SUMMARY
Chromatin diminution is the programmed elimination of specific DNA sequences during development. It occurs in diverse species, but the function(s) of diminution and the specificity of sequence loss remain largely unknown. Diminution in the nematode Ascaris suum occurs during early embryonic cleavages and leads to the loss of germline genome sequences and the formation of a distinct genome in somatic cells. We found that ~43 Mb (~13%) of genome sequence is eliminated in A. suum somatic cells, including ~12.7 Mb of unique sequence. The eliminated sequences and location of the DNA breaks are the same in all somatic lineages from a single individual, and between different individuals. At least 685 genes are eliminated. These genes are preferentially expressed in the germline and during early embryogenesis. We propose that diminution is a mechanism of germline gene regulation that specifically removes a large number of genes involved in gametogenesis and early embryogenesis.
INTRODUCTION
Metazoans must both ensure the stability of their genomes, and also carefully regulate the expression of germline genes in somatic tissues. Failure of either of these processes has severe consequences. However, there are examples of programmed genome instability that are integral to the biology of an organism. Well-known examples include vertebrate immunoglobulin gene rearrangement that enables antibody diversification (Jung et al., 2006) and the extensive remodeling of the somatic genome that occurs during development of the macronucleus in ciliates (Chalker and Yao, 2011). Chromatin diminution is another form of genome rearrangement with DNA loss that occurs during the development of diverse Metazoa including some nematodes, copepod crustaceans, insects, lampreys/hagfish, and zebra finches (Bachmann-Waldmann et al., 2004; Goday and Esteban, 2001; Goday and Pimpinelli, 1993; Kloc and Zagrodzinska, 2001; Muller et al., 1996; Muller and Tobler, 2000; Nemetschke et al., 2010; Smith et al., 2009; Tobler et al., 1992; Tobler et al., 1985; Zufall et al., 2005). DNA elimination occurs either during gametogenesis or during differentiation of the somatic lineage early in embryonic development. In some species, DNA elimination may be involved in sex determination (Goday and Esteban, 2001; Goday and Pigozzi, 2010; Nemetschke et al., 2010), but in most organisms that undergo programmed chromatin diminution, the sequences lost and the role(s) of diminution remain unknown.
Chromatin diminution was first described in an ascaridid nematode by Theodor Boveri in 1887 (Boveri, 1887). Diminution in nematodes is restricted to a number of parasitic nematodes primarily in the Ascarididae (Goday and Pimpinelli, 1993; Muller et al., 1996; Muller and Tobler, 2000; Pimpinelli and Goday, 1989) and does not occur in Caenorhabditis elegans (Emmons et al., 1979). In the pig parasite A. suum, diminution occurs during the third through fifth cleavages (4 to 16 cell stage) of development in five distinct somatic precursor cells that give rise to different cell lineages (Fig. 1). This raises the key questions: 1) Are the sequences lost and the changes that occur during diminution the same in all five of these precursor cells, and 2) What are the sequences that are lost and their functional significance?
Figure 1. A. suum early embryo development, cell lineage and chromatin diminution.
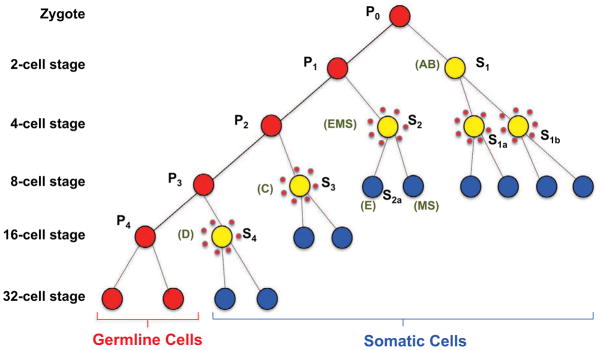
Primordial germ cells (P) are in red, cells undergoing chromatin diminution are represented by yellow filled circles surrounded by dots, and blue cells (S) are precursor somatic cells and lineages. The primordial germ cells numbers correspond to their division state. P0 is the zygote whereas P1 through P4 represent the primordial germ cell derived from each subsequent cleavage of the germ cells as illustrated. S1–S4 cells are successive precursor somatic cell derived from each division of a germ cell (EMS = intestine, body wall muscle, and pharynx; E = intestine; MS = Body wall muscle, neurons, somatic gonad, coelomocytes, and pharynx; AB = Nervous system, hypodermis, and pharynx; C = Body wall muscle, hypodermis, and neurons; and D = Body wall muscle). Adapted from Theodor Boveri (Boveri, 1899, 1910) and Fritz Müller and Heinz Tobler (Goday and Pimpinelli, 1993; Muller et al., 1996; Muller and Tobler, 2000; Pimpinelli and Goday, 1989).
During the Ascaris diminution process, chromosomes are broken and the fragments to be eliminated remain at the metaphase plate while the retained DNA is segregated into daughter cells. It has been estimated that 25% of A. suum genome is lost in somatic cells, whereas the germline genome remains intact (Muller et al., 1996; Muller and Tobler, 2000; Tobler et al., 1992; Tobler et al., 1985). The eliminated DNA includes highly repetitive satellite sequences consisting primarily of a 121 bp tandem repeat that is located in heterochromatin-like blocks at internal sites and chromosome ends (Muller et al., 1982; Niedermaier and Moritz, 2000; Streeck et al., 1982). Both internal regions and terminal heterochromatic regions are eliminated, with addition of new telomeres resulting in a ~50% increase in chromosome number (Bachmann-Waldmann et al., 2004; Huang et al., 1996; Jentsch et al., 2002; Magnenat et al., 1999; Muller et al., 1991; Niedermaier and Moritz, 2000). While a few chromosomal breakpoints where new telomere addition occurs have been partially characterized (Bachmann-Waldmann et al., 2004; Huang et al., 1996; Jentsch et al., 2002; Magnenat et al., 1999; Muller et al., 1991) and three single-copy genes that are eliminated have been identified (Etter et al., 1994; Huang et al., 1996; Spicher et al., 1994), the full complement of eliminated genes and sequences and the consequences for the somatic cells remain unknown (Goday and Pimpinelli, 1993; Muller et al., 1996; Muller and Tobler, 2000; Pimpinelli and Goday, 1989).
RESULTS and DISCUSSION
Identification of Eliminated Sequences and Breakpoints Associated with Ascaris Diminution
We deep sequenced genomic DNA libraries from the spermatids (germline) and intestine (somatic) of a single male A. suum (Table S1) to enable a genome-wide analysis of DNA changes following diminution. We used these data to independently assemble the germline and somatic genomes (Table 1), and mapped the raw reads of the germline and somatic genomes back to the germline assembly. These analyses identified sequences that are eliminated to form the somatic genome and DNA breakpoints associated with A. suum chromatin diminution (Fig. 2 and Fig. S1). Paired-end and mate-pair reads for the germline and somatic regions strongly supported the germline assembly, the identified breakpoints, and DNA loss (Tables S3 and S4). The results indicated that the major type of genome alteration is chromosome breakage, loss of DNA sequence, and the healing of retained chromosomes by telomere addition (Figs. 3 and 4 and Tables S3 and S4). We found no evidence for the loss of interstitial sequences followed by DNA fusions or other genome rearrangements. Breakpoints, DNA loss, and new telomere addition identified in the genome sequencing data were confirmed using specific PCR assays (Figs. 2 and 3). Among the sequences eliminated are at least 35 loci that may have arisen through recent duplication and rearrangement in the A. suum lineage (Fig. 4).
Table 1.
Ascaris suum germline and somatic genome assemblies
| Ascaris genomes | Features | Germline | Somatic | Jex et. al1 |
|---|---|---|---|---|
| Assemblies | Estimated genome size (Mb)2 | ~ 334 | ~ 291 | ~ 309 |
| Total number of bp assembled (bp) | 265,545,801 | 251,265,282 | 272,782,664 | |
| N50 of scaffolds (bp); N50 number | 290,558; 260 | 65,087; 1,011 | 407,899; 179 | |
| N90 of scaffolds (bp); N90 number | 48,674; 1,100 | 11,448; 4,399 | 80,017; 748 | |
| Number of scaffolds (>= 2,000 bp) | 31,538 (2,186) | 37,686 (7,692) | 29,831 (1,618) | |
| Maximum length of scaffold (bp) | 1,465,500 | 600,478 | 3,795,215 | |
| N50 of contigs (bp); N50 number | 49,549; 1,510 | 36,306; 1,925 | 23,038; 3,512 | |
| N90 of contigs (bp); N90 number | 11,178; 5,601 | 7,566; 7,407 | 5,913; 11,869 | |
|
| ||||
| Protein-coding genes | Putative coding gene number | 15,446 | 14,761 | 18,542 |
| Average gene size (bp) | 9,467 | 9,473 | 6,536 | |
| Average coding sequence length (bp) | 1,119 | 1,128 | 983 | |
| Average exon number per gene | 8.4 | 8.4 | 6 | |
| Average exon length (bp) | 201 | 201 | 153 | |
| Average intron length (bp) | 1,056 | 1,050 | 1,081 | |
|
| ||||
| Non-coding RNAs | Ribosomal RNAs (rRNAs, copies for 18s-5.8s-26s)2 | ~ 500 | ~ 500 | NA |
| Splice leader RNAs (including 5s rRNA)2 | ~ 265 | ~ 265 | NA | |
| Transfer RNAs (tRNAs) (+ tRNA pseudogenes) | 383 (+ 31) | 172 (+ 13) | 255 (+ 16) | |
| microRNAs (miRNAs) | 100 | 100 | 100 | |
|
| ||||
| Functional coverage | % of cDNA contigs >= 200 nt present (total 58,085)3 | 91.9% | 88.2% | 92.2% |
| % of cDNA bases present (total 58.1 Mb) | 97.6% | 94.6% | 98.0% | |
| % of unique small RNA reads mapped (total 20.2M)4 | 81.3% | 70.7% | 76.3% | |
| % of all small RNA reads mapped (total 690.7M)4 | 89.1% | 80.5% | 78.6% | |
See Jex et al., 2011.
Genome size and repetitve RNA copy number estimation based on coverage (see Supplementary Experimental Procedures).
For the de-novo assembled cDNA contigs, > 90% of the sequence mapped back to the genome using BLAT.
See Wang et al., 2011.
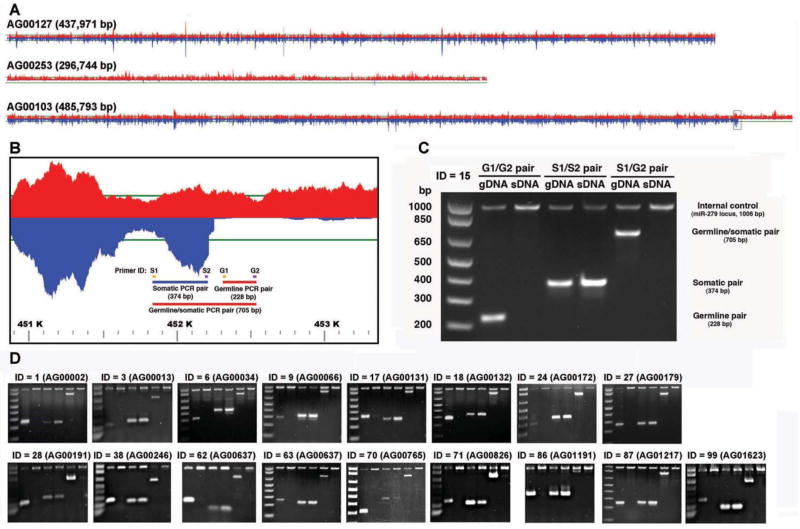
Figure 2. A. suum DNA elimination.
A. Germline and somatic read coverage for regions of the A. suum genome illustrating the retention of segments in both the germline and somatic tissue (top), a region completely eliminated in the somatic tissue (middle), and a DNA breakpoint and region eliminated in the somatic tissues (bottom). Red designates germline reads and blue are somatic reads with the horizontal green line representing 50-fold coverage.
B. Enlarged region of a scaffold (AG00103, Fig. 2A bottom) illustrating the PCR strategy used to verify DNA elimination predicted from the comparison of the germline and somatic genome sequences.
C. PCR data confirm the elimination of DNA corresponding to scaffold AG00103 in A. suum somatic tissues. Note that the germline primer pair (G1/G2) produces a PCR product in the germline DNA (gDNA), but not somatic DNA (sDNA). The somatic primer pair (S1/S2) leads to PCR products in both the germline and somatic DNA, and the primer pair spanning the breakpoint (S1/G2) produces a PCR product only in the germline. gDNA = germline testis DNA and sDNA = somatic intestine DNA, isolated from the single male worm from which the genome sequences were derived. The 1,006 bp PCR product present in all lanes represents a control PCR corresponding to a single copy locus (miR-279) present in both the germline and somatic genomes.
D. PCR data confirm the elimination of DNA in 17 additional independent loci in the A. suum somatic genome. The PCR strategy illustrated in B and C was applied to these loci.
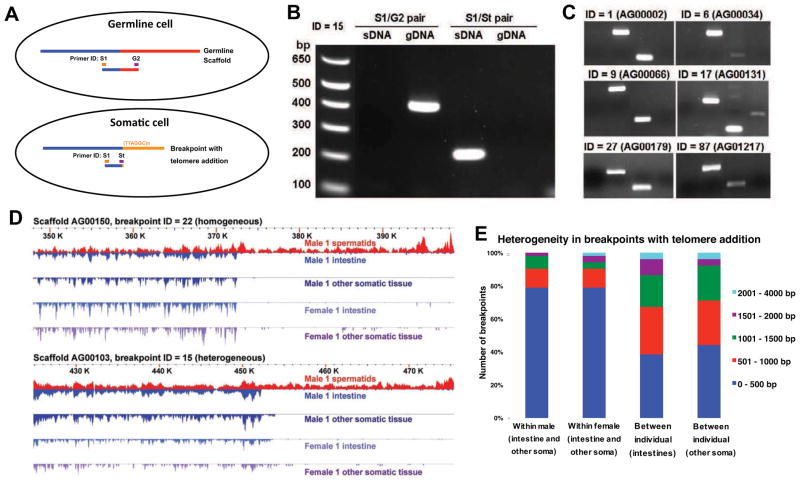
Figure 3. A. suum chromosome breaks with telomere addition in somatic cells.
A. The PCR strategy used to verify telomere sequence addition in the somatic cells. Primer St (Somatic telomere) is a hybrid primer consisting of 3′ nucleotides corresponding to the unique somatic sequence and 5′ nucleotides corresponding to telomeric sequence [(TTAGGC)n] (see Supplementary Experimental Procedures for primers sequences).
B. PCR data confirm telomere addition in the A. suum somatic tissues at breakpoint 15. Primers are defined in Fig. 3A and genomic DNA sources defined as in Fig. 2C.
C. PCR data confirm the telomere addition at 6 additional independent loci with chromosome breaks.
D. Heterogeneity in breakpoints with telomere addition. Two breakpoints with telomere addition are illustrated. Note that the exact breakpoint for one of the loci in different somatic tissues varies (intestine and carcass), particularly between individuals.
E. Overall heterogeneity in breakpoints with telomere addition. For these 52 breakpoints, we compared the genomes of pairs of somatic tissues (intestine and other somatic) from the same individual and between individuals and measured the difference in the position of the breakpoints identified.
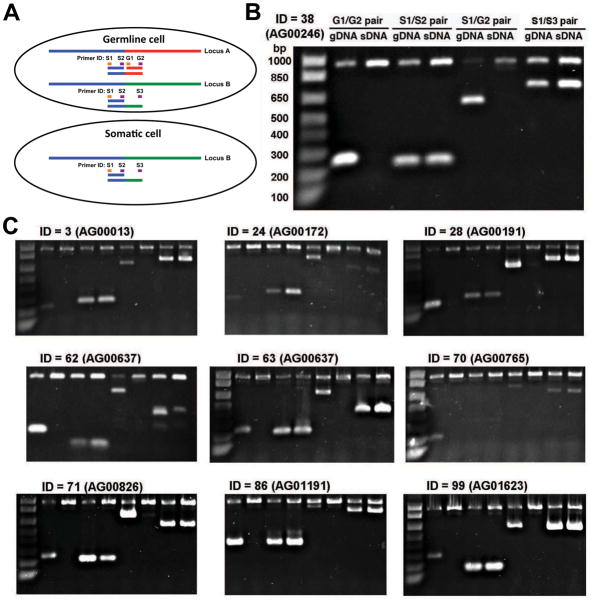
Figure 4. Loss of one member of duplicated, rearranged loci.
A. Loss of one of two similar germline loci in the somatic genome. Illustration shows two germline loci in a germline cell containing common sequences (> 97% identical) (blue line), divergent sequence (green or red lines), and the loss of one germline locus in somatic cells following chromatin diminution. Primers and PCR strategy used to verify loci in the germline and somatic genomes are shown.
B. PCR data confirm locus A is present in germline cells but lost in somatic cells. Locus B is present in both germline and somatic cells.
C. Additional PCR data for other loci demonstrating the loss of one member of duplicated, rearranged loci.
See also Table S4.
Diminution Process is Conserved in Distinct Somatic Lineages and Between Male and Female
To determine if diminution happens in the same way in all somatic cell lineages (Fig. 1), we compared the sequences lost from the male intestine to those lost from other somatic cell types (e.g., body wall muscle, pharyngeal muscle, hypodermis, and neurons) (Table S1) in the same male. The intestine is derived from a single cell (E) in the A. suum cell lineage (Fig. 1) whereas other somatic tissues are derived from additional and often multiple cell lineages that independently undergo chromatin diminution (Fig. 1, cells labeled AB, C, and D) (Boveri, 1899, 1910). We found that the DNA loss and chromosome breaks in an individual male are conserved between the intestine and other somatic cell types (Fig. 3D and Fig. S1), suggesting that the mechanism and consequences of diminution are the same in different cells. Additional comparison of the sequence lost between the male and a female worm shows a high degree of fidelity in the breaks and DNA loss between individuals (Fig. 3D and Fig. S1). However, there is some heterogeneity in the exact position, with ~80% of the breaks occurring within 500 bp of each other within an individual and ~70% within 1,000 bp between individuals (Fig. 3E and Fig. S1) consistent with earlier studies (Bachmann-Waldmann et al., 2004; Huang et al., 1996; Jentsch et al., 2002; Magnenat et al., 1999; Muller et al., 1991). Analysis of DNA sequence up to 5 kb on either side of the telomere addition sites did not reveal any specific sequence motifs or other characteristics that might mark the regions for chromosomal breakage (Fig. S2 and Supplementary Experimental Procedures).
A Large Number of Germline Genes are Eliminated During Diminution
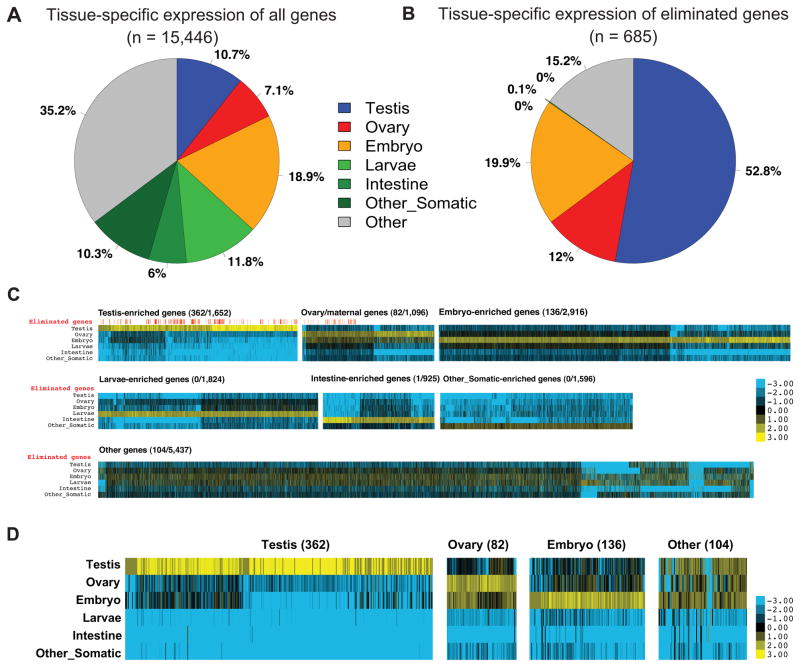
Our analysis of the DNA lost revealed that ~43 Mb (~13%) of sequence is eliminated from the germline genome during the formation of the somatic genome (Table 1). The majority of the eliminated sequence (29 Mb, 70% of the eliminated sequence) is the 121 bp satellite, repeat sequence previously described (Muller and Tobler, 2000; Muller et al., 1982; Streeck et al., 1982) (Table S2). No other major loss of repetitive sequence was observed. The remaining ~12.7 Mb of the eliminated sequence is unique and includes at least 685 predicted genes (Tables S5 and S6). We sought to identify whether the eliminated genes shared any common features in their patterns of expression. Genome-wide expression profiles were constructed by RNA-seq of poly(A)+ RNA from testis, ovary, embryo, larvae, intestine, muscle and other somatic tissues (Table S1), and the expression level was measured by reads per kilobase of template per million mapped reads (RPKM). Remarkably, these results revealed that 85% of the eliminated genes are expressed preferentially during gametogenesis or early embryogenesis, and the remaining 15% are expressed in both (Fig. 5 and Table S6). Most of these genes are expressed specifically in the testis and therefore are likely involved in spermatogenesis. However, not all genes expressed in the germline were eliminated during chromatin diminution. Functional annotations of the eliminated genes suggest that they are enriched for protein kinases, protein phosphatases, proteins associated with chromatin, RNA and nucleotide binding proteins, and translation initiation proteins (Table S6). These proteins are associated with network functions including protein synthesis, RNA post-transcriptional modification, gene expression, cell death, and cellular compromise (Table S6). Notably, at least 30 families of these eliminated genes are orthologs of well-characterized genes in C. elegans whose loss is associated with clear phenotypes in germline formation, gametogenesis, and early embryogenesis (e.g., air-1, gld-2, cgh-1, gla-3, fer-1, spe genes, and pab genes) (Table S6).
Figure 5. Eliminated A. suum genes are primarily expressed in the germline.
A. RNA expression of A. suum genes (n = 15,446) in different tissues. Gene expression enrichment was categorized by comparing RNA-seq data (Table S1) using reads per kilobase of template per million mapped reads (RPKM).
B. The 685 eliminated A. suum genes are highly expressed in the germline and early embryogenesis.
C. Expression Heatmap for all A. suum genes. Shown are expression heatmaps for different groups of genes illustrated in Fig. 5A. For each gene, the colors represent log2 values of fold changes to the average expression level (RPKM) for a gene in different stages. For each group of genes, the eliminated/total number of genes is indicated and a red vertical line above the heatmap marks the genes eliminated (see Fig. 5D).
D. The expression profiles of eliminated A. suum genes. See Fig. 5C for legend. Note that for the 104 genes in the Other group, the majority of them are expressed in testis, ovary, and the early embryo.
See also Figure S3 and Table S5–6.
Eliminated Genes Suggest Biological Functions for Diminution
We observed that ~53% (363) of the eliminated genes have paralogs in the genome (Table S6). This is consistent with a model where an ancient genome duplication in the A. suum lineage was balanced by chromatin diminution to regulate gene dosage or to provide a mechanism for the selective retention of specific genes and thus their function (Bachmann-Waldmann et al., 2004; Goday and Esteban, 2001; Goday and Pimpinelli, 1993; Muller et al., 1996; Muller and Tobler, 2000; Tobler et al., 1992; Tobler et al., 1985). A previous study demonstrated one of the two paralogs of ribosomal protein rps-19 gene was eliminated in A. suum, suggesting that the two proteins may play differential roles in translation (Etter et al., 1994). Recent data on rps-19 indicates that mutations or knockdown of this and other specific ribosomal protein genes in vertebrates leads to discrete changes in the translation of specific mRNAs, but not general translation (Horos et al., 2012; Kondrashov et al., 2011). Our observation that a major group of the eliminated genes is associated with translation (Tables S6) reinforces the idea that the translation machinery may differ between the germline and soma. Notably, we found that a number of translation initiation factors are eliminated from the germline (eIF4E, eIF5, eIF2 subunit 2, eIF3B, eIF3C, eIF3D, eIF3i, and eIF2 subunit 3). Another, non-exclusive model of the function of diminution is that in addition to the elimination of the genes, the chromosomal regions eliminated play an important role in chromatin organization that contribute to broader gene regulation. For example, these regions may repress genes in the germline or their elimination may activate key somatic genes. We found no evidence of telomeric position effect silencing of genes due to telomere addition (Huang et al., 1996) (Fig. S1) but cannot eliminate the possibility of other indirect effects.
Ascaris Chromatin Diminution, Small RNAs, and Marks for Chromosome Breaks
In ciliates, small RNAs (piRNAs) are known to play a key role in the programmed DNA rearrangements and elimination (Chalker and Yao, 2011). We looked previously for small RNAs (Wang et al., 2011) that might be associated with Ascaris chromatin diminution, in particular RNAs mapping to the 121-bp repeat element that constitutes ~30 Mb of the eliminated sequence. We characterized small RNAs before, during, and after chromatin diminution and found no correlation between the eliminated repeats and any small RNAs. Furthermore, piRNAs and PIWI Argonautes are absent in Ascaris (Wang et al., 2011). Recent studies in C. elegans suggest that small RNAs also play a role in the recognition of “self” versus “non-self” and in multigenerational epigenetic inheritance (Ashe et al., 2012; Bagijn et al., 2012; Buckley et al., 2012; Lee et al., 2012; Luteijn et al., 2012; Shirayama et al., 2012). We reexamined the expression of Ascaris small RNAs (Wang et al., 2011) correlating with chromatin diminution. We found no temporal or any other correlation of small RNAs associated with regions retained, regions eliminated, or the chromosome breakpoint regions. While preliminary analyses of Ascaris polyA+ RNA levels (RNA-seq) demonstrate that several Argonaute and other protein mRNAs increase during the period of early embryo development leading up to and during the time of diminution, their increased expression may not be correlated with diminution and could be associated with the maternal to zygotic transition or serve other functions during this complex period of development. Overall, these and other data in Ascaris [including the loss of large numbers of genes, the lack of discrete sequence elements that mark sites of DNA breaks for telomere addition (Fig. S2), and the absence of removal of interstitial DNA sequences followed by DNA fusion] suggest that the function and mechanism for DNA elimination in Ascaris may differ from the programmed rearrangements in ciliates. As we did not identify discrete sequence elements that mark the sites of DNA breaks, we suggest that epigenetic marks (and even small RNAs yet to be identified) could play an important role in defining chromosome break sites and play a role in chromatin diminution. Additional studies will be required to further examine these possibilities.
Recently, a preliminary genomic study on the programmed DNA elimination in sea lampreys also demonstrated that unique sequences were eliminated from somatic cells (Smith et al., 2012). Among the genes eliminated in the somatic cells were some involved in transcriptional programs that are likely to play a role in maintaining germline function. Thus, elimination of specific germline-expressed genes in metazoa may be a common function of chromatin diminution.
Our work is a comprehensive analysis of the germline and somatic genome from a metazoan, the DNA lost and the chromosome changes that occur, and the elimination of specific germline expressed genes suggesting a function for that Ascaris chromatin diminution and a paradigm for DNA elimination. A hallmark of most Metazoa is that germline cells are set aside early in development. Soma-specific elimination provides a unique mechanism of gene repression, reminiscent of Weismann’s original theory of the differentiation between germline and soma (Weissmann, 1893). Our comprehensive identification of the genome changes in the soma of A. suum provides the foundation for the elucidation of the features and epigenetic changes underlying the mechanisms of selective DNA breakage and DNA loss in chromatin diminution. Understanding chromosome breaks, telomere healing, and selective DNA loss in chromatin diminution is likely to offer insight into genome stability and changes in normal processes and disease.
EXPERIMENTAL PROCEDURES
Sample collection and DNA isolation
A. suum samples were collected from pig intestines at a slaughterhouse in Sandusky, OH, U.S or Ghent, Belgium. A single male A. suum (U.S.) was dissected and the spermatids, the intestine, the testis, and the remaining tissue (carcass, which includes muscle, hypodermis, pharynx, and neurons) collected, frozen in liquid nitrogen, and stored at −80 °C. A single female (U.S.) was dissected and the ovary/oviduct, uterus, intestine, and carcass collected, frozen in liquid nitrogen, and stored at −80 °C.
Genome sequencing and assembly
Sequencing
Genomic DNA libraries were constructed from A. suum germline and somatic tissues and sequenced (Table S1). DNA isolation and libraries were constructed using standard methods and Illumina protocols and sequenced on the Illumina GAIIx or HiSeq platforms except where noted in the supplemental Materials and Methods. Genomic reads for assembly, scaffolding and analysis are listed in Table S1. The fold coverage numbers in Table S1 for all these libraries are derived from high quality reads that can be mapped back to the final assemblies.
Assembly
Reads from germline or somatic sources were used to independently assemble the two genomes. To minimize the sequence heterogeneity, we only used DNA sequences from a single male for the generation of consensus sequences within the assembly. Genomic reads for scaffolding (Table S1) were only used to confirm and support the links that bridge contigs into scaffolds, and none of these sequences were incorporated into the genome assemblies. Because of the presence of some duplicated loci (Fig. 4 and Table S4), we applied a “sub-assembly” strategy to capture all changes that occur in the germline genome. First, we built an initial germline genome assembly using velvet (v1.1.03) (Zerbino and Birney, 2008). From this assembly, we defined ~12.7 Mb of eliminated sequences (see below). All mappable germline reads were divided into 2 groups: those reads to the 12.7 Mb eliminated regions and all their pairs (from the paired-end and mate-pair libraries) and those reads retained in the soma following diminution. Next, we assembled the two groups of reads independently by using velvet (Zerbino and Birney, 2008). Finally, we combined the eliminated and retained assemblies using phrap (http://www.phrap.org/, v1.080812). Each assembly step was optimized and scaffolding performed under the overall guideline of sequentially bridging gaps ≤ 200, 500, 1,500, and 3,500 bp with ≥ 20 pairs of supporting reads, and the assemblies were checked using the Tablet assembly viewer (v1.11.11.01). The somatic genome was built using velvet, with ≥ 10 pairs of paired-end Illumina reads or ≥ 3 pairs of Sanger capillary reads for scaffolding, and the assembly also checked using the Tablet viewer. Note the N50 of the somatic assembly is not as good as that of the germline. This is mainly due to the lower average sequencing depth on the large fragment paired-end library for the somatic genome (compared with the germline) available for scaffolding.
Identification of eliminated sequences, germline genome changes, and breakpoints with telomere addition
We used two libraries generated and analyzed in parallel with similar coverage (2 × 150 bp, 360 bp insert size, ~50 × coverage) from the single male spermatids and intestine to map reads back to the germline genome assembly (bowtie2, v2.0.0-beta5) (Langmead and Salzberg, 2012) to identify eliminated sequence and genome changes in the germline. We used a 100-bp window to scan the genome for regions with a somatic/germline coverage ratio < 0.1 to provide an initial set of long sequence blocks containing potentially eliminated sequences. To identify germline genome changes, we manually checked all scaffolds ≥ 3 kb with ≥ 500 bp contiguous coverage. We identified 102 loci with DNA alterations (Tables S3 and S4). We then compared their sequences (10 Kb flanking the changes) to the somatic genome assembly, identified their exact positions where DNA loss occurred, and used these positions to establish the eliminated and retained regions in scaffolds. Germline scaffolds that did not harbor any DNA breakpoints were defined either as retained or eliminated based on the coverage ratio (see Fig. 2A and Fig. S1 for examples). From the somatic assembly, we also identified DNA breakpoints with addition of telomeres. Somatic scaffolds with telomeric sequences were independently confirmed by paired-end reads (Table S3) and PCR (Fig. 3A–C). For those DNA changes without telomere addition, their somatic loci were evaluated by germline paired-end reads (Table S4) and PCR analysis (Fig. 4) to confirm their presence in the germline.
RNA preparation, RNA-seq and assembly
Samples for RNA preparations are the same as those described for DNA preparations above or previously (Wang et al., 2011). Total RNA was prepared and RNA-seq libraries were made and sequenced as described (Wang et al., 2011). For each sample, 200 μg of total RNA was used for poly(A) selection and 200 ng of poly(A)+ RNA was used to make the cDNA libraries. Two cDNA assemblies were made by using all the RNA-seq data: one is a de-novo assembly using velvet/oases (v1.1.03/v0.2) (Zerbino and Birney, 2008), which has been used for the genome functional coverage assessment and gene annotation pipeline; the other is map-based assembly using tophat/cufflinks (v1.3.0) (Trapnell et al., 2012), which was used to facilitate gene prediction (see below).
Gene model prediction
We integrated gene evidence from multiple sources to build gene models for the A. suum germline genome. First, a two-pass MAKER (Holt and Yandell, 2011) annotation pipeline (v2.22) was used. In the 1st pass, evidence was used from the RNA-seq assembly, alignments to the Swiss-prot protein database, predictions of the ab initio gene finders SNAP (v2010-07-28) (Korf, 2004), and trained using CEGMA (v2.0) (Parra et al., 2007) gff output and GeneMark-ES (v2.3e) (Lomsadze et al., 2005). For the 2nd pass, first-pass MAKER gff files were used to train Augustus (v2.5.5) (Stanke and Waack, 2003), retrain the SNAP models, and MAKER was rerun with the addition of these two programs. Second, we annotated genomic regions without MAKER genes by RNA-seq data using tophat/cufflinks (v1.3.0) (Trapnell et al., 2012). Last, regions without MAKER and tophat/cufflinks genes were further annotated with transferred annotations from a published A. suum assembly (Jex et al., 2011) by using RATT (Otto et al., 2011). The final gene set consists of 11,446 genes from MAKER, 2,947 genes from tophat/cufflink and 1,053 genes from RATT.
Gene expression analysis
To profile the tissue expression of all A. suum genes, we used 8 different developmental stages/tissues of A. suum, including testis, ovary, embryo, larvae, intestine, muscle, male carcass and female carcass (Table S1). For each predicted gene, their expression level (RPKM) was calculated using tophat/cufflinks (Trapnell et al., 2012). Genes with a RPKM ≥ 2 and the RPKM ≥ 1.5 fold higher in one particular tissue compared to all other tissues were defined as enriched gene expression in a particular tissue (maternal genes RPKM >= 2/3 × higher than the embryo) (Figs. 5 and Table S5). This is a relatively high stringency cutoff due to the existence of neighbor/similar tissues in development, such as ovary/embryo/larvae and muscle/male carcass/female carcass in this analysis. The expression profiles for tissue specific genes and other genes were clustered by using Cluster (v3.0) (Eisen et al., 1998) and visualized in heatmaps using treeview (http://jtreeview.sourceforge.net/) (Fig. 5). For these analyses, the average expression in muscle, male carcass, and female carcass was used to estimate the baseline expression level for other somatic tissues. Groups of enriched genes are also illustrated in dotplots for enriched tissue versus all other tissues (Fig. S3).
Supplementary Material
Highlights.
Over 685 (5%) of Ascaris suum genes are eliminated during chromatin diminution
Eliminated genes are mainly expressed during gametogenesis and early embryogenesis
The genome changes are conserved in different somatic cell lineages and sexes
Diminution may be a mechanism of gene regulation that silences germline genes
Acknowledgments
We thank Richard Komuniecki, Bruce Bamber, Amanda Korchnak, Vera Hapiak, Jeff Myers, Peter Geldhof, Jenna Mann, and Routh Packing Co. for their support and hospitality in collecting A. suum material; Paul Megee, Mark Johnston, David Bentley, Jay Hesselberth, Jun Yu, and Lee Niswander, for their suggestions and comments on the manuscript; Nancy Holroyd, Alejandro Sanchez and other members of the Sanger parasite genomes group for sequencing the Belgian Ascaris; Jill Castoe, Katrina Diener, Kenneth Jones, Davis Farrell, and Bifeng Gao at UC for sequencing and bioinformatics support. We also thank the Edinburgh Compute and Data Facility for access to resources. This work was supported in part by grants to R.E.D. (NIH AI0149558, AI078087, and AI098421), to M.M. (NIH AI081803 and GM097435), a UK BBSRC PhD studentship to G.K., and School of Biological Sciences PhD fellowship and Overseas Research Student Award from the University of Edinburgh to S.K.
Footnotes
Supplemental Information includes 4 figures, 6 tables, and Supplemental Experimental Procedures.
Accession numbers and URLs
Genomic and RNA-seq reads were deposited at the SRA (SRP013430, SRP013609 and ERP000532), GSS (DU976516.1 - DU978593.1, ED033836.1 - ED479302.1, ED562826.1 - ED567026.1), and GEO (GSE38470) databases. The germline and somatic genomes are also available in NCBI BioProject database with accession number PRJNA62057. Ascaris suum genome browser: http://ascaris.nematodegenomes.org/. Data is also deposited at http://nematode.net and http://www.wormbase.org/.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann-Waldmann C, Jentsch S, Tobler H, Muller F. Chromatin diminution leads to rapid evolutionary changes in the organization of the germ line genomes of the parasitic nematodes A. suum and P. univalens. Mol Biochem Parasitol. 2004;134:53–64. doi: 10.1016/j.molbiopara.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Ueber Differenzierung der Zellkerne wahrend der Furchung des Eies von Ascaris megalocephala. Anat Anz. 1887;2:688–693. [Google Scholar]
- Boveri T. Festschr fur C von Kupffer. Jena: Fisher; 1899. Die Entwicklung von Ascaris megalocephala mit besonderer Rucksicht auf die Kernverhaltnisse; pp. 383–430. [Google Scholar]
- Boveri T. Zugleichein Beitrag zur Frage qualitativ-ungleicher Chromosomen-Teilung Festschr fiir R Hertwig. III. Jena: Fischer; 1910. Die Potenzen der Ascaris-Blastomeren bei abgeanderter Furchung; pp. 131–214. [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012 doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. DNA elimination in ciliates: transposon domestication and genome surveillance. Annu Rev Genet. 2011;45:227–246. doi: 10.1146/annurev-genet-110410-132432. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons SW, Klass MR, Hirsh D. Analysis of the constancy of DNA sequences during development and evolution of the nematode Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 1979;76:1333–1337. doi: 10.1073/pnas.76.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter A, Bernard V, Kenzelmann M, Tobler H, Muller F. Ribosomal heterogeneity from chromatin diminution in Ascaris lumbricoides. Science. 1994;265:954–956. doi: 10.1126/science.8052853. [DOI] [PubMed] [Google Scholar]
- Goday C, Esteban MR. Chromosome elimination in sciarid flies. Bioessays. 2001;23:242–250. doi: 10.1002/1521-1878(200103)23:3<242::AID-BIES1034>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Goday C, Pigozzi MI. Heterochromatin and histone modifications in the germline-restricted chromosome of the zebra finch undergoing elimination during spermatogenesis. Chromosoma. 2010;119:325–336. doi: 10.1007/s00412-010-0260-2. [DOI] [PubMed] [Google Scholar]
- Goday C, Pimpinelli S. The occurrence, role and evolution of chromatin diminution in nematodes. Parasitology today (Personal ed) 1993;9:319–322. doi: 10.1016/0169-4758(93)90229-9. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horos R, Ijspeert H, Pospisilova D, Sendtner R, Andrieu-Soler C, Taskesen E, Nieradka A, Cmejla R, Sendtner M, Touw IP, et al. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012;119:262–272. doi: 10.1182/blood-2011-06-358200. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Stoffel R, Tobler H, Mueller F. A newly formed telomere in Ascaris suum does not exert a telomere position effect on a nearby gene. Mol Cell Biol. 1996;16:130–134. doi: 10.1128/mcb.16.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, Tobler H, Muller F. New telomere formation during the process of chromatin diminution in Ascaris suum. The International journal of developmental biology. 2002;46:143–148. [PubMed] [Google Scholar]
- Jex AR, Liu S, Li B, Young ND, Hall RS, Li Y, Yang L, Zeng N, Xu X, Xiong Z, et al. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Kloc M, Zagrodzinska B. Chromatin elimination--an oddity or a common mechanism in differentiation and development? Differentiation. 2001;68:84–91. doi: 10.1046/j.1432-0436.2001.680202.x. [DOI] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr, Mello CC. C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, Ketting RF. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. Embo J. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnenat L, Tobler H, Muller F. Developmentally regulated telomerase activity is correlated with chromosomal healing during chromatin diminution in Ascaris suum. Mol Cell Biol. 1999;19:3457–3465. doi: 10.1128/mcb.19.5.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Bernard V, Tobler H. Chromatin diminution in nematodes. Bioessays. 1996;18:133–138. doi: 10.1002/bies.950180209. [DOI] [PubMed] [Google Scholar]
- Muller F, Tobler H. Chromatin diminution in the parasitic nematodes Ascaris suum and Parascaris univalens. Int J Parasitol. 2000;30:391–399. doi: 10.1016/s0020-7519(99)00199-x. [DOI] [PubMed] [Google Scholar]
- Muller F, Walker P, Aeby P, Neuhaus H, Felder H, Back E, Tobler H. Nucleotide sequence of satellite DNA contained in the eliminated genome of Ascaris lumbricoides. Nucleic Acids Res. 1982;10:7493–7510. doi: 10.1093/nar/10.23.7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Wicky C, Spicher A, Tobler H. New telomere formation after developmentally regulated chromosomal breakage during the process of chromatin diminution in Ascaris lumbricoides. Cell. 1991;67:815–822. doi: 10.1016/0092-8674(91)90076-b. [DOI] [PubMed] [Google Scholar]
- Nemetschke L, Eberhardt AG, Hertzberg H, Streit A. Genetics, chromatin diminution, and sex chromosome evolution in the parasitic nematode genus Strongyloides. Current biology. 2010;20:1687–1696. doi: 10.1016/j.cub.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Niedermaier J, Moritz KB. Organization and dynamics of satellite and telomere DNAs in Ascaris: implications for formation and programmed breakdown of compound chromosomes. Chromosoma. 2000;109:439–452. doi: 10.1007/s004120000104. [DOI] [PubMed] [Google Scholar]
- Otto TD, Dillon GP, Degrave WS, Berriman M. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 2011;39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S, Goday C. Unusual kinetochores and chromatin diminution in Parascaris. Trends in genetics. 1989;5:310–315. doi: 10.1016/0168-9525(89)90114-5. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proceedings of the National Academy of Sciences. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Baker C, Eichler EE, Amemiya CT. Genetic consequences of programmed genome rearrangement. Curr Biol. 2012;22:1524–1529. doi: 10.1016/j.cub.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicher A, Etter A, Bernard V, Tobler H, Muller F. Extremely stable transcripts may compensate for the elimination of the gene fert-1 from all Ascaris lumbricoides somatic cells. Developmental biology. 1994;164:72–86. doi: 10.1006/dbio.1994.1181. [DOI] [PubMed] [Google Scholar]
- Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19ii(Suppl 2):215–225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- Streeck RE, Moritz KB, Beer K. Chromatin diminution in Ascaris suum: nucleotide sequence of the eliminated satellite DNA. Nucleic Acids Res. 1982;10:3495–3502. doi: 10.1093/nar/10.11.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler H, Etter A, Muller F. Chromatin diminution in nematode development. Trends in genetics. 1992;8:427–432. doi: 10.1016/0168-9525(92)90326-y. [DOI] [PubMed] [Google Scholar]
- Tobler H, Muller F, Back E, Aeby P. Germ line - soma differentiation in Ascaris: A molecular approach. Experentia. 1985;41:1311–1319. [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Czech B, Crunk A, Wallace A, Mitreva M, Hannon GJ, Davis RE. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011;21:1462–1477. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann A. The Germ-Plasm: A Theory of Heredity. London: Walter Scott, LTD; 1893. [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufall RA, Robinson T, Katz LA. Evolution of developmentally regulated genome rearrangements in eukaryotes. J Exp Zool B Mol Dev Evol. 2005;304:448–455. doi: 10.1002/jez.b.21056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.