Abstract
The biological function of most proteins relies on reversible post-translational modifications, among which phosphorylation is most prominently studied and well recognized. Recently, a growing amount of evidence indicates that acetylation-deacetylation reactions, when applied to crucial mediators, can also robustly affect the function of target proteins and thereby have wide-ranging physiological impacts. Sirtuin 1 (SIRT1), which functions as a nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylase, deacetylates a wide variety of metabolic molecules in response to the cellular energy and redox status and as such causes significant changes in metabolic homeostasis. This review surveys the evidence for the emerging role of SIRT1-mediated deacetylation in the control of metabolic homeostasis.
Keywords: SIRT1, NAD, deacetylation, metabolic homeostasis
Introduction
Post-translational modification (PTM) refers to a chemical modification of a protein after its translation. During PTM, biochemical groups such as acetyl, phosphate, methyl, ubiquitin, and various lipids and carbohydrates can be attached to or removed from specific amino acid residues in the proteins. PTM represents an important mechanism to diversify and regulate protein function. Although phosphorylation is probably the most extensively studied and prevalent PTM in numerous biological pathways, the list of proteins that undergo acetylation-deacetylation is increasing rapidly. First identified in histones 40 years ago, acetylation-deacetylation has been known to occur and functionally alter over 80 transcription factors and many other nuclear regulators [1,2]. Illustrating the fact that acetylation was not a rare event, a proteomics study using mouse liver mitochondria identified 277 acetylation sites in 133 mitochondrial proteins [3], in which the acetylation was not or poorly described before. Most recently, another proteomics survey using three human cancer cell lines revealed 3600 acetylation sites on 1750 proteins, indicating that acetylation-deacetylation reactions, probably as prevalent as phosphorylation-dephosphorylation events, affect almost all major cellular pathways [4].
Sirtuin 1 (SIRT1), a mammalian ortholog of the yeast silent information regulator 2 (Sir2), has been referenced as a longevity protein or an aging regulator more and more frequently since growing evidence implies that the Sir2 family proteins mediate lifespan extension of yeast, worms, flies, and mammals [5-7]. In mammals, SIRT1 belongs to a small gene family with seven members, designated Sirtuin 1 through 7. Among them, SIRT1 is by far the best characterized [8-10]. SIRT1, functioning as a protein deacetylase, transfers the acetyl group of lysines in a protein substrate to the ADP-ribose moiety of nicotinamide adenine dinucleotide (NAD+) to produce a deacetylated protein, nicotinamide (NAM), and 2′-O-acetyl-ADP ribose [11,12]. A variety of SIRT1 substrates that are involved in a broad range of physiological functions, including genomic stability [13,14], cell survival [15-17], inflammation [18,19], and metabolism [9,20], have been identified, which probably confers upon SIRT1 the magical power to promote longevity. In this review, we will mainly discuss the beneficial effects of SIRT1-mediated deacetylation of metabolic molecules on energy and metabolic homeostasis.
SIRT1 deacetylates the PGC1s, the master regulators of mitochondrial function
One of the well-established SIRT1 metabolic substrates is the peroxisome proliferator activated receptor γ (PPARγ) coactivator 1 (PGC1) α. PGC1α was initially identified as a transcriptional coactivator of PPARγ capable of inducing the transcription of uncoupling protein-1 in brown adipose tissue [21]. Later, PGC1α has been shown to coactivate a wide variety of transcription factors including PPARα, hepatocyte nuclear factor 4α (HNF4α), estrogen-related receptor α (ERRα), Forkhead box-containing protein type O 1 (FOXO1), nuclear respiratory factor (NRF) 1 and 2, many of which are involved in the transcriptional control of mitochondrial proteins [22-24]. Thus PGC1α is endowed with the master ability to direct the complex program of mitochondrial biogenesis and function.
SIRT1, activated under low-nutrient conditions or by pharmacological SIRT1 activating compounds (STACs), interacts with and deacetylates PGC1α [25-27]. This deacetylation activates PGC1α and subsequently results in a significant yet well-coordinated change of mitochondrial gene expression (Fig. 1). This translates in vivo in the regulation of mitochondrial function and fuel usage in multiple metabolic tissues. In the fasted liver, deacetylation of PGC1α by SIRT1 leads to a transcriptional switch from glycolytic to gluconeogenic genes and thus increases the hepatic glucose production [25,26]. The gene expression of fatty acid oxidation enzymes is induced as well in the fasted liver to shift the fuel usage from glucose to fatty acids [26]. Deacetylation of PGC1α by SIRT1 in skeletal muscle also activates the expression of mitochondrial fatty acid oxidation genes in response to caloric deprivation [27].
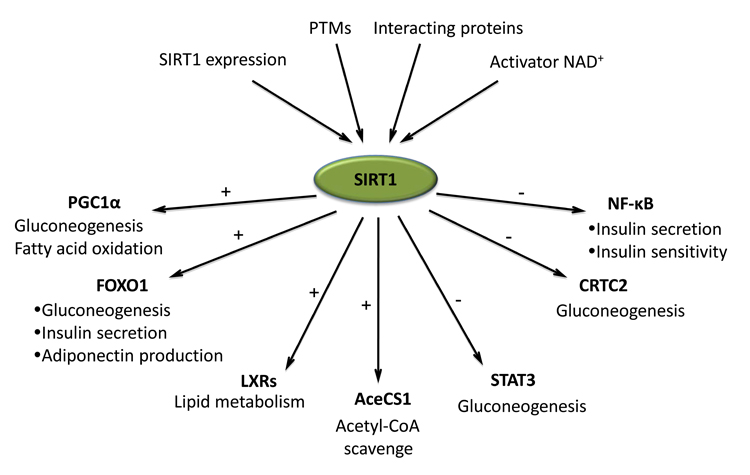
Fig. 1.
The role of protein deacetylation by SIRT1 in the control of metabolic homeostasis in response to nutrient and environmental stimuli. The deacetylase activity of SIRT1 is shaped and fine-tuned through its protein level, post-translational modifications, and association with inhibitors or cofactors. Most importantly, SIRT1 activity depends on the availability of cellular NAD+, which is orchestrated by the cellular redox status, NAD+ synthesis, and utilization in response to the nutrient and environment. The activated SIRT1 modulates many aspects of glucose and lipid homeostasis through deacetylating key metabolic molecules. The deacetylation of PGC1α by SIRT1 activates its transcriptional activity and thus induces the expression of target genes involved in gluconeogenesis and fatty acid oxidation. The SIRT1-mediated deacetylation of FOXO1 also increases its transcriptional activity and promotes the expression of gluconeogenic genes in liver, insulin gene in pancreas, and adiponectin gene in the adipose tissues. The sterol sensors LXRs are deacetylated and activated by SIRT1. The deacetylation contributes to balance cholesterol homeostasis in vivo. In addition, SIRT1 is capable to deacetylate and activate the cytoplasmic AceCS1, which scavenges acetate to usable acetyl-CoA. SIRT1 boosts and maintains gluconeogenesis in response to low nutrients by deacetylating and inhibiting other two transcription (co)factors CRTC2 and STAT3. The deacetylation of CRTC2 promotes its degradation whereas the STAT3 deacetylation renders STAT3 in the cytoplasm and prevents its inhibition on PGC1α expression. Similarly, the deacetylation of NF-κB by SIRT1 abrogates the nuclear translocation of NF-κB. This results in a decreased expression of iNOS gene in pancreas and preserves the viability and insulin secretion function of islets. In adipose tissues, the inhibition of NF-κB function via SIRT1 deacetylation decreases the inflammatory gene expression and correspondingly improves the insulin sensitivity.
Treatment with the natural SIRT1 activator, resveratrol, also reduces PGC1α acetylation and enhances mitochondrial biogenesis and oxidative phosphorylation (OXPHOS) in mice [28,29]. As a result, mice have increased aerobic capacity and are resistant to high fat diet (HFD)-induced obesity and insulin resistance [29]. The administration of a more specific and potent synthetic SIRT1 activator, SRT1720, leads to similar protective effects on metabolism in mice [30,31]. At the molecular level, PGC1α acetylation is also reduced by this STAC, leading to PGC1α activation. Similar to the changes in gene expression profile observed under low-nutrient conditions, the transcription of fatty acid oxidation genes is significantly induced in SRT1720 treated mice [30].
The genetic models in which SIRT1 expression levels are modulated also support a role of SIRT1-mediated deacetylation of PGC1α in metabolic control. Knockdown of SIRT1 in mouse liver decreases PGC1α-dependent expression of gluconeogenic and fatty acid oxidation genes [26]. These mice have hence a modest but consistently lower blood glucose level, combined with increased intracellular hepatic free fatty acid accumulation, as compared with control mice. Overexpression of SIRT1 reverses those effects, which are strikingly dependent on the presence of PGC1α [26]. Similarly, knockdown of SIRT1 in the liver of diabetic rats causes increased PGC1α acetylation and decreased gluconeogenic gene expression [32]. Liver specific deletion of SIRT1 impairs PPARα signaling and decreases fatty acid oxidation [33]. In SIRT1 deficient hepatocytes, PGC1α, as a coactivator of PPARα, is still recruited to the PPARα target promoters of fatty acid oxidation genes. However, its hyperacetylated state inhibits its ability to coactivate the transcription [33]. In contrast, in two independent studies, the moderate overexpression of SIRT1 in mice leads to elevated transcription of gluconeogenic genes [34] and a few other PGC1α target genes [35] in the hepatocytes.
Very recently, PGC1β, another member of PGC1 family, was shown to be acetylated by the acetyltransferase general control of amino acid synthesis (GCN5) and deacetylated by SIRT1 [36]. Less is, however, known about the function of PGC1β. The characterization of PGC1β deficient mice indicates that PGC1β is required for normal expression of OXPHOS genes and mitochondrial function in liver and skeletal muscle [37]. The acetylation of PGC1β by GCN5 inactivates PGC1β activity and thus attenuates the expression of endogenous PGC1β target genes such as medium chain acyl-CoA dehydrogenase (MCAD) and glucose transporter 4 (Glut4) [36]. Correspondingly, the glucose uptake in response to insulin is impaired in GCN5-overexpressing primary skeletal myotubes [36]. The deacetylation of PGC1β by SIRT1 probably has the reverse effects but this needs to be further confirmed.
SIRT1 deacetylates FOXO1, a crucial mediator of energy metabolism
FOXO transcription factors, especially FOXO1, play a significant role in the regulation of whole body energy metabolism [38]. SIRT1 has been shown to deacetylate FOXO1. The deacetylated FOXO1 was enriched in the nucleus and activated its target genes involved in gluconeogenesis in hepatocytes (Fig. 1) [39]. A detailed study revealed that the gluconeogenesis during caloric deprivation is orchestrated initially by the transcriptional cofactor CREB-regulated transcription coactivator 2 (CRTC2, will be discussed below) and sequentially by FOXO1 [40]. The activation of FOXO1 through SIRT1-mediated deacetylation is crucial to support the expression of the gluconeogenic program in the later stage of fasting [40]. Knockdown of SIRT1 in the liver of diabetic rats causes increased FOXO1 acetylation and decreased gluconeogenic gene expression [32]. Considering that PGC1α serves as a coactivator for FOXO1, the deacetylation of both PGC1α and FOXO1 by SIRT1 might have synergistic effects on their common gluconeogenic target genes.
In pancreatic β cells, FOXO1 promotes cell survival and preserves their insulin secretion function, on which the SIRT1-mediated deacetylation also has a remarkable impact (Fig. 1) [41]. By replacing the six lysines (the acetylation sites) in FOXO1 with either arginine (preventing acetylation) or glutamine (mimicking acetylation), the Accili group [41] showed that acetylation is required for the nuclear translocation of FOXO1 in pancreatic βTC-3 cells in response to oxidative stress. The acetylated FOXO1 binds to the promyelocytic leukemia (Pml) nuclear bodies, where it is deacetylated by SIRT1 to become transcriptional active. Subsequently, the activated FOXO1 induces the expression of NeuroD and musculoaponeurotic fibrosarcoma oncogene homolog (MafA), two critical transcription factors that activate the expression of the insulin gene [42]. It will be interesting to address whether the transient acetylation of FOXO1 observed in pancreatic cells is also a prerequisite for the nuclear translocation of FOXO1 in hepatocytes.
In adipocytes, the deacetylation of FOXO1 by SIRT1 enhances the transcription of adiponectin (Fig. 1) [34,43]. A FOXO1 mutant that can not be acetylated increases adiponectin expression whereas the FOXO1 mutant mimicking constitutive acetylation fails to activate the transcription of adiponectin [34]. In line with this observation in vitro, overexpression of SIRT1 in mice leads to an increase of adiponectin in the plasma and white adipose tissue and thus improves the insulin sensitivity [34].
SIRT1 deacetylates NF-κB, a key regulator of inflammation
The transcriptional factor nuclear factor-κB (NF-κB) is a key regulator of inflammation [44]. SIRT1 has been shown to deacetylate the p65 subunit of NF-κB and thus inhibit its transcriptional activity [45]. Very recently, the deacetylation of NF-κB by SIRT1 was found to protect pancreatic β cells against cytokine toxicity [46]. SIRT1 protein level decreases in cytokine-treated pancreatic RIN cells and rat islets. SIRT1 overexpression or activation by resveratrol reduces the acetylation of NF-κB and prevents its nuclear translocation to activate the expression of its target gene nitric oxide synthase (iNOS) in response to cytokines [46]. As a result, islet viability is increased and insulin secretion is preserved (Fig. 1) [46].
A growing amount of evidence indicates that chronic, low-grade inflammation can cause peripheral insulin resistance [47]. Genetic or chemical inhibition of NF-κB has been shown to improve insulin resistance [48,49]. The inhibition of NF-κB via SIRT1-mediated deacetylation seems to have similar effects (Fig. 1). In adipocytes, SIRT1 also deacetylates NF-κB and inhibits its binding to the target gene promoters [19]. Consistent with this, treatment of HFD-fed mice with the SIRT1 activator SRT1720 decreases NF-κB mediated inflammatory gene expression within adipose tissue depots. Correspondingly, insulin signaling is improved and the insulin-stimulated glucose uptake and Glut4 translocation is enhanced. In contrast, knockdown of SIRT1 has the reverse effects [19].
SIRT1 deacetylates LXRs, sterol sensors for cholesterol metabolism
The nuclear receptors liver X receptor (LXR) α and β serve as sterol sensors to regulate the cholesterol metabolism and hepatic lipid homeostasis [50-52]. Cholesterol is crucial to maintain cell membrane structure and also serves as a precursor for the synthesis of bile acids, steroids, and vitamin D. However, excessive cholesterol in the body accumulates in macrophages, favoring the development of atherosclerosis [53]. Reverse cholesterol transport (RCT), the process that helps to remove cholesterol from macrophages by redirecting cholesterol to the liver for subsequent elimination via the bile acid synthesis, is mainly regulated at the transcription levels [54]. The crucial cholesterol transporters for RCT, such as ATP binding cassette A1 (ABCA1) and sterol 27 hydroxylase (Cyp27A1), are transcriptionally regulated by the LXRs [55,56]. LXRs also regulate the expression of cytochrome P450 cholesterol 7α-hydroxylase (Cyp7A1), the rate-limiting enzyme of bile acid synthesis for the final step of RCT [57].
Interestingly, a single lysine K432 in LXRα and K433 in LXRβ are deacetylated by SIRT1. The deacetylation promotes the transcriptional activity of LXRs and facilitates their ubiquitination and subsequent degradation (Fig. 1) [58]. As such, SIRT1-mediated deacetylation enhances LXR cycling on their target promoters [59,60]. The deficiency of SIRT1 in primary macrohages decreases the induction of the LXR target gene ABCA1 upon LXR ligand treatment. Correspondingly, cholesterol export was impaired in SIRT1 deficient primary macrophages [58]. In line with this blunted cholesterol efflux observed in vitro, plasma HDL-cholesterol levels are significantly lower in SIRT1 +/− mice [58]. Specific knockdown of SIRT1 in liver also results in the lower levels of serum cholesterol [26]. At the molecular level, the absence of SIRT1 in those mice leads not only to decreased expression levels of ABCA1 but also Cyp7A1 [26], suggesting that the LXR-mediated bile acid synthesis, in addition to choleseterol efflux, is also affected by SIRT1.
It is important to note in this context that several other studies suggest that the actions of SIRT1 on cholesterol homeostasis may be complex. Activation of SIRT1 by the administration of STACs (either resveratrol or SRT1720) has no impact on blood HDL cholesterol levels [28-30]. Strikingly, the systemic HDL cholesterol levels are also reduced in mice overexpressing SIRT1 [61]. The fact that calorie restriction, which is known to activate SIRT1, improves atherosclerosis in both mice and humans, and also promotes cholesterol homeostasis in humans [62,63], however, are in line with the potential positive effects of SIRT1 on RCT observed in the knockout or knockdown models. To address the complex impact of the SIRT1-mediated regulation of LXRs and cholesterol metabolism, further studies at the molecular level in mice that are either challenged by a hypercholesterolemic diet or genetically predisposed to atherosclerosis will be required. Specific inactivation of SIRT1 in macrophages, liver, or intestine will also help to elucidate the contribution of SIRT1 to RCT in individual tissues.
SIRT1 deacetylates other metabolic molecules
CRTC2
As mentioned above, the transcription cofactor CRTC2 is critical for the induction of gluconeogenic genes during the early stage of caloric deprivation [40]. Interestingly, the transcriptional activity and protein stability of CRTC2 are also regulated by reversible acetylation [40]. Upon fasting, the acetyltransferases p300 and CREB binding protein (CBP) associate with and acetylate CRTC2. This acetylation promotes CRTC2 activity and stimulates the expression of its target gluconeogenic genes [40]. In the later stages of fasting, the activated SIRT1 deacetylates CRTC2 and promotes its ubiquitin-dependent degradation (Fig. 1). FOXO1 and PGC1α, activated by SIRT1 deacetylation, become then the major players to further support the expression of the gluconeogenic program [39,40].
STAT3
The transcription factor signal transducer and activator of transcription 3 (STAT3), a negative regulator of gluconeogenesis, is recently shown to be also deacetylated by SIRT1 [64]. STAT3 suppresses PGC1α expression and thus inhibits the gluconeogenesis in the liver [65,66]. The deacetylation of STAT3 by SIRT1 inhibits its phosphorylation. Thereby, STAT3 is not able to translocate into the nucleus to repress PGC1α expression and its suppression on gluconeogenesis is relieved (Fig. 1) [64].
AceCS1
In mammals, two acetyl-CoA synthetases (AceCS), the cytoplasmic AceCS1 and the mitochondrial AceCS2, are identified. These AceCS enzymes scavenge excessive acetate and turn it into usable acetyl-CoA [67,68]. SIRT1 has been shown to deacetylate and activate AceCS1 (Fig. 1) [69]. Overexpression of SIRT1, along with AceCS1 in the cells, led to a synergistic increase of acetate incorporation into lipids [69]. The activation of AceCS1 through SIRT1-mediated deacetylation might also facilitate other acetyl-CoA requiring metabolic reactions in cytoplasm.
SIRT1 serves as a sensor of cellular energy and redox states through NAD+ availability
As mentioned above, the protein deacetylation by SIRT1 requires NAD+ as a co-substrate. In fact, NAD+ is indispensible to the deacetylase activity of the Sir2 family proteins whereas the reaction by-product NAM blunts their activities [70-72]. The strict dependence of SIRT1 activity on NAD+ links its enzymatic activity to the availability of NAD+ in cells (Fig. 1).
One main role of NAD+ in organisms is to transfer electrons in redox reactions, through which the energy from nutrients is released for further utilization. For example, glycolysis and the citric acid cycle transfer energy from nutrients to NAD+ by its reduction to NADH, which is then transferred into the mitochondria. The NADH is then oxidized in turn by the electron transport chain, which pumps protons across the mitochondrial membrane and generates ATP through OXPHOS [73]. Therefore, it is believed that SIRT1 is capable of sensing the nutrient and redox status in cells through the fluctuation of NAD+/NADH ratio [23,74,75].
Most recently, nutrient deprivation or energy stress was shown to increase the NAD+ pool through the activation of AMP-activated protein kinase (AMPK) [76,77]. As indicated by its name, AMPK is activated by the increased AMP/ATP ratio in cells. The activated AMPK boosts ATP production processes and inhibits ATP use, thus restoring energy balance. Therefore, AMPK serves as a critical regulator of mitochondrial biogenesis and function in response to energy deprivation caused through low nutrients, exercise, and oxidative stress [78-81].
In skeletal myoblasts, AMPK, activated under low nutrition conditions or by 5-aminoimidazole-4-carboxamide-1-β-D-riboside (AICAR), induces the transcription of nicotinamide phosphoribosyltransferase (Nampt) [77]. Nampt is the rate-limiting enzyme in the mammalian NAD+ biosynthesis. It initiates NAD+ biosynthesis by catalyzing the conversion of NAM into nicotinamide mononucleotide (NMN), which is then converted to NAD+ via nicotinamide mononucleotide adenylyltransferase (Nmnat) [82-86]. Increased level of Nampt contributes to the synthesis of NAD+, which helps activate the function of SIRT1 [77].
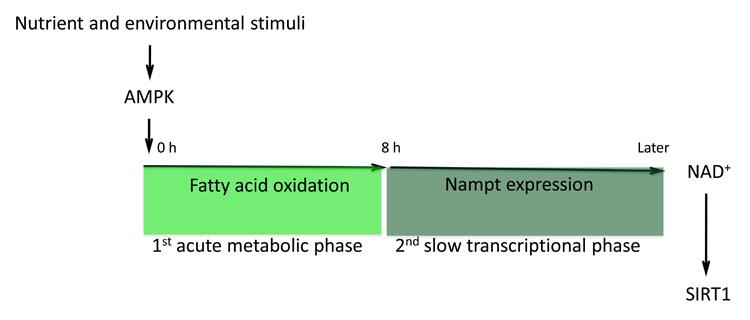
A more detailed study revealed that AMPK enhances NAD+/NADH ratio in a two-step model, the first step being independent of Nampt [76]. AMPK, activated by AICAR, increases NAD+ level, NAD+/NADH ratio, and simultaneously SIRT1-mediated PGC1α deacetylation in myotubes as early as 4 hours after AICAR treatment, before a potential contribution of Nampt induction to NAD+ synthesis. A significant increase of NAD+ pool was also evident 3 hours after exercise in tibialis anterior muscle. Strikingly, acute blockage of Nampt activity with a specific inhibitor FK866 did not affect the elevated NAD+/NADH ratio or PGC1α deacetylation in the first 8 hours after AMPK activation, suggesting a second Nampt-independent mechanism responsible for the initial boost in NAD+. Inhibition of fatty acid oxidation, however, completely abolished AMPK-induced acute increase of NAD+/NADH ratio and PGC1α deacetylation [76], suggesting that fatty acid oxidation pathway is crucial for AMPK to elevate NAD+/NADH ratio at least at the early stage of its activation. A time course dissection of Nampt expression showed that Nampt protein level began to increase only ~12 hours after AMPK activation (C. Canto and J. Auwerx, unpublished data), long after the “metabolic” induction of NAD+. Consistent with this two-step mechanism, chronic knockdown of Nampt for 48 hours impaired SIRT1-mediated PGC1α deacetylation [76]. Altogether, these data suggest a two-step induction of NAD+ levels by AMPK activation, composed of a fast “metabolic” phase that relies on fatty acid oxidation and a slower Nampt-dependent “transcriptional” phase, which is important to sustain the NAD+ pool in the later stage of AMPK activation (Fig. 2).
Fig. 2.
A two-step induction of NAD+ levels by AMPK activation. AMPK, activated by the nutrient deprivation or energy stress, enhances NAD+ levels through two steps. The initial acute metabolic phase relies on the boost in fatty acid oxidation. The second slow transcription phase depends on the induction of Nampt expression. Altogether, a higher level of NAD+ is obtained, which activates SIRT1.
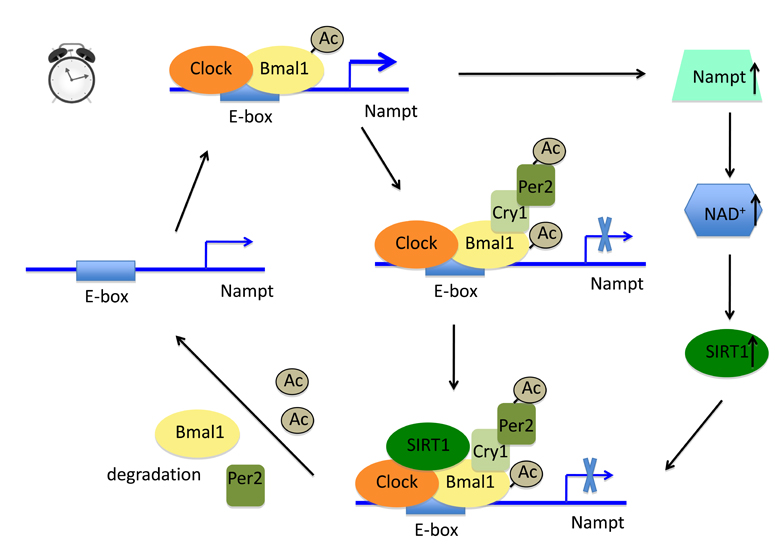
A new twist of this NAD+ availability and SIRT1 function is the recently identified circadian feedback loop between SIRT1 activity and Nampt transcription (Fig. 3) [87,88]. SIRT1 activity, intracellular NAD+ levels, and Nampt transcription oscillate in a circadian manner [87-90]. SIRT1 was first found to associate with the central circadian regulators Clock/Bmal1 in a circadian manner and as such be responsible for the deacetylation of Bmal1 and its corepressor Period 2 (Per 2) [89,90]. The deacetylation facilitates the efficient degradation of Bmal1 and Per2, and thus ensures to re-prime the promoter regions of Clock/Bmal1 target genes [89,90]. In a further twist on this story, SIRT1 was thereafter identified, along with Clock/Bmal1, to rhythmically occupy on the Nampt promoter to control the circadian expression of Nampt [87,88]. Deletion of Clock in mice abolishes the oscillation of Nampt transcription [88]. Conversely, disruption of Nampt function by FK866 blunts the SIRT1 activity and thus abrogates SIRT1-mediated deacetylation oscillation of Bmal1 [88]. Thus, SIRT1 activity and Nampt transcription are tightly linked together under the control of circadian rhythm.
Fig. 3.
A circadian feedback loop between SIRT1 activity and Nampt transcription. The core circadian regulators Clock/Bmal1 bind to the E-box of Nampt gene. Clock acetylates Bmal1 and local histone tails to promote the transcription of Nampt. The resulting Nampt enzyme boosts the synthesis of NAD+, which activates SIRT1. Meanwhile, other two Clock/Bmal1 targets Cry1 and Per2 are induced by Clock/Bmal1. The Cry1/Per2 heterodimer, serving as corepressors of Clock/Bmal1, turns off the transcription of Clock/Bmal1 target genes, including Nampt. The activated SIRT1 is subsequently recruited to the transcriptional machinery by Clock and deacetylates Bmal1 and Per2. This deacetylation promotes the degradation of both proteins. As such, the promoter region of Nampt is re-primed and the gene is ready for the next round of Clock-dependent induction.
Concluding remarks and future perspectives
Protein deacetylation by SIRT1, triggered under energy deprivation, energy stress or regulated through circadian clock, modulates many aspects of glucose and lipid homeostasis. This points to a complex regulatory role of SIRT1-mediated deacetylation in whole body metabolism. Because of the pleiotropic effect of germline SIRT1 deficiency on the development of animals [91], tissue-specific SIRT1 deficient animals will become very valuable tools to dissect the functional significance of deacetylation by SIRT1 in metabolic tissues. So far only liver-specific genetic ablation of SIRT1 is reported. Other metabolic tissue-specific or conditional deletions of the SIRT1 gene will definitely provide novel insights into the physiological actions of SIRT1.
The ability to sense the cellular NAD+ availability enables SIRT1 to modulate metabolism and other important physiological events in response to a variety of nutritional and environmental stimuli, though it is not likely that NAD+ acts as the sole switch controlling SIRT1 activity. Transcriptional control of SIRT1 expression, PTMs including sumoylation and phosphorylation, association of cofactors (such as active regulator of SIRT1 (AROS)) or inhibitors (such as deleted in breast cancer 1 (DBC1)), and subcellular localization all contribute to shape, fine tune, and terminate the SIRT1 response (Fig. 1) [20,92,93]. Untangling the crosstalk among those regulatory knots and identification of novel modulators will help to understand the comprehensive regulatory SIRT1 circuits.
The nutrient availability not only has an impact on the regulation of the SIRT1 deacetylase, but equally affects the regulation of protein acetyltransferases, as highlighted by a recent study showing that the acetyltransferases GCN5 and steroid receptor 3 (SRC3) were robustly induced in the muscle after HFD feeding but significantly reduced upon fasting [94]. Both GCN5 and SRC3 regulate the glucose metabolism through increasing the acetylation state of PGC1α [94,95], acting from the opposite angle as SIRT1. Thus, both acetyltransferases and deacetylases, as ying-yang players controlling acetylation-deacetylation, serve as real energy sensors to coordinate the metabolic homeostasis. Further work to delineate the functions of diverse acetyltransferases that are responsible to acetylate SIRT1 metabolic substrates will complete the flip side picture of SIRT1 action on metabolism.
Remarkably, the functional output of SIRT1-mediated deacetylation varies among the substrates. In some cases, the deacetylation activates the function of the substrates (such as PGC1α, FOXO1) whereas in other cases the deacetylation inhibits their actions (STAT3, NF-κB) (Fig. 1). Although all the physiological consequences of the deacetylation favor to reach a better-balanced energy homeostasis, it is still puzzling to see the multitude of mechanisms involved at the molecular level. It is tempting to speculate that other PTMs such as phosphorylation, ubiquitination, and methylation might have synergistic or antagonistic effects along with the deacetylation. In this context it is interesting to point out that a PGC1α mutant, lacking the two AMPK phosphorylation sites, becomes resistant to deacetylation by SIRT1 and not able to activate its downstream genes [76]. Therefore, similar to the epigenetic regulatory code that affects histone and chromatin function, a code might exist in non-histone proteins to direct the function of key cellular mediators. Further work to unveil the interplay between SIRT1-mediated deacetylation and other PTMs triggered by other sensors will shed light on not only the SIRT1 field but also contribute to understand the epigenetic code.
Finally, identification of novel SIRT1 targets, definition of the function of other sirtuin members, and their possible cross-regulation with SIRT1 are also needed, as this will further enhance our knowledge of sirtuin biology. All the progress will pave the way for a better understanding of metabolic diseases and possible development of novel therapies that target members of the sirtuin gene family.
Acknowledgement
We thank E. Jeninga and C. Canto for their helpful discussions and comments. The work in the laboratory of the authors is supported by grants of the Ecole Polytechnique Fédérale de Lausanne (EPFL), National Science Foundation of Switzerland, NIH (DK59820), and the European Research Council Ideas programme (Sirtuins; ERC-2008-AdG-23118). J. Y. is supported by a FEBS long-term fellowship.
References
- 1.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Yang XJ, Seto E. Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 5.Canto C, Auwerx J. Caloric restriction, sirt1 and longevity. Trends Endocrinol Metab. 2009;20:325–331. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michan S, Sinclair D. Sirtuins in mammals: Insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: The ‘magnificent seven’, function, metabolism and longevity. Ann Med. 2007;39:335–345. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007;21:1745–1755. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- 9.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Feige JN, Auwerx J. Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr Opin Cell Biol. 2008;20:303–309. doi: 10.1016/j.ceb.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved nad+-dependent protein deacetylase activity in the sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein sir2 and its homologs are nad-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. Sirt1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. Sirt1 promotes DNA repair activity and deacetylation of ku70. Exp Mol Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 15.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 16.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of sirt1 cooperates with sirt1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 17.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of foxo transcription factors by the sirt1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 18.Yang SR, Wright J, Bauter M, Seweryniak K, Kode A, Rahman I. Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via rela/p65 nf-kappab in macrophages in vitro and in rat lungs in vivo: Implications for chronic inflammation and aging. Am J Physiol Lung Cell Mol Physiol. 2007;292:L567–576. doi: 10.1152/ajplung.00308.2006. [DOI] [PubMed] [Google Scholar]
- 19.Yoshizaki T, Milne JC, Imamura T, Schenk S, Sonoda N, Babendure JL, Lu JC, Smith JJ, Jirousek MR, Olefsky JM. Sirt1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol Cell Biol. 2009;29:1363–1374. doi: 10.1128/MCB.00705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann N Y Acad Sci. 2009;1173(Suppl 1):E10–19. doi: 10.1111/j.1749-6632.2009.04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 22.Finley LW, Haigis MC. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res Rev. 2009;8:173–188. doi: 10.1016/j.arr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the pgc-1alpha and sirt1 pathways. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of pgc-1alpha and sirt1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 26.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through sirt1/pgc-1alpha. Embo J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating sirt1 and pgc-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific sirt1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 31.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of sirt1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erion DM, Yonemitsu S, Nie Y, Nagai Y, Gillum MP, Hsiao JJ, Iwasaki T, Stark R, Weismann D, Yu XX, Murray SF, Bhanot S, Monia BP, Horvath TL, Gao Q, Samuel VT, Shulman GI. Sirt1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc Natl Acad Sci U S A. 2009;106:11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of sirt1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. Sirt1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelly TJ, Lerin C, Haas W, Gygi SP, Puigserver P. Gcn5-mediated transcriptional control of the metabolic coactivator pgc-1beta through lysine acetylation. J Biol Chem. 2009;284:19945–19952. doi: 10.1074/jbc.M109.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vianna CR, Huntgeburth M, Coppari R, Choi CS, Lin J, Krauss S, Barbatelli G, Tzameli I, Kim YB, Cinti S, Shulman GI, Spiegelman BM, Lowell BB. Hypomorphic mutation of pgc-1beta causes mitochondrial dysfunction and liver insulin resistance. Cell Metab. 2006;4:453–464. doi: 10.1016/j.cmet.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gross DN, van den Heuvel AP, Birnbaum MJ. The role of foxo in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- 39.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor foxo1 via sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, Gu W, Accili D. Foxo1 protects against pancreatic beta cell failure through neurod and mafa induction. Cell Metab. 2005;2:153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 42.Olbrot M, Rud J, Moss LG, Sharma A. Identification of β-cell-specific insulin gene transcription factor ripe3b1 as mammalian mafa. Proc Natl Acad Sci U S A. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao L, Shao J. Sirt1 regulates adiponectin gene expression through foxo1-c/enhancer-binding protein alpha transcriptional complex. J Biol Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 44.Tak PP, Firestein GS. Nf-kappab: A key role in inflammatory diseases. J Clin Invest. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of nf-kappab-dependent transcription and cell survival by the sirt1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JH, Song MY, Song EK, Kim EK, Moon WS, Han MK, Park JW, Kwon KB, Park BH. Overexpression of sirt1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-kappab signaling pathway. Diabetes. 2009;58:344–351. doi: 10.2337/db07-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, Wynshaw-Boris A, Poli G, Olefsky J, Karin M. Ikk-beta links inflammation to obesity-induced insulin resistance. Nat Med. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 49.Shoelson SE, Lee J, Yuan M. Inflammation and the ikk beta/i kappa b/nf-kappa b axis in obesity- and diet-induced insulin resistance. Int J Obes Relat Metab Disord. 2003;27(Suppl 3):S49–52. doi: 10.1038/sj.ijo.0802501. [DOI] [PubMed] [Google Scholar]
- 50.Cummins CL, Mangelsdorf DJ. Liver x receptors and cholesterol homoeostasis: Spotlight on the adrenal gland. Biochem Soc Trans. 2006;34:1110–1113. doi: 10.1042/BST0341110. [DOI] [PubMed] [Google Scholar]
- 51.Fayard E, Schoonjans K, Auwerx J. Xol inxs: Role of the liver x and the farnesol x receptors. Curr Opin Lipidol. 2001;12:113–120. doi: 10.1097/00041433-200104000-00004. [DOI] [PubMed] [Google Scholar]
- 52.Schoonjans K, Brendel C, Mangelsdorf D, Auwerx J. Sterols and gene expression: Control of affluence. Biochim Biophys Acta. 2000;1529:114–125. doi: 10.1016/s1388-1981(00)00141-4. [DOI] [PubMed] [Google Scholar]
- 53.Linsel-Nitschke P, Tall AR. Hdl as a target in the treatment of atherosclerotic cardiovascular disease. Nat Rev Drug Discov. 2005;4:193–205. doi: 10.1038/nrd1658. [DOI] [PubMed] [Google Scholar]
- 54.Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol. 2003;65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- 55.Zelcer N, Tontonoz P. Liver x receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116:607–614. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Attie AD, Kastelein JP, Hayden MR. Pivotal role of abca1 in reverse cholesterol transport influencing hdl levels and susceptibility to atherosclerosis. J Lipid Res. 2001;42:1717–1726. [PubMed] [Google Scholar]
- 57.Kalaany NY, Gauthier KC, Zavacki AM, Mammen PP, Kitazume T, Peterson JA, Horton JD, Garry DJ, Bianco AC, Mangelsdorf DJ. Lxrs regulate the balance between fat storage and oxidation. Cell Metab. 2005;1:231–244. doi: 10.1016/j.cmet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 58.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. Sirt1 deacetylates and positively regulates the nuclear receptor lxr. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 59.Dennis AP, O’Malley BW. Rush hour at the promoter: How the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–151. doi: 10.1016/j.jsbmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 60.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 61.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. Sirt1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 62.Guo Z, Mitchell-Raymundo F, Yang H, Ikeno Y, Nelson J, Diaz V, Richardson A, Reddick R. Dietary restriction reduces atherosclerosis and oxidative stress in the aorta of apolipoprotein e-deficient mice. Mech Ageing Dev. 2002;123:1121–1131. doi: 10.1016/s0047-6374(02)00008-8. [DOI] [PubMed] [Google Scholar]
- 63.Fontana L, Meyer TE, Klein S, Holloszy JO. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci U S A. 2004;101:6659–6663. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q. Stat3 inhibition of gluconeogenesis is downregulated by sirt1. Nat Cell Biol. 2009;11:492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inoue H, Ogawa W, Asakawa A, Okamoto Y, Nishizawa A, Matsumoto M, Teshigawara K, Matsuki Y, Watanabe E, Hiramatsu R, Notohara K, Katayose K, Okamura H, Kahn CR, Noda T, Takeda K, Akira S, Inui A, Kasuga M. Role of hepatic stat3 in brain-insulin action on hepatic glucose production. Cell Metab. 2006;3:267–275. doi: 10.1016/j.cmet.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 66.Inoue H, Ogawa W, Ozaki M, Haga S, Matsumoto M, Furukawa K, Hashimoto N, Kido Y, Mori T, Sakaue H, Teshigawara K, Jin S, Iguchi H, Hiramatsu R, LeRoith D, Takeda K, Akira S, Kasuga M. Role of stat-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med. 2004;10:168–174. doi: 10.1038/nm980. [DOI] [PubMed] [Google Scholar]
- 67.Fujino T, Kondo J, Ishikawa M, Morikawa K, Yamamoto TT. Acetyl-coa synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem. 2001;276:11420–11426. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- 68.Yamashita H, Kaneyuki T, Tagawa K. Production of acetate in the liver and its utilization in peripheral tissues. Biochim Biophys Acta. 2001;1532:79–87. doi: 10.1016/s1388-1981(01)00117-2. [DOI] [PubMed] [Google Scholar]
- 69.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-coa synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human sirt1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 71.Lin SJ, Defossez PA, Guarente L. Requirement of nad and sir2 for life-span extension by calorie restriction in saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 72.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein sir2 is an nad-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 73.Rich PR. The molecular machinery of keilin’s respiratory chain. Biochem Soc Trans. 2003;31:1095–1105. doi: 10.1042/bst0311095. [DOI] [PubMed] [Google Scholar]
- 74.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of nad- an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010 doi: 10.1210/er.2009-0026. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Belenky P, Bogan KL, Brenner C. Nad+ metabolism in health and disease. Trends Biochem Sci. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 76.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. Ampk regulates energy expenditure by modulating nad+ metabolism and sirt1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating sirt1 through ampk-mediated regulation of nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kahn BB, Alquier T, Carling D, Hardie DG. Amp-activated protein kinase: Ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 79.Fulco M, Sartorelli V. Comparing and contrasting the roles of ampk and sirt1 in metabolic tissues. Cell Cycle. 2008;7:3669–3679. doi: 10.4161/cc.7.23.7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hardie DG. Ampk: A key regulator of energy balance in the single cell and the whole organism. Int J Obes (Lond) 2008;32(Suppl 4):S7–12. doi: 10.1038/ijo.2008.116. [DOI] [PubMed] [Google Scholar]
- 81.Canto C, Auwerx J. Pgc-1alpha, sirt1 and ampk, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Preiss J, Handler P. Enzymatic synthesis of nicotinamide mononucleotide. J Biol Chem. 1957;225:759–770. [PubMed] [Google Scholar]
- 83.Hogeboom GH, Schneider WC. Cytochemical studies. Vi. The synthesis of diphosphopyridine nucleotide by liver cell nuclei. J Biol Chem. 1952;197:611–620. [PubMed] [Google Scholar]
- 84.Emanuelli M, Carnevali F, Saccucci F, Pierella F, Amici A, Raffaelli N, Magni G. Molecular cloning, chromosomal localization, tissue mrna levels, bacterial expression, and enzymatic properties of human nmn adenylyltransferase. J Biol Chem. 2001;276:406–412. doi: 10.1074/jbc.M008700200. [DOI] [PubMed] [Google Scholar]
- 85.Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, Andris F. Pre-b-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in nad biosynthesis. Eur J Immunol. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 86.Revollo JR, Grimm AA, Imai S. The nad biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 87.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through nampt-mediated nad+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the nad+ salvage pathway by clock-sirt1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The nad+-dependent deacetylase sirt1 modulates clock-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. Sirt1 regulates circadian clock gene expression through per2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 91.McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian sir2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kwon HS, Ott M. The ups and downs of sirt1. Trends Biochem Sci. 2008;33:517–525. doi: 10.1016/j.tibs.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 93.Zschoernig B, Mahlknecht U. Sirtuin 1: Regulating the regulator. Biochem Biophys Res Commun. 2008;376:251–255. doi: 10.1016/j.bbrc.2008.08.137. [DOI] [PubMed] [Google Scholar]
- 94.Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O’Malley BW, Auwerx J. The genetic ablation of src-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of pgc-1{alpha} Proc Natl Acad Sci U S A. 2008;105:17187–17192. doi: 10.1073/pnas.0808207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. Gcn5 acetyltransferase complex controls glucose metabolism through transcriptional repression of pgc-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]