening of genetic diversity by distant crosses.
Acknowledgments
We thank Dr Marie Kubalakova (Laboratory of Molecular Cytogenetics and Cytometry, Institute of Experimental Botany, Olomouc, Czech Republic) for supplying pSc 119.2 and pAs1 clones and for her valuable suggestions and Dr Tomasz Książczyk (Laboratory of Cytogenetics and Molecular Biology, Institute of Plant Genetics, Polish Academy of Sciences, Poznan, Poland) for supplying 5S and 25S rDNA sequences and for his clues. The technical assistance of Mrs. Grażyna Cicha, Ms. Jolanta Belter and Mrs. Joanna Maszner is gratefully acknowledged.
References
- Apolinarska B, Wiśniewska H, Wojciechowska B. Aegilops-rye amphiploids and substitution rye used for introgression of genetic material into rye (Secale cereale L.) J Appl Genet. 2010;51:413–420. doi: 10.1007/BF03208871. [DOI] [PubMed] [Google Scholar]
- Arseniuk E, et al. Triticale diseases—a review. In: Guedes-Pinto H, et al., editors. Triticale: today and tomorrow. The Netherlands: Kluwer; 1996. pp. 499–525. [Google Scholar]
- Badaeva ED, Friebe B, Gill BS. Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosome of diploid species. Genome. 1996;39:293–306. doi: 10.1139/g96-040. [DOI] [PubMed] [Google Scholar]
- Badaeva ED, Friebe B, Gill BS. Genome differentiation in Aegilops. 2. Physical mapping of 5S and 18S–26S ribosomal RNA gene families in diploid species. Genome. 1996;39:1150–1158. doi: 10.1139/g96-145. [DOI] [PubMed] [Google Scholar]
- Badaeva ED, Amosova AV, Samatadze TE, Zoshchuk SA, Szostak NG, Chikida NN, Zelenin AV, Raupp WJ, Friebe B, Gill BS. Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Syst Evol. 2004;246:45–76. doi: 10.1007/s00606-003-0072-4. [DOI] [Google Scholar]
- Belyayev A, Raskina O (2012) Evolution at range margins: monitoring the genome of Aegilops speltoides. Chromosome biology, Genome evolution and Speciation. Research Conference, 23–25 April 2012, Gatersleben, Germany
- Belyayev A, Raskina O, Nevo E. Detection of alien chromosomes from S-genome species in the addition/substitution lines of bread wheat and visualization of A-, B- and D-genomes by GISH. Hereditas. 2001;135:119–122. doi: 10.1111/j.1601-5223.2001.00119.x. [DOI] [PubMed] [Google Scholar]
- Benavente E, Fernández-Calvín B, Orellana J. Relationship between the levels of wheat-rye metaphase I chromosomal pairing and recombination revealed by GISH. Chromosoma. 1996;105:92–96. doi: 10.1007/BF02509518. [DOI] [PubMed] [Google Scholar]
- Castilho A, Heslop-Harrison JS. Physical mapping of 5S and 18S–25S rDNA and repetitive DNA sequences in Aegilops umbellulata. Genome. 1995;38:91–96. doi: 10.1139/g95-011. [DOI] [PubMed] [Google Scholar]
- Chee PW, Lavin M, Talbert LE. Molecular analysis of evolutionary patterns of U genome wild wheats. Genome. 1995;38:290–297. doi: 10.1139/g95-036. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Jouve N. Evolutionary trends of different repetitive DNA sequences during speciation in the genus Secale. J Hered. 2002;93:339–345. doi: 10.1093/jhered/93.5.339. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Schwarzacher T, Jouve N. Identification of different chromatin classes in wheat using in situ hybridization with simple sequence repeat oligonucleotides. Theor Appl Genet. 2000;101:711–717. doi: 10.1007/s001220051535. [DOI] [Google Scholar]
- Cuadrado A, Cardoso M, Jouve N. Physical organization of simple sequence repeats (SSRs) in Triticeae: structural, functional and evolutionary implications. Cytogenet Genome Res. 2008;120:210–219. doi: 10.1159/000121069. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Dvoŕák J. Ribosomal RNA multigene loci: nomads of Triticeae genomes. Genetics. 1995;140:1367–1377. doi: 10.1093/genetics/140.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Lipton FGH, Miller TE (1995) Wheats. Triticum spp. (Gramineae-Triticinae). In: Smartt J, Simmonds NW (eds) Evolution of plant crops, 2nd edn. Longman Scientific and Technical Press pp 184–192
- Gerlach WL, Dyer TA. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980;11:4851–4865. doi: 10.1093/nar/8.21.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill BS, Kimber G. Recognition of translocation and allen chromosome transfers in wheat by the Giemsa-C-banding technique. Crop Sci. 1977;17:264–266. doi: 10.2135/cropsci1977.0011183X001700020008x. [DOI] [Google Scholar]
- Hedge SG, Valkoun J, Waines G. Genetic diversity in wild and weedy Aegilops, Amblyopyrum, and Secale species — a preliminary survey. Crop Sci. 2002;42:608–614. doi: 10.2135/cropsci2002.0608. [DOI] [Google Scholar]
- Kihara H. Genomanalyse bei Triticum und Aegilops. IX. Systematischer Aufbau der Gattung Aegilops auf genomeanalytischer Grundlage. Cytologia (Tokio) 1963;14:135–144. doi: 10.1508/cytologia.14.135. [DOI] [Google Scholar]
- Książczyk T, Taciak M, Zwierzykowski Z. Variability of ribosomal DNA sites in Festuca pratensis, Lolium perenne, and their intergeneric hybrids, revealed by FISH and GISH. J Appl Genet. 2010;51:449–460. doi: 10.1007/BF03208874. [DOI] [PubMed] [Google Scholar]
- Kwiatek M, Błaszczyk L, Wiśniewska H, Apolinarska B. Aegilops-Secale amphiploids: chromosome categorisation, pollen viability and identification of fungal disease resistance genes. J Appl Genet. 2012;53:37–40. doi: 10.1007/s13353-011-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Delourme A consensus linkage map for rapeseed (Brassica napus L.): construction and integration of three individual maps from DH populations. Theor Appl Genet. 2001;103:491–507. doi: 10.1007/s001220100560. [DOI] [Google Scholar]
- Łukaszewski AJ, Gustafson JP. Translocations and modifications of chromosomes in triticale × wheat hybrids. Theor Appl Genet. 1983;64:239–248. doi: 10.1007/BF00303771. [DOI] [PubMed] [Google Scholar]
- Mahelka V, Kopeckỳ D, Paštová L. On the genome constitution and evolution of intermediate wheatgrass (Thinopyrum intermedium: Poaceae, Triticeae) BMC Evol Biol. 2011;11:127. doi: 10.1186/1471-2148-11-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár I, Cifuentes M, Schneider A, Benavente E, Molnár-Láng M. Association between simple sequence repeat-rich chromosome regions and intergenomic translocation breakpoints in natural populations of allopolyploid wild wheats. Ann Bot. 2011;107:65–76. doi: 10.1093/aob/mcq215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai Y, Friebe B, Hatchett JH, Yamamoto M, Gill BS. Molecular cytogenetic analysis of radiation-induced wheat-rye terminal and intercalary chromosomal translocations and the detection of rye chromatin specifying resistance to Hessian fly. Chromosoma. 1993;102:88–95. doi: 10.1007/BF00356025. [DOI] [Google Scholar]
- Nagaki K, Tsujimoto H, Isono K, Sasakuma T. Molecular characterization of a tandem repeat, Afa family, and its distribution among Triticeae. Genome. 1995;38:479–486. doi: 10.1139/g95-063. [DOI] [PubMed] [Google Scholar]
- Pedersen C, Langridge P. Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome. 1997;40:589–593. doi: 10.1139/g97-077. [DOI] [PubMed] [Google Scholar]
- Pijnacker LP, Ferwerda MA. Giemsa C-banding of potato chromosomes. Can J Genet Cytol. 1984;26:415–419. [Google Scholar]
- Raskina O, Belyayev A, Nevo E. Repetitive DNAs of wild emmer wheat (Triticum dicoccoides) and their relation to S-genome species: molecular cytogenetic analysis. Genome. 2002;45:391–401. doi: 10.1139/g01-142. [DOI] [PubMed] [Google Scholar]
- Raskina O, Barber JC, Nevo E, Belyayev A. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genome. Cytogenet Genome Res. 2008;120:351–357. doi: 10.1159/000121084. [DOI] [PubMed] [Google Scholar]
- Resta P, Zhang HB, Dubcovsky J, Dvorak J. The origins of the genomes of Triticum biunciale, T. ovatum, T. neglectum, T. columnare and T. rectum (Poaceae) based on variation in repeated nucleotide sequences. Am J Bot. 1996;83:1556–1565. doi: 10.2307/2445829. [DOI] [Google Scholar]
- Salina EA, Lim YK, Badaeva ED, Shcherban AB, Adonina IG, Amosova AV, Samatadze TE, Vatolina TY, Zoshchuk SA, Leitch AR. Phylogenetic reconstruction of Aegilops section Sitopsis and the evolution of tandem repeats in the diploids and derived wheat polyploids. Genome. 2006;49:1023–1035. doi: 10.1139/G06-050. [DOI] [PubMed] [Google Scholar]
- Salina EA, Sergeeva EM, Adonina IG, Shcherban AB, Afonnikov DA, Belcram H, Huneau C, Chalhoub B. Isolation and sequence analysis of the wheat B genome subtelomeric DNA. BMC Genomics. 2009;10:414–428. doi: 10.1186/1471-2164-10-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Linc G, Molnár-Láng M. Fluorescence in situ hybridization polymorphism using two repetitive DNA clones in different cultivars of wheat. Plant Breed. 2003;122:396–400. doi: 10.1046/j.1439-0523.2003.00891.x. [DOI] [Google Scholar]
- Schneider A, Linc G, Monar I, Molnár-Láng M. Molecular cytogenetic characterization of Aegilops biuncialis and its use for the identification of 5 derived wheat — Aegilops biuncialis disomic addition lines. Genome. 2005;48:1070–1082. doi: 10.1139/g05-062. [DOI] [PubMed] [Google Scholar]
- Schneider A, Molnár I, Molnár-Láng M. Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica. 2008;163:1–19. doi: 10.1007/s10681-007-9624-y. [DOI] [Google Scholar]
- Sepsi A, Molnar I, Szalay D, Molnar-Lang M. Characterization of a leaf rust-resistant wheat — Thinopyrum ponticum partial amphiploid BE-1, using sequential multicolor GISH and FISH. Theor Appl Genet. 2008;116:825–834. doi: 10.1007/s00122-008-0716-4. [DOI] [PubMed] [Google Scholar]
- Shcherban AB, Badaeva ED, Amosova AV, Adonina IG, Salina EA. Genetic and epigenetic changes of rDNA in a synthetic allotetraploid, Aegilops sharonensis × Ae. umbellulata. Genome. 2008;51:261–271. doi: 10.1139/G08-006. [DOI] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci U S A. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert LE, Kimber G, Magyar GM, Buchanan CB. Repetitive DNA variation and pivotal-differential evolution of wild wheats. Genome. 1993;36:14–21. doi: 10.1139/g93-003. [DOI] [PubMed] [Google Scholar]
- Unfried I, Gruendler P. Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res. 1990;18:4011. doi: 10.1093/nar/18.13.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Slageren MW (1994) Wild wheats: a monograph of Aegilops L., and Amblyopyrum (Jaub. & Spach) Eig (Poaceae). Agricultural University of Wageningen, the Netherlands, and International Center for Agricultural Research in Dry Areas, Aleppo, Syria
- Wojciechowska B. Hybrids of tetraploid Aegilops sp. with Secale cereale. Genet Pol. 1996;37A:174–178. [Google Scholar]
- Wojciechowska B, Pudelska H. Production, morphology and fertility of the amphiploids Aegilops variabilis × Secale cereale and Ae. kotschyi × S. cereale. Cereal Res Commun. 1999;27:79–82. [Google Scholar]
- Wojciechowska B, Pudelska H. Production and morphology of the hybrids Aegilops kotschyi × Secale cereale and Ae. biuncialis × S. cereale. J Appl Genet. 2002;43:279–285. [PubMed] [Google Scholar]
- Wojciechowska B, Pudelska H. Hybrids and amphiploids of Aegilops ovata L. with Secale cereale L.: production, morphology and fertility. J Appl Genet. 2002;43:415–421. [PubMed] [Google Scholar]
- Wojciechowska B, Pudelska H. Production and characterization of amphiploids of Aegilops kotschyi and Ae. biuncialis with Secale cereale, and of backcross hybrids of Ae. biuncialis × S. cereale amphiploids with 2× and 4× S. cereale. J Appl Genet. 2005;46:157–161. [PubMed] [Google Scholar]
- Zhang P, Li W, Fellers J, Friebe B, Gill BS. BAC-FISH in wheat identifies chromosome landmarks consisting of different types of transposable elements. Chromosoma. 2004;112:288–299. doi: 10.1007/s00412-004-0273-9. [DOI] [PubMed] [Google Scholar]
- Zohary D, Feldman M. Hybridization between amphiploids and the evolution of polyploids in the wheat Aegilops–Triticum group. Evolution. 1962;16:44–61. doi: 10.2307/2406265. [DOI] [Google Scholar]


