Abstract
The polarization of macrophages into a regulatory-like phenotype and the production of IL-10 plays an important role in the resolution of inflammation. We show in this study that PGE2, in combination with LPS, is able to promote an anti-inflammatory phenotype in macrophages characterized by high expression of IL-10 and the regulatory markers SPHK1 and LIGHT via a protein kinase A–dependent pathway. Both TLR agonists and PGE2 promote the phosphorylation of the transcription factor CREB on Ser133. However, although CREB regulates IL-10 transcription, the mutation of Ser133 to Ala in the endogenous CREB gene did not prevent the ability of PGE2 to promote IL-10 transcription. Instead, we demonstrate that protein kinase A regulates the phosphorylation of salt-inducible kinase 2 on Ser343, inhibiting its ability to phosphorylate CREB-regulated transcription coactivator 3 in cells. This in turn allows CREB-regulated transcription coactivator 3 to translocate to the nucleus where it serves as a coactivator with the transcription factor CREB to induce IL-10 transcription. In line with this, we find that either genetic or pharmacological inhibition of salt-inducible kinases mimics the effect of PGE2 on IL-10 production.
Introduction
Macrophages form an important component of the innate immune response and are required to fulfill a range of different roles depending on their specific surroundings. The detection of pathogens by macrophages occurs via pattern recognition receptors (PRRs). These are germline-encoded receptors that are able to recognize specific classes of pathogen-derived ligands, often referred to as pathogen-associated molecular patterns. PRR activation triggers the induction of both pro- and anti-inflammatory cytokines. Importantly, different types of PRRs, acting either alone or in concert, promote distinct cytokine profiles, thus allowing the immune response to be tailored to the type of pathogen (1). In addition, the response of the macrophage to PRR activation can be further modulated by other inputs, such as ligands for certain G-protein coupled receptors (GPCRs) or ITAM-coupled receptors (2, 3). These interactions allow macrophages to mount the correct response during the different phases of the immune response. For instance, macrophages are one of the first cells to detect invading pathogens and help trigger the immune response via the production of proinflammatory mediators. In contrast, during the resolution of inflammation, macrophages play an anti-inflammatory role and are required for the removal of apoptotic cells.
The ability of macrophages to undertake very different functions has led to the concept of macrophage polarization, whereby in response to a specific set of signals, distinct phenotypes can be induced in macrophages to allow them to undertake their required functions (4–8). In vivo, macrophage polarization is likely to be very plastic and macrophages can assume a wide spectrum of phenotypes. Several broad classifications of polarized macrophages have, however, been proposed (6–8). M1, or classically activated, macrophages can be induced by a combination of LPS and IFN-γ. These cells are proinflammatory and produce high amounts of TNF and IL-12 but low amounts of the anti-inflammatory cytokine IL-10. Stimulation with IL-4 gives rise to M2a, or alternatively activated macrophages, which play a role in responses to extracellular parasites and tissue repair. Stimulation of macrophages with LPS in combination with immune complexes results in macrophages that produce high levels of IL-10 but low levels of IL-12. These have been termed “regulatory” or M2b macrophages and are proposed to play a role in the resolution of inflammation.
Increasingly there is evidence that a failure to appropriately control macrophage function may have a role in disease. For example, excessive M1 polarization in adipose tissue has been linked to metabolic disease (9), whereas unwanted M2a polarization may give rise to fibrosis (8). M2-like macrophages also have similarities to tumor-associated macrophages, which, through their ability to suppress inflammation, help protect the tumor from attack by the immune system (10). Various signals, including PGs, TGF-β, and IL-10, have been suggested to aid in the development of tumor-associated macrophages (10). Given the potential roles of macrophages in disease, the modulation of macrophage polarization in vivo could represent a novel therapeutic strategy. In support of this, transfer of regulatory macrophages into mice has been reported to be protective in models of experimental autoimmune encephalomyelitis and endotoxic shock (11–13). Understanding how intracellular signaling controls this macrophage response is therefore an important question (6).
A characteristic of regulatory macrophages is their ability to produce high levels of IL-10 relative to other macrophage subsets. IL-10 is a potent repressor of proinflammatory cytokine production by macrophages and acts as a key anti-inflammatory mediator (14, 15). The importance of IL-10 for maintaining balance in the immune system in vivo is underscored by the finding that IL-10 knockout mice can spontaneously develop colitis (16). Consistent with this, inactivating mutations in either the IL-10 or IL-10R genes in humans results in a severe form of early-onset colitis (17, 18). The transcriptional regulation of IL-10 in macrophages is complex, and roles for Sp1, NF-κB, C/EBP, and CREB have been proposed (reviewed in Ref. 14), although how this gene is highly induced in regulatory macrophages is not clear.
Protein kinase A (PKA) is activated by increases in the intracellular levels of cAMP, which can be triggered by a number of stimuli in macrophages. Several studies have shown that agents that elevate cAMP can enhance IL-10 transcription (19–24). One example of this is the PG PGE2. PGs are lipid-derived molecules that play important roles in modulating the immune system. As early as 1986, a repressive effect of PGE2 on macrophage TNF and IL-1 production was shown (25, 26), and this has subsequently been confirmed by other studies. Of note, PGE2 has been shown to have a synergistic effect with LPS on IL-10 production (27). However, the molecular mechanism behind this has not been established. We show in this study that PGE2 activates a PKA–salt-inducible kinase (SIK)–CREB-regulated transcription coactivator (CRTC)3-dependent pathway, which in combination with TLR agonists, gives rise to macrophages that produce high levels of IL-10 and are polarized toward a regulatory-like macrophage phenotype.
Materials and Methods
Materials
MRT67307 was provided by MRC Technology. Kin112 was a gift from N. Gray (Harvard University, Cambridge, MA). The specificity of these inhibitors has been described previously (28). PGE2 was purchased from Biomol International. 8-Br-cAMP, 6-Bnz-cAMP, and 8-pCPT-MeO-cAMP were all purchased from BioLog Life Science Institute. The myristoylated PKI peptide was from Enzo Life Sciences, rolipram and IBMX were from Sigma-Aldrich, and the EP2 and EP4 agonists, Butaprost and CAY10580, were from Cayman Chemicals.
Clones
The CREB-dependent luciferase reporter and Renilla luciferase constructs were obtained from Promega. SIK2 (XM_041314) was cloned by standard techniques as a GFP-tagged construct into the pBabe expression vector. CRTC3 (NM_022769.3) was cloned as a mCherry-tagged construct, also into the pBabe vector.
Mice
C57BL/6 wild-type mice were obtained either from Charles River Laboratories or bred in-house. IL-10 knockout, MSK1/2 double knockout, and CREB Ser133Ala knockin mice have been described previously (16, 29, 30). IL-10 and MSK1/2 knockouts had been backcrossed onto C57BL/6 for a minimum of 12 generations and CREB Ser133Ala mice for 6 generations. ATF1 Ser63Ala knockin mice were generated by Taconic Artemis by standard methods using C57BL/6 embryonic stem cells. Macrophage-specific conditional LKB1 knockout mice were generated by crossing a floxed LKB1 allele (31) to LysMCre transgenic mice (32). Deletion of LKB1 in bone marrow–derived macrophages from these mice was confirmed by quantitative PCR (qPCR) (data not shown).
Cell culture
Primary bone marrow–derived macrophages (BMDMs) were isolated as described previously (33). Briefly, to obtain BMDMs, bone marrow cells were maintained on bacterial grade plates for 1 wk in DMEM supplemented with 10% heat-inactivated FBS (Biosera), 2 mM L-glutamine, 100 U/ml penicillin G, 100 mg/ml streptomycin, 0.25 mg/ml amphotericin (Invitrogen), and 5 ng/ml mCSF (R&D Systems). Adherent cells were then replated on tissue culture grade plates in fresh media and used 24 h after replating. RAW264.7, U20S, and Hek293 cells were cultured in DMEM supplemented with 10% heat-inactivated FBS, 2 mM L-glutamine, 100 U/ml penicillin G, and 100 mg/ml streptomycin. Hek293 cells were transfected using polyethyleneimine (34).
Small interfering RNA transfection
Small interfering RNA (siRNA) transfection in RAW264.7 cells was carried out using the Lonza Amaxa Nucleofection optimized RAW264.7 protocol and Nucleofector Kit V. Briefly, cells were spun down and resuspended in nucleofection reagent, combined with 100 nM siRNA and electroporated in accordance with manufacturer’s protocols. BMDMs were differentiated for 7 d, as previously described, and then, siRNA transfection was performed via nucleofection using the Lonza Amaxa Optimized Mouse Macrophage Nucleofection system (catalog number VPA-1009). Smartpool siGENOME siRNAs were purchased from Thermo-Dharmacon.
qPCR
Cells were lysed and total RNA purified using the Qiagen microRNeasy system, according to the manufacturer’s protocols. Total RNA (0.5–1 μg) was reverse transcribed using iScript (Bio-Rad) and qPCR was carried out using SYBRgreen based detection methods. Levels of 18s or GAPDH were used as normalization controls, and fold induction calculated using the equation:
where E is the efficiency of the PCR, ct is the threshold cycle, u is the mRNA of interest, r is the reference gene (18s RNA or GAPDH), s is the sample, and c is the average of the unstimulated wild-type control samples. Primer sequences are shown in Table I.
Table I. Primer sequences used for qPCR.
| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| Areg | CGACAAGAAAACGGGACTG | AACTGGGCATCTGGAACC |
| Arginase 1 (Arg1) | CTCCAAGCCAAAGTCCTTAGAG | AGGAGCTGTCATTAGGGACATC |
| ATF1 | CAGACAACAAAGACAGATGACC | AACAGCAACACGGTTCTCC |
| CREB | ATCAGTTATCCAGTCTCCACAAGTCC | GTGATGGCAGGGGCTGAAGTC |
| CRTC1 | ACTCAAAGAAGGCGGGTTCC | TGGGTGGCAGGGATCAGG |
| CRTC2 | TGCGACTGGCTTATACAAGG | GAGTGCTCCGAGATGAATCC |
| CRTC3 | AGCCATCACTTCATCAAGC | ATTCCCATCAAACTGTCTCC |
| IkBa | ACACGTGTCTGCACCTAG | TCAGACGCTGGCCTCCAAAC |
| IL-10 | CCCTTTGCTATGGTGTCCTTTC | GATCTCCCTGGTTTCTCTTCCC |
| IL-12p35 | TATCTCTATGGTCAGCGTTCC | TGGTCTTCAGCAGGTTTCG |
| IL-12p40 | TCATCAGGGACATCATCAAACC | TGAGGGAGAAGTAGGAATGGG |
| LIGHT | CTGCATCAACGTCTTGGAGA | GATACGTCAAGCCCCTCAAG |
| nor1 | GCCATCTCCTCCGATCTGTATG | GAGGCCGTCAGAAGGTTGTAG |
| nur77 | CCTGTTGCTAGAGTCTGCCTTC | CAATCCAATCACCAAAGCCACG |
| IL-6 | TTCCATCCAGTTGCCTTCTTG | AGGTCTGTTGGGAGTGGTATC |
| TNF | CAGACCCTCACACTCAGATCATC | GGCTACAGGCTTGTCACTCG |
| SPHK1 | ACAGCAGTGTGCAGTTGATGA | GGCAGTCATGTCCGGTGATG |
| YM1 | AGAAGGGAGTTTCAAACCTGGT | GTCTTGCTCATGTGTGTAAGTGA |
Microarray analysis
Affymetrix mouse gene 1.1 ST arrays were used to generate a gene expression dataset from BMDMs treated with LPS or LPS + PGE2 for 1 h and untreated control. Four biological replicates were analyzed per group. The arrays were generated by the Finnish Microarray and Sequencing Centre (Turku, Finland), according to the manufacturer’s protocols. Briefly, RNA was extracted as described above, and RNA quality was analyzed by Agilent’s Bioanalyzer electrophoresis station. Using specific protocols, hybridization to Affymetrix mouse gene 1.1 ST array plates (P/N 901622) was carried out in GeneTitan, and Affymetrix GeneChip Command Console 3.1 was used to control GeneTitan hybridization process and in summarizing probe cell intensity data (.CEL file generation).
The data analysis was carried out in the High Performance Computing facility of the College of Life Sciences using Affymetrix Power Tools, R (version 2.13.1) – Bioconductor and Partek GS 6.5 (version 6.11.0321) software. For quality control and probe sets annotations, the latest annotations files (release 32, dated 23-06-2011) downloaded from Affymetrix were used. Using Affymetrix Power Tools, the probe level expression data were summarized to transcript cluster level and normalized using two different normalization methods—robust multi-array average and probe logarithmic intensity error–guanine-cytosine background-adjusted. The dataset quality was analyzed from the spike in probe sets, principal component analysis, and hierarchical clustering analysis and by generating various quality control plots such as mean raw signal intensity plots, mean absolute deviation of the residuals plots, and probe cell intensity plots. Differential expression analysis was carried out using the ‘limma’ package. The potential CREB binding genes in the differentially expressed genes were arrived by querying them against the CREB target gene database that was locally created from the paper of Zhang et al. (35).
Array data has been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/ accession number GSE41833).
Immunoblotting
BMDMs were lysed directly into SDS sample buffer and aliquots run on 10% polyacrylamide gels using standard methods. RAW264.7 and Hek293 cells were lysed into a triton lysis buffer, and 30 μg soluble protein extract was run on the gels. For immunoprecipitation of SIK2, total SIK2 Ab coupled to protein G–Sepharose beads was used to immunoprecipitate SIK from 0.5 mg soluble cell extract. Proteins were transferred onto nitrocellulose membranes, and specific proteins were detected by immunoblotting. Abs against phospho-Tyr705 STAT3, total STAT3, phospho-ERK1/2, phospho-p38, total ERK1/2, total p38α, phospho-Ser133 CREB (which also recognizes ATF1 phosphorylated on Ser63), total CREB, phospho-Thr581 MSK1, total IκBα, p105, and phospho-class II HDAC (HDAC4 p-Ser246/HDAC5 p-Ser259/HDAC7 p-Ser155) were from Cell Signaling Technology. The CRTC3 Ab (EPR3440) was from Abcam. The phospho-Ser343 SIK2 was from a custom Ab program. The total MSK1 and GFP Abs were generated in-house (30). HRP-conjugated secondary Abs were from Pierce (Cheshire, U.K.), and detection was performed using the ECL reagent from Amersham Biosciences (Buckinghamshire, U.K.). Blots were quantified using Quantity One (Bio-Rad). All phospho-blots were quantified with respect to their total protein, or total ERK1/2 protein, and calculated as a ratio of unstimulated wild-type samples with WT 0 being equal to 1.
Cytokine measurements
IL-10, TNF, IL-6, IL-12p40, and IL-12p70 were measured using a multiplex-based assay from Bio-Rad according to the manufacturer’s protocols.
Phosphoproteomics
Phosphoproteomics was carried out in RAW264.7 cells that had been labeled using the stable isotope labeling of amino acids in cell culture (SILAC) method. After stimulation, cells were lysed, and protein levels were quantified. Equal amounts of soluble protein from each condition were reduced, alkylated and digested with trypsin. Phospho-peptides were then enriched by sequential hydrophilic chromatography followed by Fe3+-immobilized metal affinity chromatography (33). Phospho-peptides were measured by tandem mass spectroscopy on Thermo Fisher Scientific LTQ Orbitrap Velos instrument set to perform top-15 data-dependent collision-induced dissociation analysis in the 350–1600 m/z range using a resolution of 60,000 for the precursor scan and a minimal intensity for sequencing of 10,000 counts. Monoisotopic precursor selection was used, and +1 as well as unassigned charge states were excluded from sequencing. Dynamic exclusion was set to a repeat count of 2 within 30 s, with exclusion duration of 90 s and an exclusion mass width of 10 ppm. The data were analyzed using MaxQuant (34, 36).
Immunofluorescence
RAW264.7 cells or BMDMs were fixed for 10 min in 3.7% paraformaldehyde, permeabilized for 10 min using 1% Nonidet P-40 in PBS, and blocked for 30 min using 3% goat serum in PBS. RAW264.7 cells were then stained with anti-CRTC3 (1:200; Abcam), followed by anti-rabbit Alexa 488 (1:500; Invitrogen). Abs were diluted in blocking buffer and incubated with the cells for 1 h at 37°C. Cells were mounted in Prolong Gold Antifade Reagent containing DAPI (Invitrogen). Images of fields of cells were collected on a Carl Zeiss LSM 700 confocal microscope with an α Plan-Apochromat ×100 objective (numerical aperture = 1.46). All images were collected using the same settings for laser power, pinhole, gain etc. Fields were selected by uniform random sampling using only the DAPI channel to select cells. Nuclear intensity was quantified using Volocity software (PerkinElmer): the nuclei were identified using the DAPI channel, and the average nuclear CRTC3 intensity for each field was collected.
Luciferase assays
Hek293 cells were transiently transfected using pGL4.29[luc2P/CRE/Hygro] and Renilla pRL-SV40 vectors (Promega) with GFP, GFP-SIK2, or GFP-SIK2 S343A expression vectors. Cells were serum starved 16 h prior to stimulation with 20 μM forskolin for 4 h. Luciferase activities were measured by using the dual luciferase assay system (Promega), according to the manufacturer’s instructions. Both Firefly and Renilla luciferase activities were measured in the same samples. Renilla luciferase activity was used to normalize Firefly luciferase activity.
Results
PGE2 induces IL-10 via a cAMP–PKA-dependent pathway
Previous studies have shown that PGE2 can inhibit the TLR-stimulated production of proinflammatory cytokines. Consistent with this, we found that PGE2 was able to inhibit the production of TNF, IL-6, and IL-12 by BMDMs treated with the TLR4 agonist LPS (Supplemental Fig. 1A). Sequences for primers used are shown in Table I. PGE2 also inhibited TNF and IL-12 gene transcription. For IL-6, the situation was more complex, as although PGE2 was able to promote IL-6 transcription at early time points, it repressed the induction of IL-6 mRNA at later time points (Supplemental Fig. 1B). In contrast to its effect on proinflammatory cytokines, PGE2 had a positive effect on the production of IL-10 in response to LPS. Although PGE2 alone had little effect on IL-10 production by BMDMs, in combination with LPS, it was able to increase both IL-10 mRNA induction and secretion relative to stimulation with LPS alone (Fig. 1A, 1B). This effect was not restricted to TLR4, because PGE2 was also able to increase IL-10 secretion following stimulation with either the TLR7/8 agonist R848 or the TLR1/2 agonist Pam3CSK4 (Supplemental Fig. 1C). LPS treatment induces the phosphorylation of STAT3 on Tyr705, and this has previously been shown to be due to an autocrine feedback involving the IL-10 secreted in response to LPS (37). In line with the increased IL-10 production by LPS in combination with PGE2, increased STAT3 phosphorylation was observed in cells stimulated with a combination of LPS and PGE2 relative to cells treated with LPS alone (Supplemental Fig. 1D).
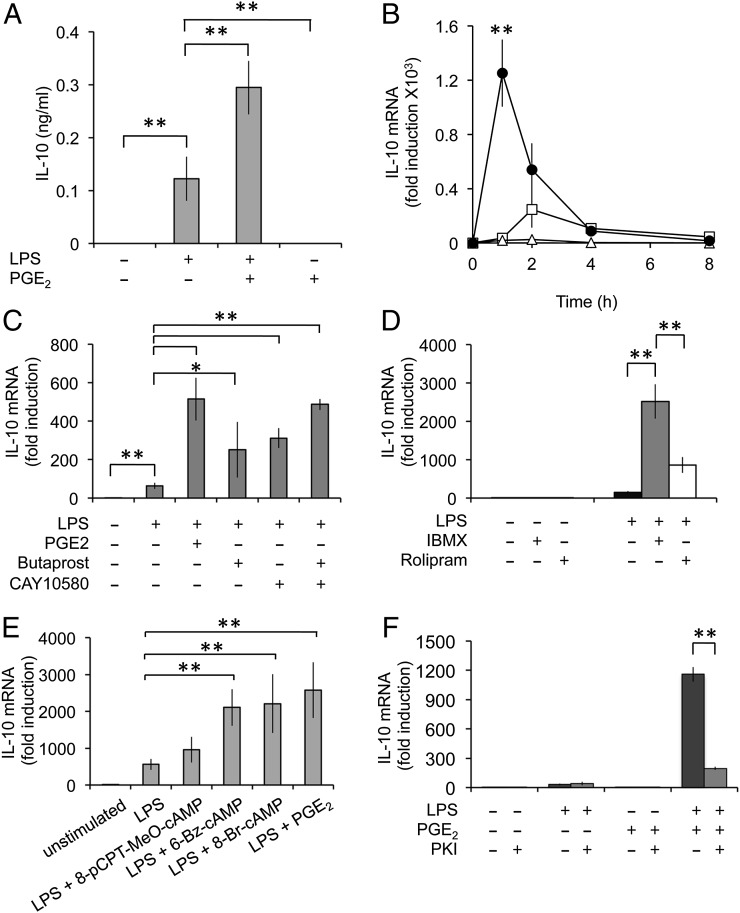
FIGURE 1.
PGE2 promotes the LPS-induced transcription of IL-10 and regulatory macrophage markers. BMDMs were isolated from wild type mice and stimulated for the indicated times with either 100 ng/ml LPS, 10 μM PGE2, or a combination of LPS and PGE2. (A) IL-10 levels secreted into the media were measured after 8 h as described in the methods. (B) Total RNA was extracted at the indicated times and the levels of IL-10 mRNA determined by qPCR. Results are expressed as fold change relative to the unstimulated control. (C) BMDMs were stimulated with either 100 ng/ml LPS or 100 ng/ml LPS in combination with 10 μM PGE2, 1 μM Butaprost (EP2 agonist), or 1 μM CAY10580 (EP4 agonist), and IL-10 mRNA levels were determined by qPCR after 1 h. (D) BMDMs were stimulated with either 100 ng/ml LPS or 100 ng/ml LPS in combination with 100 μM IBMX or 10 μM rolipram, and IL-10 mRNA levels were determined by qPCR after 1 h. (E) BMDMs were stimulated with either 100 ng/ml LPS or 100 ng/ml LPS in combination with 10 μM PGE2, 250 μM 8-pCPT-MeO-cAMP, 250 μM 6-Bz-cAMP, or 100 μM 8-Br-cAMP, and IL-10 mRNA levels were determined by qPCR after 1 h. (F) Where indicated, BMDMs were preincubated with 10 μM myristoylated PKI peptide for 1 h before stimulation with LPS and/or PGE2 for an additional hour. IL-10 mRNA levels were then measured by qPCR. For each panel, results represent the average and SD of independent cultures from four mice. *p < 0.05, **p < 0.01 (Student t test).
We next examined the signaling pathway by which PGE2 regulated IL-10 mRNA induction. PGE2 acts via members of the EP receptor family. These are GPCRs, and two of the four EP receptors (EP2 and EP4) couple via Gs and therefore can elevate cAMP levels allowing activation of PKA (38). Similar to PGE2, treatment of macrophages with either the EP2 agonist Butaprost or the EP4 agonist CAY10580 in combination increased IL-10 mRNA induction relative to LPS alone (Fig. 1C). A combination of EP2 and EP4 agonists had a greater effect than either agonist alone (Fig. 1C). In cells, cAMP is rapidly turned over via the action of phosphodiesterases (PDEs), and inhibition of PDEs should also mimic the effects of PGE2 by increasing cAMP levels. To test this, cells were treated with IBMX, a pan-PDE inhibitor. Similar to PGE2, IBMX had little effect on IL-10 mRNA induction on its own, but did have a synergetic effect in combination with LPS (Fig. 1D). PDE4 inhibitors have been proposed as anti-inflammatory drugs, so we also tested the effect of the PDE4-specific inhibitor rolipram. Similar to IBMX, rolipram also increased LPS-induced IL-10 mRNA levels although the effect was less pronounced than for IBMX (Fig. 1D). cAMP can signal downstream through both PKA and exchange proteins activated by cAMP (EPAC)–dependent pathways. The actions of these two pathways can be distinguished through the use of cAMP analogs specific for the individual pathways. Treatment of the cells with 8-Br-cAMP, which activates both PKA and EPAC, was able to mimic the effects of PGE2 on LPS-induced IL-10 mRNA levels. A similar result was obtained with the PKA-specific analog 6-Bz-cAMP (Fig. 1E). The EPAC-specific compound 8-pCPT-MeO-cAMP, however, had no effect on IL-10 mRNA levels. Consistent with the role for PKA, treatment of the cells with a cell-permeable (myristolyated) form of the PKA inhibitory peptide, PKI, blocked the effect of PGE2 on LPS-induced IL-10 mRNA induction (Fig. 1F).
IL-10 production sets up a feedback loop to repress the production of proinflammatory cytokines. As discussed above, PGE2 inhibits proinflammatory cytokine production, raising the possibility that PGE2 could repress LPS-induced cytokine production via enhancing IL-10 secretion. To examine this, BMDMs were isolated from wild-type and IL-10 knockout mice. In both genotypes, PGE2 strongly repressed TNF and IL-12p70 production, indicating that the primary effect of PGE2 on these cytokines was IL-10 independent (Supplemental Fig. 1E). PGE2 was also able to repress the induction of both IL-6 and IL-12p40 secretion in response to LPS in both wild-type and IL-10 knockout cells. The degree of inhibition however was greater in wild-type cells compared with IL-10 knockouts, suggesting that PGE2 repressed IL-6 and IL-12p40 via both IL-10–dependent and –independent mechanisms (Supplemental Fig. 1E).
PGE2 induces a regulatory-like phenotype in macrophages
The ability to produce high levels of IL-10 but low amounts of IL-12 has previously been associated with the polarization of macrophages into a regulatory phenotype (7). To examine the effect of PGE2 on macrophage polarization, we analyzed the effect of PGE2 on the expression of macrophage polarization markers. PGE2, either alone or in combination with LPS, did not induce the transcription of Ym1 (Fig. 2A) or MR1 (data not shown), markers of alternatively activated (M2a) macrophages (7, 8). A combination of LPS and PGE2 was, however, able to induce the mRNA for Arg1, another potential marker for alternatively activated macrophages in mice (Fig. 2B). LIGHT (TNFSF14) and SPHK1 have been reported as markers of regulatory (M2b) macrophages (39). Although neither LPS nor PGE2 alone effectively stimulated the transcription of these genes, a combination of both LPS and PGE2 together was able to induce these genes (Fig. 2A).
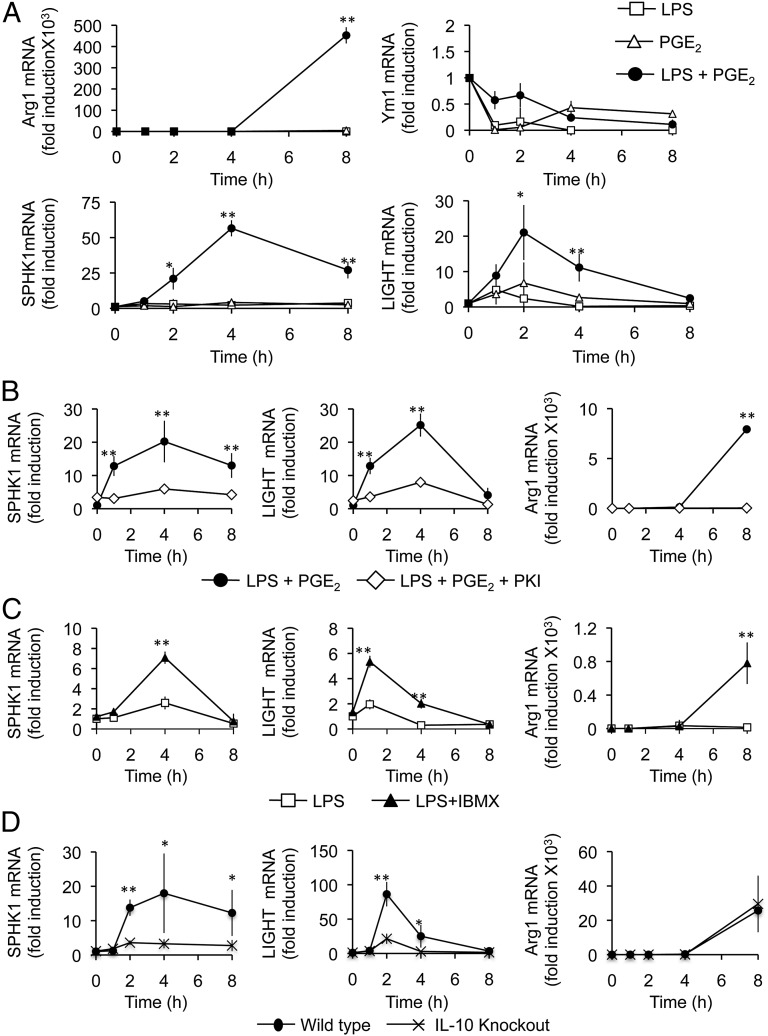
FIGURE 2.
PGE2 promotes the expression of regulatory macrophage markers. (A) BMDMs were stimulated with 100 ng/ml LPS, 10 μM PGE2, or a combination of LPS and PGE2 for the indicated times, and the levels of Ym1, Arg1, LIGHT, and SPHK1 mRNAs were determined. (B) BMDMs were preincubated with 10 μM myristoylated PKI peptide for 1 h as indicated before stimulation with a combination of LPS and PGE2 for the indicated times. The levels of SPHK1, LIGHT, and Arg1 mRNAs were determined by qPCR. (C) BMDMs were stimulated for the indicated times with either LPS or LPS and IBMX, and the levels of SPHK1, LIGHT, and Arg1 mRNAs were determined by qPCR. (D) BMDMs were isolated from either wild-type or IL-10 knockout mice and stimulated for the indicated time with a combination of LPS and PGE2. The levels of SPHK1, LIGHT, and Arg1 mRNAs were determined by qPCR. Results represent the average and SD of independent cultures from four mice. *p < 0.05, **p < 0.01 (Student t test).
The induction of LIGHT, SPHK1, and Arg1 in response to a combination of LPS and PGE2 was blocked by PKI (Fig. 2B), whereas treatment of the cells with the PDE inhibitor IBMX in combination with LPS also induced the mRNA for these genes (Fig. 2C), indicating a role for PKA in the induction of these genes. Because IL-10 feedback can modulate transcription in macrophages, we examined the induction of the regulatory marker genes in IL-10 knockout macrophages. The induction of both LIGHT and SPHK1 mRNA was dependent on IL-10. In contrast, the effect of PGE2 on Arg1 induction was independent of IL-10 (Fig. 2D).
PGE2 promotes IL-10 mRNA induction independently of CREB Ser133 phosphorylation
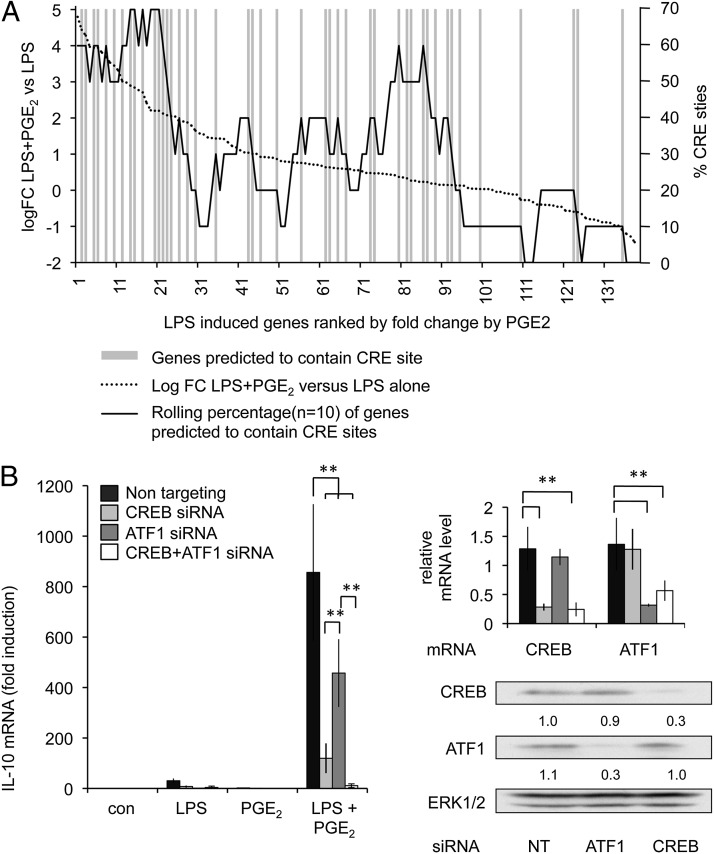
The above data demonstrate that PGE2 synergizes with LPS to promote the transcription of IL-10 as well as genes associated with an anti-inflammatory phenotype in macrophages. To examine the ability of PGE2 to affect changes in mRNA levels induced by LPS, we carried out microarray profiling on BMDMs stimulated with either LPS or a combination of LPS and PGE2 for 1 h. At this time point, although PGE2 was able to repress a small number of LPS-induced genes (including IL-12 and TNF), most genes were either unaffected or induced more strongly when PGE2 was added in combination with LPS relative to LPS alone (Supplemental Fig. 2). The genes that were upregulated when PGE2 was added included several genes, such as nur77 and Areg, that have been shown in other systems to be CREB dependent (40, 41). This would suggest that PGE2 could act via a PKA–CREB-dependent pathway. We therefore compared the upregulated genes with a database of predicted and validated CREB target genes (http://natural.salk.edu/CREB/). This demonstrated that there was an enrichment in potential CREB target genes in the genes that were more strongly upregulated by a combination of LPS and PGE2 relative to LPS alone (Fig. 3A). Therefore, PGE2 is likely to exert some of its early effects on LPS-induced transcription in a CREB-dependent manner. Binding sites for CREB have been previously identified in the IL-10 promoter (14), whereas knockout of MSK1 and 2, which phosphorylate CREB downstream of TLR4, reduces IL-10 induction in response to LPS (37). To determine whether CREB was required for PGE2 to act in concert with LPS to promote IL-10 transcription, we used siRNA to knockdown CREB, or the related transcription factor ATF1, in RAW264.7 cells (Fig. 3B). This showed that knockdown of CREB inhibited the transcription of IL-10 in response to a combination of LPS and PGE2. Knockdown of ATF1 had less effect; however, a combined knockdown of CREB and ATF1 was more effective than knockdown of CREB alone (Fig. 3B).
FIGURE 3.
PGE2 synergizes with LPS to promote CREB-dependent transcription. (A) BMDMs were stimulated with 100 ng/ml LPS or a combination of LPS and 10 μM PGE2 for 1 h. Total RNA was isolated and subjected to microarray analysis as described in the Materials and Methods. Data were then filtered to identify genes that were upregulated at least 4-fold by LPS or LPS and PGE2. The upregulated genes were compared with a list of potential CREB target genes (http://natural.salk.edu/CREB/). Genes were ranked by the fold change between stimulation with LPS alone and a combination of LPS and PGE2. Genes in this ranking that were predicted to contain cAMP response element (CRE) sites in their promoters are indicated by the gray bars. A rolling average (n = 10) of the percentage of CRE-containing genes was also calculated. (B) RAW264.7 cells were transfected with either a nontargeting siRNA control or siRNAs against CREB or ATF1 as indicated. The levels of CREB and ATF1 knockdown were determined by qPCR or immunoblotting (right panels). Levels of CREB and ATF1 protein were quantified relative to total ERK1/2 protein and represented as a ratio of the nontargeting control. Cells were stimulated with the indicated combinations of 100 ng/ml LPS or 10 μM PGE2 for 1 h. IL-10 mRNA levels were then measured by qPCR (left panels). Data represent the average and SD of four stimulations. **p < 0.01 (Student t test) relative to the nontargeting oligonucleotide transfections alone.
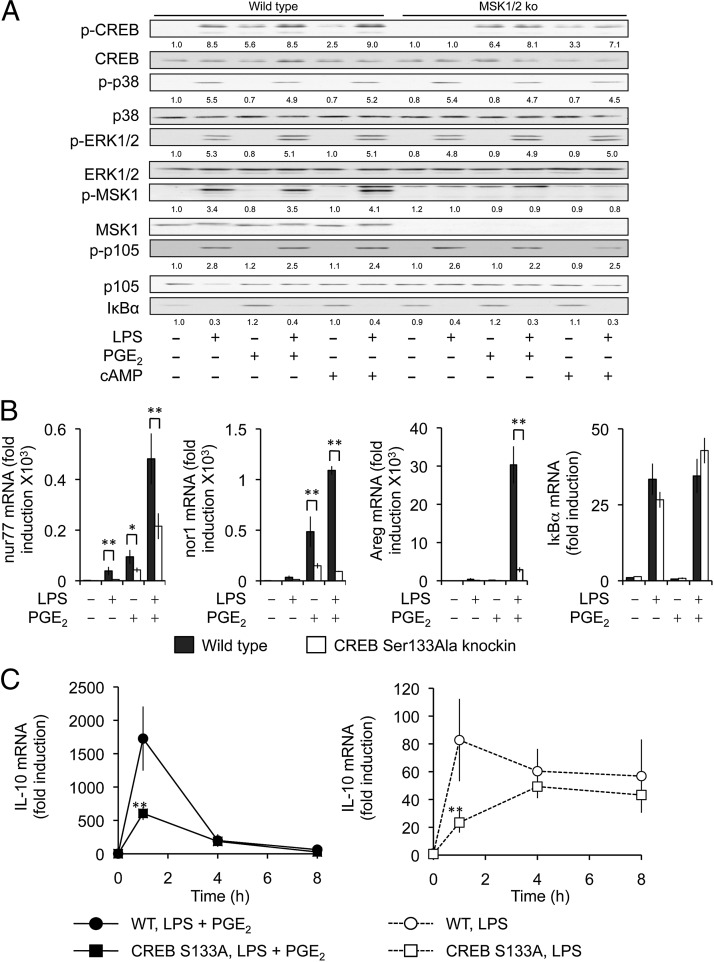
The classical mechanism by which CREB is activated involves the phosphorylation of Ser133, which creates a binding site for CBP or p300 (42). Ser133 in CREB can be phosphorylated by several kinases, including PKA downstream of cAMP and MSK1 or 2 downstream of TLR signaling (37, 42–44). One explanation of the effect of PGE2 on IL-10 transcription would be that it further increases LPS-induced CREB phosphorylation. We therefore examined whether PGE2 or 8-Br-cAMP could induce a synergistic effect on either the phosphorylation of CREB on Ser133 or ATF1 on Ser63 (the site analogous to Ser133 in CREB). LPS-induced the phosphorylation of CREB and ATF1, and this was prevented by double knockout of MSK1 and 2 (Fig. 4A). PGE2 or cAMP also induced CREB and ATF1 phosphorylation, and in line with this being catalyzed by PKA, it was unaffected by MSK1/2 knockout. Surprisingly, a combination of LPS and either PGE2 or 8-Br-cAMP did not result in a stronger phosphorylation of CREB or ATF1 than with either agonist alone. This was not due to an inhibition of TLR signaling by PGE2, because PGE2 did not affect the activation of MSK1/2, or their upstream activators ERK1/2 or p38 MAPK. PGE2 also did not affect LPS-induced NF-κB signaling as judged by the phosphorylation of p105 or degradation of IκBα (Fig. 4A).
FIGURE 4.
PGE2 acts via the alternative CREB coactivator CRTC3 in macrophages. (A) BMDMs from wild-type or MSK1/2 double-knockout mice were isolated and stimulated for 30 min with 100 ng/ml LPS, 10 μM PGE2, or 100 μM 8-Br-cAMP as indicated. Cells were then lysed, and the levels of the indicated proteins were determined by immunoblotting. Levels of phosphorylated proteins were quantified relative to their respective total protein, with the exception of phospho-MSK1 and IκBα, which are quantified respective to total ERK1/2, and represented as a ratio of the unstimulated wild-type control. (B) BMDMs from wild-type or CREB Ser133Ala knockin mice were isolated and stimulated with the indicated combinations of LPS and PGE2 for 1 h. Total RNA was then isolated and the levels of nur77, nor1, Areg, and IκBα were determined by qPCR. (C) Wild-type or CREB Ser133Ala BMDMs were stimulated with LPS or a combination of LPS and PGE2 for the indicated times and the levels of IL-10 mRNA determined by qPCR. Because of the much higher fold induction in the presence of PGE2, data for LPS alone were also plotted separately in the left panel. Results represent the average and SD of independent cultures from four mice per genotype. For differences between the wild-type and CREB Ser133Ala BMDMs, *p < 0.05, **p < 0.01 (Student t test).
To examine the role of Ser133 phosphorylation in IL-10 transcription more directly, we isolated BMDMs from mice that had a Ser to Ala knockin mutation of either Ser133 in CREB or Ser63 in ATF1. The Ser63Ala ATF1 knockin mutation had no effect on the induction of IL-10 in response to LPS or a combination of LPS and PGE2 (Supplemental Fig. 3). The mutation of Ser63 in ATF1 also did not affect the transcription of the classical CREB dependent gene nur77, suggesting that CREB was the dominant isoform for these responses (Supplemental Fig. 3). In contrast, the mutation of Ser133 in CREB greatly reduced the transcription of the classical CREB dependent immediate early genes nur77, nor1, and Areg in response to either LPS or PGE2 (Fig. 4B). This was not due to a general impairment in transcription as the induction of the CREB-independent gene IκBα was normal (Fig. 4B). Interestingly, the effect of the CREB knockin on the ability of PGE2 to have a synergistic effect with LPS on CREB-dependent gene induction was gene specific. For nur77 and Areg, a combination of PGE2 and LPS gave a stronger induction than either agonist alone in both wild-type and CREB Ser133Ala knockin cells, although the absolute levels of nur77 and Areg mRNA were lower in the knockin BMDMs compared with wild-type cells. In the CREB Ser133Ala cells, LPS and PGE2 together did not, however, increase nor1 mRNA induction more than PGE2 alone (Fig. 4B). The mutation of Ser133 in CREB reduced the ability of LPS to stimulate IL-10 transcription at early time points, although less effect was seen at later times (Fig. 4C, right panel). This is consistent with the finding that prolonged IL-10 transcription in response to LPS is regulated via an IFN-β feedback loop (45). The induction of IL-10 in response to a combination of LPS and PGE2 was also reduced in the CREB knockin BMDMs relative to wild-type cells (Fig. 4C). Interestingly, however, the CREB knockin did not block the synergetic effect of PGE2 on LPS-induced cytokine production, and the fold induction seen between LPS and a combination of LPS and PGE2 was similar in the two genotypes.
CRTC3 is required for PGE2 to regulate IL-10 mRNA induction
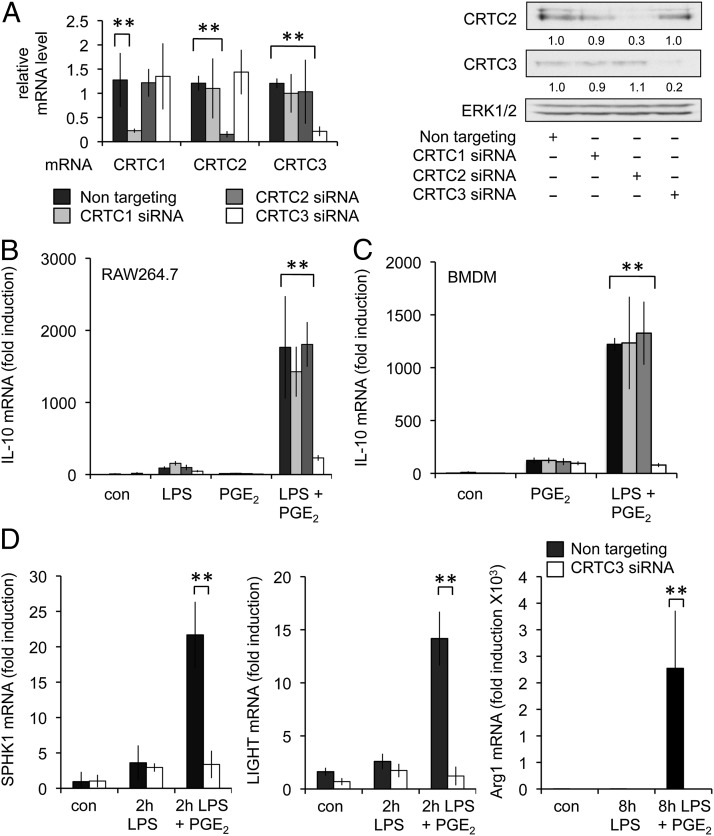
The above results would suggest that PGE2 regulates the IL-10 promoter via a mechanism dependent on CREB but at least in part independently of Ser133 phosphorylation. Because the CRTC proteins have been proposed to bind to CREB independently of Ser133 phosphorylation, we examined the role they played in the induction of IL-10 (46, 47). Three isoforms of CRTC exist in mammalian cells, and siRNA was used to knockdown each isoform in RAW264.7 cells. qPCR for CRTC1, 2, or 3 demonstrated that the knockdowns were successful and isoform specific (Fig. 5A). To date, we have not yet identified a good Ab for mouse CRTC1; however, immunoblotting for CRTC2 and CRTC3 confirmed that knockdown of CRTC2 and CRTC3 protein was successful (Fig. 5A). Analysis of IL-10 mRNA levels revealed that knockdown of CRTC1 or CRTC2 had no effect on the induction of IL-10 mRNA. Knockdown of CRTC3, however, was able to block the synergistic effect of PGE2 on LPS-induced IL-10 mRNA levels (Fig. 5B). Similar results were also obtained in primary BMDMs (Fig. 5C). CRTC3 knockdown also blocked the induction of SPHK1, LIGHT, and Arg1 by LPS/PGE2 stimulation (Fig. 5D).
FIGURE 5.
CRTC3 is required for PGE2 to promote IL-10 and regulatory marker gene expression. (A) RAW264.7 cells were transfected with siRNA against CRTC1, 2, or 3 or a nontargeting siRNA control. The effectiveness of the knockdowns was determined by qPCR for CRTC1, 2, and 3 mRNA as well as by immunoblotting for CRTC2 and CRTC3. Levels of CRTC2 and CRTC3 protein were quantified relative to total ERK1/2 protein and represented as a ratio of the nontargeting control. (B) After transfection with the indicated siRNA, RAW264.7 cells were stimulated with the indicated combinations of 100 ng/ml LPS or 10 μM PGE2 for 1 h. IL-10 mRNA levels were then measured by qPCR. Error bars represent the SD from three transfections per condition. (C) Wild-type BMDMs were transfected with siRNA against CRTC1, 2, or 3 or a nontargeting siRNA control. Twenty-four to 48 hours after transfection cells were stimulated with either LPS or LPS and PGE2 for 1 h, and the levels of IL-10 mRNA were determined. (D) Wild-type BMDMs were transfected with siRNA against CRTC3 or a nontargeting siRNA. Cells were then stimulated with either LPS or LPS and PGE2 for 2 or 8 h as indicated. The induction of SPHK1, LIGHT, and Arg1 was then determined by qPCR. In (C) and (D), error bars represent the SD of results from four independent preparations of BMDMs. **p < 0.01 (Student t test) relative to the nontargeting oligonucleotide transfections alone.
SIKs regulate CRTC3 localization and IL-10 induction
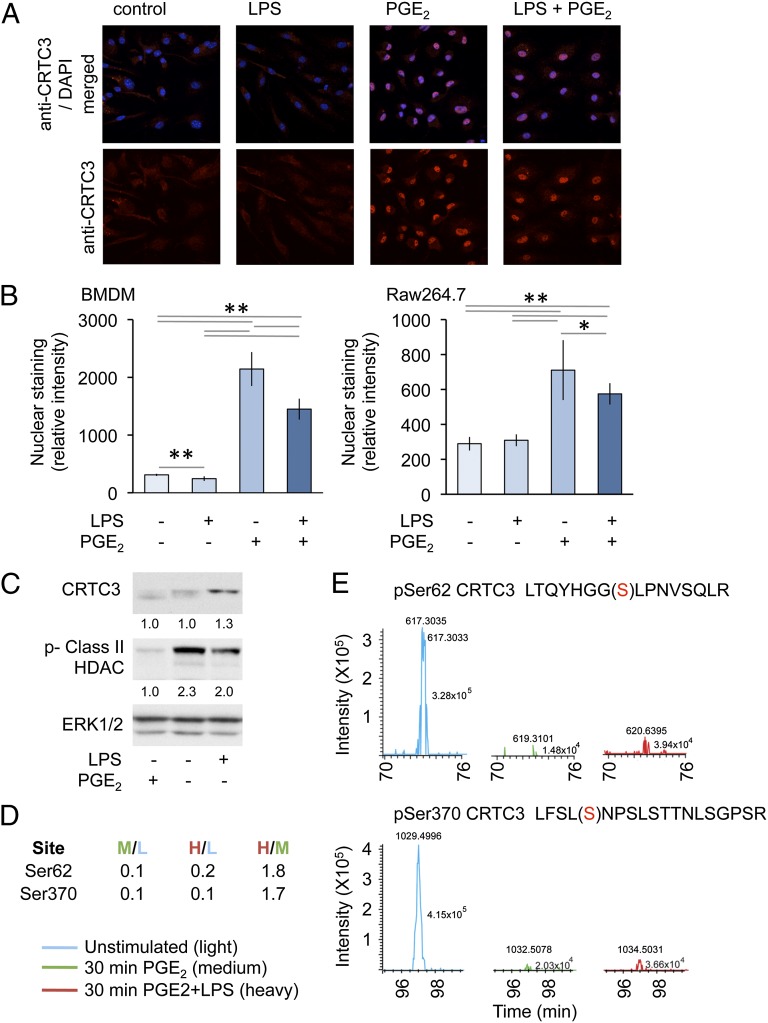
CRTC3 was found to be predominantly localized to the cytoplasm in unstimulated BMDMs or RAW264.7 cells (Fig. 6A, 6B), and this localization was not affected by LPS stimulation. PGE2 was able to promote the relocalization of CRTC3 from the cytoplasm to the nucleus. This relocalization was also observed when cells were stimulated with a combination of both LPS and PGE2 (Fig. 6A, 6B). By analogy with what has been proposed for CRTC2, CRTC3 localization would be controlled via the phosphorylation of specific 14-3-3 binding sites that promote the retention of CRTC3 in the cytoplasm. Stimulation of RAW264.7 cells with PGE2 resulted in the dephosphorylation of CRTC3, as judged by a downward band-shift on SDS polyacrylamide gels; a result consistent with the ability of PGE2 to promote the dephosphorylation of CRTC3 and therefore promote its nuclear localization (Fig. 6C). To examine CRTC3 phosphorylation in more detail, we used a SILAC-based phosphoproteomic approach. Analysis of these data revealed two peptides, corresponding to Ser62 and Ser370, that were dephosphorylated in response to stimulation with PGE2 (Fig. 6D, 6E). By analogy with CRTC2 (46), Ser162 would also be predicted to be a SIK phosphorylation site, however in CRTC3 this site lies in a large tryptic peptide that would be unlikely to be detected by mass spectrometry methods used. These sites are phosphorylated by members of the SIK family, and mutation of these sites to prevent their phosphorylation results in the localization of CRTC3 to the nucleus and increases CREB dependent transcription (Supplemental Fig. 4).
FIGURE 6.
PGE2 regulates CRCT3 localization and phosphorylation. (A and B) BMDMs isolated from wild-type mice or RAW264.7 cells were plated onto coverslips. Cells were stimulated for 1 h with 100 ng/ml LPS or 10 μM PGE2 as indicated. Cells were then fixed and stained with an anti-CRTC3 Ab (red) and DAPI (blue). Images were obtained by confocal microscopy. Original magnification ×100; the view shown is 127.9 μ wide. Representative images from BMDMs are shown (A). Nuclear staining was quantified as described in the methods. Error bars represent SD from 10 independent images; *p < 0.05, **p < 0.01 (Student t test) (B). (C) RAW264.7 cells were stimulated with either LPS or PGE2 for 30 min, and the levels of CRTC3, ERK1/2, and phosphorylated class II HDACs were determined by immunoblotting. Levels of CRTC3 and phosphorylated HDAC protein were quantified relative to total ERK1/2 protein and represented as a ratio of the unstimulated control. (D and E) RAW264.7 cells were labeled with SILAC medium as described in Materials and Methods. Cells grown in light media were left untreated, whereas cells grown in medium media were treated with 10 μM PGE2, and cells grown in heavy media were stimulated with a combination of 10 μM PGE2 and 100 ng/ml LPS. After 30 min, cells were lysed and subjected to a phosphoproteomic screen as described in Materials and Methods. Mass spectroscopy spectra for peptides corresponding to CRTC3 phosphorylated on Ser62 and Ser370 are shown (E) and the ratios of these peptides in the different conditions are given in (D).
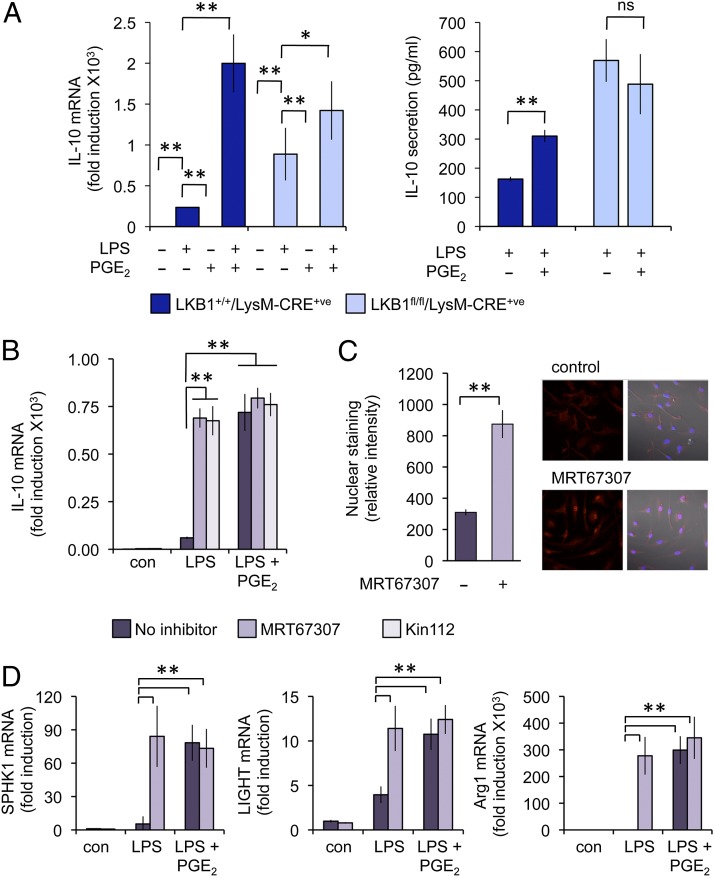
Because members of the SIK family have been suggested as potential kinases for the 14-3-3 binding sites on CRTC2, we examined the roles of these kinases in IL-10 induction. SIKs are members of the AMPK-related kinase group. Like other AMPK family members, SIKs require phosphorylation of their T loop by LKB1 to be active (48). We therefore generated a conditional knockout of LKB1 in macrophages using a LysM CRE. Analysis of BMDMs from these mice revealed that LPS-induced higher levels of IL-10 mRNA compared with wild-type cells, which would be consistent with a constitutive activation of CRTC3 in these cells. Importantly, PGE2 was not able to significantly increase IL-10 mRNA induction in response to LPS in LKB1-deficient cells (Fig. 7A). Although this would be consistent with a role for SIKs in regulating IL-10 transcription, other AMPK-related kinases in addition to SIKs are inactivated in LKB1 knockouts. To address this issue we employed two novel and structurally different inhibitors of SIKs. MRT67307 was originally described as a TBK1 inhibitor, however it also effectively inhibits SIKs. Importantly, Kin112 inhibits SIKs but not other members of the AMPK family (28). MRT67307 and Kin112 did not induce IL-10 transcription on their own; however, they were able to synergistically induce IL-10 mRNA induction in combination with LPS. Importantly, this occluded the effect of PGE2 on LPS-induced IL-10 transcription, suggesting that PGE2 and SIK inhibitors acted via a common mechanism (Fig. 7B). In line with its ability to promote LPS-induced IL-10 transcription, MRT67307 was able to induce the nuclear localization of CRTC3 in BMDMs (Fig. 7C). Because PGE2 promotes the induction of regulatory macrophage marker genes via CRTC3 (Figs. 2, 5), we also examined the effect of SIK inhibitors on these genes. A combination of MRT67307 and LPS resulted in a higher induction of LIGHT, SPHK1, and Arg1 mRNA relative to LPS alone, and MRT67307 occluded the ability of PGE2 to promote the transcription of these genes (Fig. 7D).
FIGURE 7.
PGE2 regulates CRTC3-induced IL-10 transcription via SIKs. (A) BMDMs were isolated from LKB1+/+/LysM-CRE+ or LKB1fl/fl/LysM-CRE+ (myeloid cell knockout) mice. Cells were stimulated with 100 ng/ml LPS or LPS and 10 μM PGE2 for 1 h, and the levels of IL-10 were determined by qPCR (A) and the secretion of IL-10 was measured at 8 h. Error bars represent the SD of three stimulations. (B) BMDMs were isolated from wild-type mice and pretreated for 1 h with 2 μM MRT67307 or 10 μM Kin112 as indicated. Cells were then stimulated with either LPS or LPS and PGE2 for 1 h, and IL-10 mRNA levels were measured by qPCR. Error bars represent the SD of results from four independent preparations of BMDMs. (C) BMDMs were left untreated or treated with MRT67307 for 1 h, and CRTC3 localization was determined by immunofluorescence (original magnification ×100; the view shown is 127.9 μ wide); representative images are shown, and the nuclear staining quantified as described in Materials and Methods. Error bars represent the SD of the quantification of 10 fields of view. (D) Where indicated, cells were treated with 2 μM MRT67307 for 1 h and then stimulated with either LPS or LPS and PGE2. SPHK1 and LIGHT mRNA levels were then measured after 4 h of stimulation and Arg1 levels after 8 h of stimulation. Error bars represent the SD of results from four independent preparations of BMDMs. *p < 0.05, **p < 0.01 (Student t test).
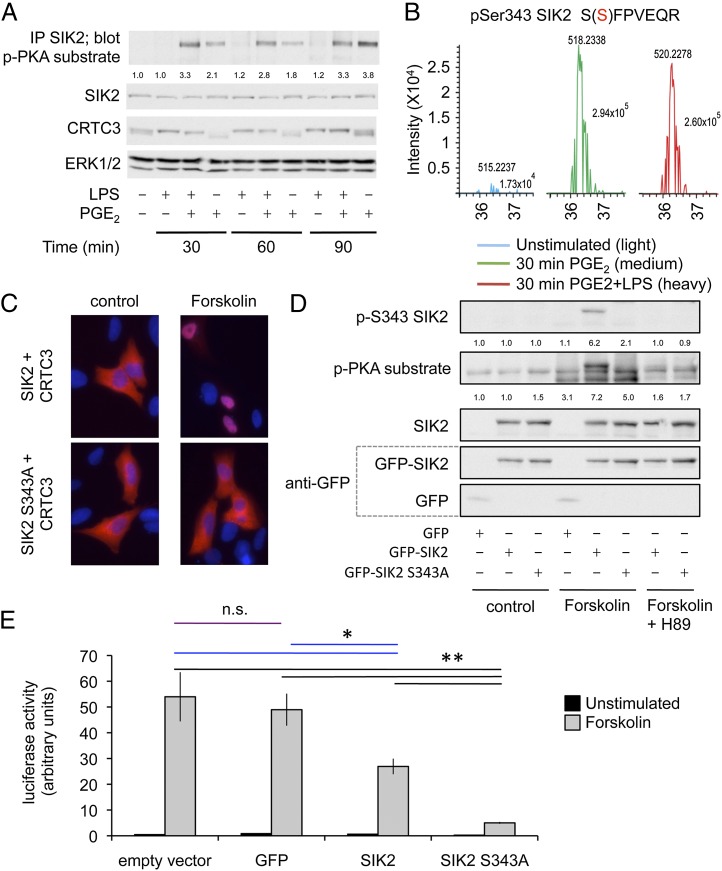
Because SIK2 has been suggested to regulate CRTCs in neurons (49), we examined the ability of PKA to phosphorylate SIK2 in macrophages. Immunoprecipitation of SIK2 followed by blotting with a phospho-PKA site substrate Ab showed that SIK2 becomes phosphorylated in response to PGE2 stimulation. To identify which sites in SIK2 were phosphorylated, we again used SILAC-based phosphoproteomics. In these experiments, phospho-peptides were detected for SIK2 and SIK3 but not SIK1. Of the peptides detected, one corresponding to phosphorylation on Ser343 was highly induced by PGE2 treatment of the macrophages (Fig. 8B). The phosphorylation of the other SIK-derived peptides detected was not strongly induced by PGE2 (Table II). To examine the role that Ser343 in SIK2 plays in the regulation of CRTC3, we overexpressed wild-type or Ser343Ala SIK2 in HeK293 cells. Following stimulation with forskolin to activate PKA, CRTC3 localized to the nucleus in cells expressing wild-type SIK2. In contrast, in the presence of cells expressing S343A SIK2, CRTC3 remained predominantly cytoplasmic (Fig. 8C). We also examined the ability of PKA to phosphorylate the overexpressed SIK2 in Hek293 cells. Wild-type SIK was phosphorylated in response to forskolin treatment, as judged by blotting with either a phospho-PKA substrate Ab or a phospho-S343 SIK2 Ab. As would be expected, the phospho-PKA Ab recognized other bands in the cell lysate; however, it was not able to recognize the S343A SIK2 mutant (Fig. 8D). The phospho-S343 SIK Ab was also unable to recognize the S343A mutant, demonstrating the specificity of this Ab (Fig. 8D). Unfortunately, the phospho-S343 was not sensitive enough to detect SIK in macrophages. Finally, we looked at the ability of wild-type and S343A SIK2 to regulate a CREB-dependent luciferase reporter. Expression of GFP did not affect the ability of forskolin to activate a CREB-dependent reporter. Expression of wild-type SIK2 did reduce the induction of the luciferase reporter, which would be consistent with increased levels of SIK2 protein, making it more difficult for PKA to completely inhibit CRTC activation. Importantly, however, mutation of Ser343 to Ala in SIK2 greatly reduced the ability of forskolin to induce the CREB-dependent luciferase reporter (Fig. 8C).
FIGURE 8.
PGE2 regulates the ability of SIK to inhibit CRTC activity. (A) RAW264.7 cells were stimulated for the indicated times with 100 ng/ml LPS, 10 μM PGE2, or both LPS and PGE2. SIK2 was immunoprecipitated and the immunoprecipitate blotted with a phospho-PKA substrate Ab. In parallel the levels of SIK2, CRTC3 and ERK1/2 were determined by immunoblotting. Levels of phospho-PKA protein were quantified relative to total SIK2 protein and represented as a ratio of the unstimulated control. (B) RAW264.7 cells were labeled with SILAC medium and phosphoproteomics carried out on unstimulated, PGE2-, and LPS + PGE2-stimulated cells as described in Materials and Methods. Data for the phospho-peptide corresponding to Ser343 on SIK2 is shown. (C) U2OS cells were transfected with mCherry-CRTC3 and either SIK2 or S343A SIK2 expression constructs. Cells were stimulated with 50 μM forskolin for 30 min and fixed, and CRTC3 localization was examined. DAPI (DNA) staining is shown in blue and mCherry-CRTC3 in red (original magnification ×100; image width is 110.2 μ). (D) Hek293 cells were transfected with the indicated expression constructs. Cells were either left unstimulated, stimulated with 50 μM forskolin for 30 min, or incubated with 10 μM H89 for 1 h and then stimulated with forskolin. Lysates were then blotted with Abs against either GFP, SIK2, phospho-Ser343 SIK2, or with a phospho-PKA substrate Ab. Levels of phospho-PKA and phospho-Ser343 SIK2 protein were quantified relative to total SIK2 protein and represented as a ratio of the unstimulated control. (E) Hek293 cells were transfected with a CREB-dependent luciferase reporter in combination with either an empty expression vector or expression vectors for GFP, wild-type GPF-tagged SIK2, or Ser343Ala GFP-tagged SIK2. Cells were either left unstimulated or stimulated with 50 μM forskolin to induce PKA activation for 4 h. Error bars represent the SEM of four stimulations. The results are representative of two independent experiments. For the forskolin stimulation, n.s. represents a nonsignificant difference (p > 0.05, Student t test). *p < 0.05, **p < 0.01 (Student t test).
Table II. SIK phospho-peptides identified in Raw264.7 cells.
| Ratios |
||||||
|---|---|---|---|---|---|---|
| Uniport | Kinase | Phospho-peptide | Site | M/L | H/L | H/M |
| Q8CFH6 | SIK2 | S(p-S)FPVEQR | Ser343 | 10.2 | 8.9 | 0.9 |
| Q8CFH6 | SIK2 | RP(p-S)TIAEQTVAK | Ser358 | 0.2 | 0.1 | 0.7 |
| Q8CFH6 | SIK2 | RA(p-S)DTSLTQGIVAFR | Ser587 | 0.2 | 1.0 | 1.0 |
| Q6P4S6 | SIK3 | RH(p-T)VGVADPR | Thr411 | 1.3 | 1.1 | 0.9 |
| Q6P4S6 | SIK3 | RA(p-S)DGGANIQLHAQQLLK | Ser493 | 0.8 | 1.2 | 1.3 |
| Q6P4S6 | SIK3 | RF(p-S)DGAASIQAFK | Ser616 | 2.7 | 2.4 | 0.9 |
Raw264.7 cells were labeled in SILAC medium. Cells were then either left unstimulated (cells grown in light media, L) or stimulated with either 10 mM PGE2 (cells grown in medium media, M) or a combination of PGE2 and 100 ng/ml LPS (cells grown in heavy media, H). Phosphoproteomic analysis was carried out as described in Materials and Methods, and the phospho-peptides identified for SIK2 and SIK3 are listed along with their ratios in the different conditions.
Discussion
PGE2 has been shown to affect macrophage function in a variety of systems. For instance, PGE2 from bone marrow stromal cells has been shown to promote IL-10 production from macrophages in vivo, and this plays a protective role in sepsis (50). In the lung, the production of PGE2 following the ingestion of apoptotic cells by phagocytes leads to enhanced IL-10 production and inhibition of phagocytosis resulting in increased susceptibility to Streptococcus pneumoniae (51), whereas PGE2 production is involved in sensitizing mice to an i.v. model of Streptococcus pyogenes (52). PGE2 produced by tumor cells has been suggested to play a role in the induction of IL-10 in macrophages (53). Elevated levels of cAMP and PGD2 have also been proposed to promote a proresolution macrophage phenotype in a zymosan-induced peritonitis model (54). Although a role for PKA was found in some of these studies, the molecular mechanism by which PGE2 modulates IL-10 production was not established. We show in this study that PGE2 in combination with LPS promotes the induction of high levels of IL-10. Increases in mRNA levels can be a result of either increased transcription or increased stability of the mRNA. Given the roles for the transcriptional regulators CREB and CRTC3 downstream of PGE2, the most likely explanation of the increase in IL-10 mRNA levels would be an increase in gene transcription. Our data would be consistent with a model in which PGE2 activates PKA, resulting in the phosphorylation of SIK2, which in turn inhibits the cellular ability of SIKs to phosphorylate CRTC3. This allows the SIK2 sites on CRTC3 to become dephosphorylated, resulting in the translocation of CRTC3 to the nucleus. Once in, the nucleus CRTC3 can interact with CREB to promote the transcription of CREB-dependent genes, which in macrophages includes IL-10. Our data would indicate that Ser343 is the critical site on SIK2 for regulation via PKA in macrophages. Ser358 and Ser587 in SIK2, or the corresponding Ser577 site in SIK1, have previously been suggested as PKA sites in other cell types (55–57). Although we detected these sites in macrophages (Table II), we did not observe any increase in their phosphorylation in response to PGE2, indicating that they are unlikely to be involved in our system.
It should be noted that activation of CREB alone is insufficient to induce IL-10 transcription. PGE2 promotes CREB Ser133 phosphorylation and CRTC3 relocalization as well as the induction of several CREB-dependent immediate early genes such as nur77 and nor1 but is a very weak stimulus for IL-10 transcription on its own. This would indicate that other CREB-independent inputs are necessary for IL-10 transcription. Given the complexity of the IL-10 promoter (14, 15), it is not unexpected that CREB is necessary but not sufficient for IL-10 transcription. Interestingly, IL-6 also has a potential CREB site in its promoter, and this may explain the biphasic effect of PGE2 on IL-6 transcription: at early time points, PGE2 promoted IL-6 transcription but at later time points it was repressed. This could be explained by the early effects of PGE2 being mediated by its ability to increase CREB-dependent transcription, whereas at later time points, it acts to repress IL-6 via a CREB-independent mechanism. This CREB-independent mechanism was not addressed in this study; however, it could relate to the ability of PGE2 to directly repress the expression of other proinflammatory cytokines, including TNF and IL-12. This could be explained by the recent report that PKA can directly target the p105 NF-κB subunit (58).
In addition to promoting IL-10 transcription, PGE2 in combination with LPS also induced a regulatory or M2b-like phenotype in macrophages, as indicated by induction of the marker genes SPHK1 and LIGHT. Arg1 was also upregulated by LPS and PGE2. Arg1 has been proposed as a marker of alternatively activated/M2a macrophages in the mouse. However, although it is strongly induced by IL-4, it can also be upregulated by other stimuli and has been reported not to be an effective M2a marker in human macrophages (8).
The modulation of macrophage phenotypes represents an interesting therapeutic strategy (6, 13). For instance, promoting an anti-inflammatory or regulatory phenotype in macrophages may be beneficial in diseases involving chronic inflammation. Alternatively, the reverse is true in cancer, where tumor-associated macrophages are correlated with an adverse outcome. The pathways involved in regulating macrophage polarization may therefore yield useful drug targets. Interestingly, other GPCRs in macrophages may act via similar pathways. Notably adenosine, which can act via Gs coupled GPCRs, has been suggested to synergize with TLR agonists to induce IL-10 (59). Interestingly, one of the mechanisms of action suggested for methotrexate, a commonly used drug in autoimmunity, is to elevate extracellular adenosine levels (60). In addition PDE4 inhibitors, which would promote PKA signaling in macrophages, have also generated interest as immunosuppressive agents and one such inhibitor, roflumilast, has been approved for use in some forms of chronic obstructive pulmonary disease (61). Thus, targeting the PKA-SIK-CRTC3/CREB pathway may represent a promising new therapeutic strategy.
Supplementary Material
Acknowledgments
We thank the Medical Research Council Protein Phosphorylation Unit DNA Sequencing Service (coordinated by Nicholas Helps) for DNA sequencing and Alan Ashworth for the LKB1 knockout mice. We thank Ed McIver and Joanne Hough at MRC Technology for developing MRT67307, Nathanael Gray for Kin112, and Thomas Houslay for statistical analysis.
This work was supported by the Medical Research Council, Arthritis Research UK, and the pharmaceutical companies supporting the Division of Signal Transduction Therapy Unit (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KGaA, Janssen Pharmaceutica, and Pfizer).
The online version of this article contains supplemental material.
The array data presented in this article have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE41833.
- BMDM
- bone marrow–derived macrophage
- CRE
- cAMP response element
- CRTC
- CREB-regulated transcription coactivator
- PDE
- phosphodiesterase
- PKA
- protein kinase A
- PRR
- pattern recognition receptor
- qPCR
- quantitative PCR
- SIK
- salt-inducible kinase
- SILAC
- stable isotope labeling of amino acids in cell culture
- siRNA
- small interfering RNA.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Takeuchi O., Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140: 805–820 [DOI] [PubMed] [Google Scholar]
- 2.Ivashkiv L. B. 2009. Cross-regulation of signaling by ITAM-associated receptors. Nat. Immunol. 10: 340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lattin J., Zidar D. A., Schroder K., Kellie S., Hume D. A., Sweet M. J. 2007. G-protein‑coupled receptor expression, function, and signaling in macrophages. J. Leukoc. Biol. 82: 16–32 [DOI] [PubMed] [Google Scholar]
- 4.Liddiard K., Rosas M., Davies L. C., Jones S. A., Taylor P. R. 2011. Macrophage heterogeneity and acute inflammation. Eur. J. Immunol. 41: 2503–2508 [DOI] [PubMed] [Google Scholar]
- 5.Lawrence T., Natoli G. 2011. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 11: 750–761 [DOI] [PubMed] [Google Scholar]
- 6.Sica A., Mantovani A. 2012. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosser D. M., Edwards J. P. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8: 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon S., Martinez F. O. 2010. Alternative activation of macrophages: mechanism and functions. Immunity 32: 593–604 [DOI] [PubMed] [Google Scholar]
- 9.Lumeng C. N., Bodzin J. L., Saltiel A. R. 2007. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117: 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruffell B., Affara N. I., Coussens L. M. 2012. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 33: 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikita J., Dubourdieu-Cassagno N., Deloire M. S., Vekris A., Biran M., Raffard G., Brochet B., Canron M. H., Franconi J. M., Boiziau C., Petry K. G. 2011. Altered M1/M2 activation patterns of monocytes in severe relapsing experimental rat model of multiple sclerosis: amelioration of clinical status by M2 activated monocyte administration. Mult. Scler. 17: 2–15 [DOI] [PubMed] [Google Scholar]
- 12.Gerber J. S., Mosser D. M. 2001. Reversing lipopolysaccharide toxicity by ligating the macrophage Fcγ receptors. J. Immunol. 166: 6861–6868 [DOI] [PubMed] [Google Scholar]
- 13.Fleming B. D., Mosser D. M. 2011. Regulatory macrophages: setting the threshold for therapy. Eur. J. Immunol. 41: 2498–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saraiva M., O’Garra A. 2010. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10: 170–181 [DOI] [PubMed] [Google Scholar]
- 15.Ouyang W., Rutz S., Crellin N. K., Valdez P. A., Hymowitz S. G. 2011. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 29: 71–109 [DOI] [PubMed] [Google Scholar]
- 16.Kühn R., Löhler J., Rennick D., Rajewsky K., Müller W. 1993. Interleukin-10‑deficient mice develop chronic enterocolitis. Cell 75: 263–274 [DOI] [PubMed] [Google Scholar]
- 17.Glocker E. O., Frede N., Perro M., Sebire N., Elawad M., Shah N., Grimbacher B. 2010. Infant colitis—it’s in the genes. Lancet 376: 1272. [DOI] [PubMed] [Google Scholar]
- 18.Glocker E. O., Kotlarz D., Boztug K., Gertz E. M., Schäffer A. A., Noyan F., Perro M., Diestelhorst J., Allroth A., Murugan D., et al. 2009. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361: 2033–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S. H., Serezani C. H., Okunishi K., Zaslona Z., Aronoff D. M., Peters-Golden M. 2011. Distinct protein kinase A anchoring proteins direct prostaglandin E2 modulation of Toll-like receptor signaling in alveolar macrophages. J. Biol. Chem. 286: 8875–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez Y., Municio C., Alonso S., Sánchez Crespo M., Fernández N. 2009. The induction of IL-10 by zymosan in dendritic cells depends on CREB activation by the coactivators CREB-binding protein and TORC2 and autocrine PGE2. J. Immunol. 183: 1471–1479 [DOI] [PubMed] [Google Scholar]
- 21.Avni D., Philosoph A., Meijler M. M., Zor T. 2010. The ceramide-1-phosphate analogue PCERA-1 modulates tumour necrosis factor-α and interleukin-10 production in macrophages via the cAMP-PKA-CREB pathway in a GTP-dependent manner. Immunology 129: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng W., Wang Y., Zhang J., Wang X., Li C., Chang Z. 2000. Effects of CTx and 8-bromo-cAMP on LPS-induced gene expression of cytokines in murine peritoneal macrophages. Biochem. Biophys. Res. Commun. 269: 570–573 [DOI] [PubMed] [Google Scholar]
- 23.Eigler A., Siegmund B., Emmerich U., Baumann K. H., Hartmann G., Endres S. 1998. Anti-inflammatory activities of cAMP-elevating agents: enhancement of IL-10 synthesis and concurrent suppression of TNF production. J. Leukoc. Biol. 63: 101–107 [DOI] [PubMed] [Google Scholar]
- 24.Siegmund B., Eigler A., Moeller J., Greten T. F., Hartmann G., Endres S. 1997. Suppression of tumor necrosis factor-α production by interleukin-10 is enhanced by cAMP-elevating agents. Eur. J. Pharmacol. 321: 231–239 [DOI] [PubMed] [Google Scholar]
- 25.Kunkel S. L., Chensue S. W., Phan S. H. 1986. Prostaglandins as endogenous mediators of interleukin 1 production. J. Immunol. 136: 186–192 [PubMed] [Google Scholar]
- 26.Kunkel S. L., Wiggins R. C., Chensue S. W., Larrick J. 1986. Regulation of macrophage tumor necrosis factor production by prostaglandin E2. Biochem. Biophys. Res. Commun. 137: 404–410 [DOI] [PubMed] [Google Scholar]
- 27.Strassmann G., Patil-Koota V., Finkelman F., Fong M., Kambayashi T. 1994. Evidence for the involvement of interleukin 10 in the differential deactivation of murine peritoneal macrophages by prostaglandin E2. J. Exp. Med. 180: 2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark K., Mackenzie K. F., Petkevicius K., Kristariyanto Y., Zhang J., Choi H. G., Peggie M., Plater L., Pedrioli P. G., McIver E., et al. 2012. Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proc. Natl. Acad. Sci. USA 109: 16986–16991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wingate A. D., Martin K. J., Hunter C., Carr J. M., Clacher C., Arthur J. S. 2009. Generation of a conditional CREB Ser133Ala knockin mouse. Genesis 47: 688–696 [DOI] [PubMed] [Google Scholar]
- 30.Wiggin G. R., Soloaga A., Foster J. M., Murray-Tait V., Cohen P., Arthur J. S. 2002. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell. Biol. 22: 2871–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakamoto K., McCarthy A., Smith D., Green K. A., Grahame Hardie D., Ashworth A., Alessi D. R. 2005. Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO J. 24: 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Förster I. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8: 265–277 [DOI] [PubMed] [Google Scholar]
- 33.Darragh J., Ananieva O., Courtney A., Elcombe S., Arthur J. S. 2010. MSK1 regulates the transcription of IL-1ra in response to TLR activation in macrophages. Biochem. J. 425: 595–602 [DOI] [PubMed] [Google Scholar]
- 34.Dzamko N., Inesta-Vaquera F., Zhang J., Xie C., Cai H., Arthur S., Tan L., Choi H., Gray N., Cohen P., et al. 2012. The IκB kinase family phosphorylates the Parkinson’s disease kinase LRRK2 at Ser935 and Ser910 during Toll-like receptor signaling. PLoS ONE 7: e39132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X., Odom D. T., Koo S. H., Conkright M. D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., et al. 2005. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA 102: 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox J., Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- 37.Ananieva O., Darragh J., Johansen C., Carr J. M., McIlrath J., Park J. M., Wingate A., Monk C. E., Toth R., Santos S. G., et al. 2008. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat. Immunol. 9: 1028–1036 [DOI] [PubMed] [Google Scholar]
- 38.Sugimoto Y., Narumiya S. 2007. Prostaglandin E receptors. J. Biol. Chem. 282: 11613–11617 [DOI] [PubMed] [Google Scholar]
- 39.Edwards J. P., Zhang X., Frauwirth K. A., Mosser D. M. 2006. Biochemical and functional characterization of three activated macrophage populations. J. Leukoc. Biol. 80: 1298–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasper L. H., Lerach S., Wang J., Wu S., Jeevan T., Brindle P. K. 2010. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. EMBO J. 29: 3660–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darragh J., Soloaga A., Beardmore V. A., Wingate A. D., Wiggin G. R., Peggie M., Arthur J. S. 2005. MSKs are required for the transcription of the nuclear orphan receptors Nur77, Nurr1 and Nor1 downstream of MAPK signalling. Biochem. J. 390: 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayr B., Montminy M. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2: 599–609 [DOI] [PubMed] [Google Scholar]
- 43.Arthur J. S. 2008. MSK activation and physiological roles. Front. Biosci. 13: 5866–5879 [DOI] [PubMed] [Google Scholar]
- 44.Park J. M., Greten F. R., Wong A., Westrick R. J., Arthur J. S., Otsu K., Hoffmann A., Montminy M., Karin M. 2005. Signaling pathways and genes that inhibit pathogen-induced macrophage apoptosis—CREB and NF-κB as key regulators. Immunity 23: 319–329 [DOI] [PubMed] [Google Scholar]
- 45.Pattison M. J., MacKenzie K. F., Arthur J. S. 2012. Inhibition of JAK kinases in macrophages increases LPS induced cytokine production by blocking IL-10 mediated feedback. J. Immunol. 189: 2784–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Screaton R. A., Conkright M. D., Katoh Y., Best J. L., Canettieri G., Jeffries S., Guzman E., Niessen S., Yates J. R., III, Takemori H., et al. 2004. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119: 61–74 [DOI] [PubMed] [Google Scholar]
- 47.Conkright M. D., Canettieri G., Screaton R., Guzman E., Miraglia L., Hogenesch J. B., Montminy M. 2003. TORCs: transducers of regulated CREB activity. Mol. Cell 12: 413–423 [DOI] [PubMed] [Google Scholar]
- 48.Lizcano J. M., Göransson O., Toth R., Deak M., Morrice N. A., Boudeau J., Hawley S. A., Udd L., Mäkelä T. P., Hardie D. G., Alessi D. R. 2004. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasaki T., Takemori H., Yagita Y., Terasaki Y., Uebi T., Horike N., Takagi H., Susumu T., Teraoka H., Kusano K., et al. 2011. SIK2 is a key regulator for neuronal survival after ischemia via TORC1-CREB. Neuron 69: 106–119 [DOI] [PubMed] [Google Scholar]
- 50.Németh K., Leelahavanichkul A., Yuen P. S., Mayer B., Parmelee A., Doi K., Robey P. G., Leelahavanichkul K., Koller B. H., Brown J. M., et al. 2009. Bone marrow stromal cells attenuate sepsis via prostaglandin E2-dependent reprogramming of host macrophages to increase their interleukin-10 production. Nat. Med. 15: 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Medeiros A. I., Serezani C. H., Lee S. P., Peters-Golden M. 2009. Efferocytosis impairs pulmonary macrophage and lung antibacterial function via PGE2/EP2 signaling. J. Exp. Med. 206: 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldmann O., Hertzén E., Hecht A., Schmidt H., Lehne S., Norrby-Teglund A., Medina E. 2010. Inducible cyclooxygenase released prostaglandin E2 modulates the severity of infection caused by Streptococcus pyogenes. J. Immunol. 185: 2372–2381 [DOI] [PubMed] [Google Scholar]
- 53.Heusinkveld M., de Vos van Steenwijk P. J., Goedemans R., Ramwadhdoebe T. H., Gorter A., Welters M. J., van Hall T., van der Burg S. H. 2011. M2 macrophages induced by prostaglandin E2 and IL-6 from cervical carcinoma are switched to activated M1 macrophages by CD4+ Th1 cells. J. Immunol. 187: 1157–1165 [DOI] [PubMed] [Google Scholar]
- 54.Bystrom J., Evans I., Newson J., Stables M., Toor I., van Rooijen N., Crawford M., Colville-Nash P., Farrow S., Gilroy D. W. 2008. Resolution-phase macrophages possess a unique inflammatory phenotype that is controlled by cAMP. Blood 112: 4117–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horike N., Takemori H., Katoh Y., Doi J., Min L., Asano T., Sun X. J., Yamamoto H., Kasayama S., Muraoka M., et al. 2003. Adipose-specific expression, phosphorylation of Ser794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. J. Biol. Chem. 278: 18440–18447 [DOI] [PubMed] [Google Scholar]
- 56.Katoh Y., Takemori H., Min L., Muraoka M., Doi J., Horike N., Okamoto M. 2004. Salt-inducible kinase-1 represses cAMP response element-binding protein activity both in the nucleus and in the cytoplasm. Eur. J. Biochem. 271: 4307–4319 [DOI] [PubMed] [Google Scholar]
- 57.Henriksson E., Jones H. A., Patel K., Peggie M., Morrice N., Sakamoto K., Göransson O. 2012. The AMPK-related kinase SIK2 is regulated by cAMP via phosphorylation at Ser358 in adipocytes. Biochem. J. 444: 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wall E. A., Zavzavadjian J. R., Chang M. S., Randhawa B., Zhu X., Hsueh R. C., Liu J., Driver A., Bao X. R., Sternweis P. C., et al. 2009. Suppression of LPS-induced TNF-α production in macrophages by cAMP is mediated by PKA-AKAP95-p105. Sci. Signal. 2: ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Németh Z. H., Lutz C. S., Csóka B., Deitch E. A., Leibovich S. J., Gause W. C., Tone M., Pacher P., Vizi E. S., Haskó G. 2005. Adenosine augments IL-10 production by macrophages through an A2B receptor-mediated posttranscriptional mechanism. J. Immunol. 175: 8260–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan E. S., Cronstein B. N. 2010. Methotrexate—how does it really work? Nat Rev Rheumatol 6: 175–178 [DOI] [PubMed] [Google Scholar]
- 61.Page C. P., Spina D. 2012. Selective PDE inhibitors as novel treatments for respiratory diseases. Curr. Opin. Pharmacol. 12: 275–286 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.