Abstract
Recently the function of the sirtuin family, named after their homology to the Saccharomyces cerevisiae gene silent information regulator 2 (Sir2), has received a lot of attention, as their beneficial impact on longevity was linked to their effects on metabolic control. All sirtuins require nicotinamide adenine dinucleotide (NAD+) for their deacetylase or ADP-ribosyl transferase activity, linking their function tightly to cellular energy levels. SIRT1, the founding member of the sirtuin family, modulates many aspects of glucose and lipid homeostasis in almost all key metabolic tissues. Other members including SIRT2, SIRT3, and SIRT4 are also implicated in various metabolic processes. Here, we review the recent data related to the role of sirtuins in the control of metabolic homeostasis and possible underlying molecular mechanisms.
Keywords: sirtuin, NAD+, deacetylase, ADP-ribosyl transferase, metabolic homeostasis, metabolic disorders
Introduction
Sirtuins in mammals were named after their homology to the Saccharomyces cerevisiae gene silent information regulator 2 (Sir2).1, 2 The sirtuins represent a small gene family with seven members in mammals, i. e. SIRT1-SIRT7. They share significant sequence homology and contain conserved catalytic and nicotinamide adenine dinucleotide (NAD+)-binding domains. Growing evidence has indicated that SIRT1, the founding member of the sirtuin family, serves as a key molecule to promote longevity, whereas the role of other sirtuins in the control of longevity starts to be appreciated.3-5 It is believed that these protective actions of the sirtuins stem at least partially from their beneficial effects on energy and metabolic homeostasis. Therefore, the elucidation of the role of the various sirtuins in metabolic control is expected to have a significant impact on the prevention and treatment of metabolic disorders such as obesity, diabetes, hyperlipidemia, and atherosclerosis.
All sirtuins require NAD+ for their enzymatic activity. Most sirtuins catalyze deacetylation reactions in which the acetyl group of an acetylated protein substrate is transferred to the ADP-ribose (ADPR) moiety of NAD+ and eventually a deacetylated protein, nicotinamide (NAM), and 2′-O-acetyl-ADP ribose are produced.6, 7 Concomitantly, NAD+ activates the deacetylase activity of sirtuins, whereas NAM and the reduced form of NAD (NADH) inhibit their activity.8-10 SIRT4 lacks deacetylase activity but instead demonstrates a robust ADP-ribosyl transferase activity.11, 12 It transfers the ADPR residue of NAD+ to their protein substrate and correspondingly NAM is produced. Similar to their regulatory effects on the deacetylase sirtuins, NAD+ is indispensable to the ADP-ribosyl transferase activity of SIRT4 whereas NAM blunts its activity.11, 12 Interestingly, SIRT6 exhibits both deacetylase and ADP-ribosyl transferase activities, which are also NAD+ dependent.13-15
First, histones were identified as the deacetylation substrates of sirtuins.16, 17 Recently, sirtuins have been found to deacetylate a number of non-histone proteins including the transcription factors p53, nuclear factor-κB (NFκB), forkhead box-containing protein type O subfamily (FOXO), peroxisome proliferator activated receptor γ (PPARγ), the transcription coregulator PPARγ coactivator 1α (PGC-1α), the metabolic enzymes acetyl-CoA synthetases (AceCSs), and the structural protein α-tubulin (reviewed in2). The known ADP-ribosylation substrates of the sirtuins include the metabolic enzyme glutamate dehydrogenase (GDH), a SIRT4 target, and DNA polymerase β, a SIRT6 target.11, 14 Thus, through sensing [NAD+]/[NADH] ratio and NAM levels, sirtuins facilitate the conversion of nutritional status into modulation of gene and protein function, leading ultimately to changes in cellular metabolism.
A tight regulation of the equilibrium between energy intake, storage, and expenditure is required for metabolic homeostasis. The involvement of sirtuins in the control of energy intake in mammals remains largely unexplored. Over the past few years, evidence is, however, accumulating for a regulatory role of the sirtuins in energy storage and expenditure in different metabolic tissues. In this review, we will discuss: 1) the metabolic activities of the sirtuins, with a particular focus on the founding member of the family, SIRT1; 2) the underlying molecular mechanisms through which the sirtuins control metabolism.
SIRT1 promotes gluconeogenesis, fatty acid oxidation, and cholesterol scavenging in the liver
The liver functions as a major metabolic regulator to control glucose and lipid homeostasis. The majority of the rate-limiting enzymes involved in the hepatic energy homeostasis are regulated at the transcriptional level.18 Several nuclear receptors, including the liver X receptors (LXRs), sterol response elements-binding proteins (SREBPs), and cofactors, such as PGC-1α, have been identified as the key transcriptional regulators for the expression of the metabolic enzymes in the liver.19-21 The SIRT1 protein has been found to be induced and activated in the liver upon fasting22 and emerging evidence has implicated it as another important metabolic regulator in this tissue (Figure 1).
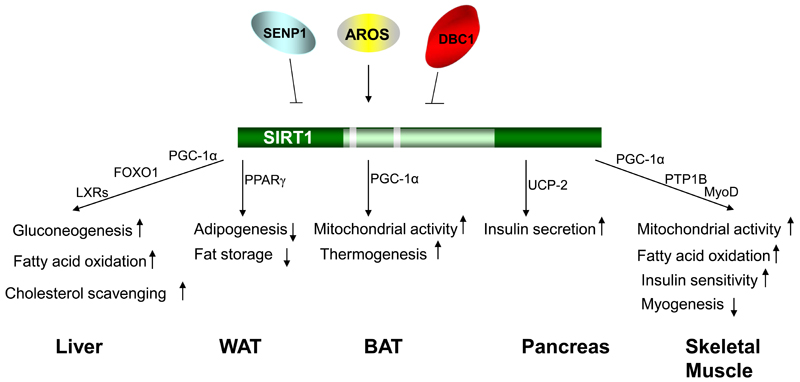
Figure 1.
The diverse functions of SIRT1 in different metabolic tissues. SIRT1 promotes gluconeogenesis through PGC-1α and possibly FOXO1 in the liver upon fasting. SIRT1 also facilitates fatty acid oxidation through PGC-1α and cholesterol scavenging through LXRs in the liver. In WAT, SIRT1 inhibits the adipogenesis process and blocks fat storage mainly by serving as a corepressor of PPARγ. In BAT, SIRT1 increases the mitochondrial activity in cells and improves the thermogenic capacity of BAT in a PGC-1α-dependent manner. SIRT1 also functions as the corepressor of PPARγ on the promoter of UCP-2 gene in the pancreas. Inhibition of UCP-2 protein expression enhances ATP production and subsequent insulin secretion in β cells. SIRT1 blocks myogenesis partially through reducing MyoD activity. In the mature skeletal muscle, SIRT1 activation leads to an increase in mitochondrial activity, fatty acid oxidation, and insulin sensitivity. SIRT1 activity itself is regulated by multiple modulator proteins. The nuclear protein AROS is suggested to promote the deacetylase activity of SIRT1, whereas another SIRT1-interacting protein DBC1 inhibits SIRT1 activity. The desumoylase SENP1 desumoylates and subsequent inactivates SIRT1.
Under low nutrient conditions, the expression of the SIRT1 protein is induced and it will interact and deacetylate PGC-1α. This deacetylation activates PGC-1α function, consequently promoting PGC-1α-mediated gluconeogenesis.22 SIRT1 has also been shown to deacetylate and activate the transcription factor FOXO1, leading to the subsequent enhanced expression of FOXO1 target genes that induce gluconeogenesis and glucose release from hepatocytes.23 Consistently, knockdown of hepatic SIRT1 with small hairpin RNA (shRNA) in mice led to decreased expression of gluconeogenic genes.24 The resulting mice demonstrated mild hypoglycemia, increased systemic glucose and insulin sensitivity because of the decreased glucose production in the liver. Overexpression of SIRT1 had the reverse effects. The majority of these effects were shown to be PGC-1α-dependent.24
The knockdown of hepatic SIRT1 also dramatically increased the level of hepatic free fatty acids (FFAs) without impacting on hepatic/serum triglycerides or serum FFA level.24 This effect is achieved through the inhibition of PGC-1α-mediated fatty acid oxidation when SIRT1 expression is reduced.
A significant accumulation of hepatic cholesterol and reduced systemic cholesterol levels were also observed upon hepatic knockdown of SIRT1 and in SIRT1 +/− mice.24, 25 Reduction of SIRT1 activity in the liver was accompanied by the decreased expression of genes important for cholesterol efflux and degradation. Interestingly, this effect of SIRT1 seems independent on PGC-1α since PGC-1α reduction had no impact on this phenotype.24 However, expression of genes that control cholesterol levels such as SR-B1 and Cyp7A1 requires PGC-1α.24 LXRs might contribute to this SIRT1 action considering the facts that both LXRα and LXRβ are deacetylated and activated by SIRT1 and that the expression of LXR target genes important for cholesterol homeostasis is decreased in SIRT1 deficient mice.25 All these observations are consistent with the fact that calorie restriction, which is known to activate SIRT1, improves atherosclerosis and cholesterol homeostasis in humans.26 However, the activation of SIRT1 by resveratrol has no impact on blood cholesterol levels,27, 28 which might be due to the potential SIRT1-independent effects of resveratrol.29
SIRT1 attenuates adipogenesis and fat storage in white adipose tissue
White adipose tissue (WAT) is the primary depot for fat in mammals. The nuclear receptor PPARγ is the master regulator of the adipogenesis process and fat storage in WAT.30, 31 Down-regulation of SIRT1 in 3T3-L1 cells enhanced adipogenesis (Figure 1) and was accompanied with higher expression levels of PPARγ target genes, including PPARγ itself, CCAAT/enhancer-binding protein (C/EBP)-α, C/EBP-δ, and fatty acid binding protein aP2.32 Conversely, overexpression of SIRT1 attenuated the adipocyte differentiation and reduced expression of fat storage-related genes in mature adipocytes. In mice, fasting-activated SIRT1 was recruited to the promoters of PPARγ target genes, together with the nuclear receptor corepressor (NCoR) and silencing mediator for retinoid and thyroid hormone receptor (SMRT), to corepress the expression of PPARγ mediated genes that drive fat storage. Consistently, mobilization of fatty acids from WAT upon fasting is compromised in SIRT1 +/− mice that have lower levels of SIRT132 and NAM treatment, which inhibits SIRT1 activity, significantly increases fat mass in the mice.33 Conversely, overexpression of SIRT1 in mice causes a large reduction in fat mass.34 Mice treated with the SIRT1 activator, resveratrol, also demonstrate a smaller amount of WAT, a decrease in fat storage and smaller adipocytes.28
SIRT1 improves mitochondrial activity and thermogenesis of brown adipose tissue
It is well established that brown adipose tissue (BAT) is responsible for non-shivering thermogenesis in rodents and human newborns.35 As well-identified BAT depots were not found in adult humans using the traditional techniques, little attention was given to the functional relevance of BAT in adults. Interestingly, recent studies, using positron emission tomography (PET), have shown that a substantial fraction of adult humans possess active BAT in the upper chest and neck regions.36 Thus, strategies to enhance BAT energy expenditure could lead to weight loss and have potential metabolic significance. Interestingly, mice treated with the SIRT1 activator resveratrol exhibit enhanced thermogenesis (Figure 1).28 Concomitantly, larger mitochondria were observed in the BAT. Both the mitochondrial size and DNA content were increased whereas lipid-droplet size was decreased, reflecting an enhanced mitochondrial activity in this tissue. At the molecular level, PGC-1α acetylation was significantly decreased in the BAT from resveratrol-treated mice and the expression levels of its related target genes were correspondingly increased. 28
A recent genomic analysis indicated that SIRT1 might function as a potential differentiation mediator for brown adipocytes.37 A microarray comparison of primary brown and white preadipocytes revealed that a substantial percentage of genes expressed in the brown preadipocytes belong to the ‘skeletal muscle’ gene ontology class. The expression levels of those myogenic genes, such as myogenin, MyoD, and Myf5, which are only expressed in brown preadipocytes, diminish during their differentiation into mature adipocytes.37 SIRT1 has been previously shown to prevent myoblast differentiation,38 and the transcription profile from myoblasts with forced expression of SIRT138 overlapped well with the one from brown preadipocytes that underwent differentiation.37 In both profiles, muscle development genes are repressed whereas mitochondrial and metabolic genes are upregulated, thus indicating a plausible role of SIRT1 in the regulation of brown adipocyte differentiation. A possible convergence of SIRT1 function with the actions of PRDM16, a key determinant of the differentiation of brown adipocytes and muscle cells,39, 40 awaits also further study.
SIRT1 enhances insulin secretion in the pancreas
Pancreatic β cells are the archetypal metabolic sensors as they adjust insulin secretion to the prevailing glucose levels. In the murine pancreas, SIRT1 is preferentially expressed in β cells.41, 42 Specific overexpression of SIRT1 in β cells enhanced glucose-stimulated insulin secretion in transgenic mice and hence improved their glucose tolerance (Figure 1).41 Conversely, knockdown of SIRT1 in β cell lines compromised insulin secretion.42 SIRT1 achieved this effect at least partially by repressing transcription of the uncoupling protein (UCP)-2.41, 42 SIRT1 directly binds to the promoter region of endogenous UCP-2 gene and overexpression of SIRT1 blunted PPARγ-mediated UCP-2 transcription.42 UCP-2 is a mitochondrial inner-membrane protein that uncouples oxygen consumption during respiration from ATP generation, by allowing proton leakage.43 The decreased UCP-2 expression mediated by SIRT1 leads hence to a better coupling of mitochondrial respiration and ATP synthesis, which enhances ATP production and thereby sensitizes the β cells to the blood glucose level.44 This is reminiscent to the phenotype of UCP-2 knockout mice, which exhibited higher ATP level in pancreatic islets and increased insulin secretion in response to glucose stimulation whereas overexpression of UCP-2 in cultured β cell lines caused impaired insulin secretion.44, 45
SIRT1 increases mitochondrial activity, fatty acid oxidation, and insulin sensitivity in the skeletal muscle
Skeletal muscle is the main tissue that possesses the mitochondrial capacity for energy expenditure in the adult human. It utilizes either glucose or lipid to produce energy in response to environmental demands. Nutrient deprivation, similar to what happens in the liver, also activates SIRT1, which subsequently deacetylates and activates PGC-1α in skeletal muscle cells (Figure 1).46 The activated PGC-1α then induces the expression of genes involved in mitochondrial fatty acid oxidation, electron transport, and oxidative phosphorylation. Activation of SIRT1 by resveratrol also increases the expression and activity of muscle PGC-1α and its downstream targets that regulate mitochondrial biogenesis and fatty acid oxidation.28 The enhanced muscle mitochondrial activity was also reflected by the fact that resveratrol-treated mice contained myofibers enriched in mitochondria and demonstrated enhanced aerobic capacity, as evidenced by their improved exercise endurance.28
Excessive nutrient uptake, on the other hand, causes the decrease of SIRT1 protein level in the gastrocnemius muscle, which is accompanied with insulin resistance.33 Forced expression of SIRT1 in C2C12 myotubes remarkably improves insulin sensitivity under insulin-resistant conditions. This effect of SIRT1 on insulin resistance is likely mediated by repressing the expression of protein tyrosine phosphatase (PTP) 1B,33 a negative regulator of the insulin signal transduction cascade.47 Consistently, insulin sensitivity of the mice that are transgenic for SIRT1 and hence express a higher amount of SIRT1 was improved.34
SIRT1 also plays a role in the myogenesis (Figure 1).38, 48 The SIRT1 protein is expressed in myoblast and its level declines gradually over the differentiation process.38 Overexpression of SIRT1 repressed muscle-specific gene transcription and retarded myogenesis. Conversely, downregulation of SIRT1 caused muscle cells to differentiate prematurely. A subset of genes repressed by SIRT1 are the target genes of the muscle transcription factor MyoD. Those genes are either involved in controlling myogenesis or exclusively expressed in skeletal muscle. MyoD turned out to be a substrate of SIRT1 and its deacetylation modification reduced its transcriptional activity, thus providing one mechanism how SIRT1 could function during myogenesis.38 More recently, SIRT1 was shown to mediate the inhibitory effect of glucose restriction, and perhaps more generally of calorie restriction, on skeletal myoblast differentiation.48
SIRT1 deacetylase activity is regulated by multiple modulators, which may also affect metabolic homeostasis
A number of modulator proteins have recently been identified, which regulate the enzymatic activity of SIRT1 (Figure 1). A protein called DBC1 (deleted in breast cancer 1) has been shown to interact with SIRT1 in cellular assays.49, 50 The leucine zipper motif of DBC1 is responsible to associate with the catalytic domain of SIRT1.49 This interaction inhibits SIRT1 to deacetylate its well-established substrate p53.49, 50 p53 is known to undergo acetylation in response to stress signals, which activates it to induce an apoptosis response.51, 52 Depletion of DBC1 by shRNA resulted in hypoacetylation of p53 and reduced apoptosis in response to stress stimuli whereas downregulation of both DBC1 and SIRT1 had no such effect, strongly supporting an inhibitory role of DBC1 on SIRT1 activity.49, 50
SIRT1 activity is also regulated by sumoylation. Lysine 734 in SIRT1 was identified as the sumoylation site.53 Mutation of this site impaired the deacetylase activity of SIRT1. Sumoylation of SIRT1 in vitro remarkably enhanced its ability to deacetylate p53. The stress-inducing agents such as UV radiation and hydrogen peroxide induce the desumoylase SENP1 to interact with SIRT1.53 This association caused the desumoylation and consequent inactivation of SIRT1. Correspondingly, the downregulation of SENP1 by a shRNA enhanced SIRT1 activity. The resulting cells hence contain higher levels of hypoacetylated p53 and display an enhanced resistance to stress-inducing apoptosis than control cells. Cells depleted of SIRT1 together with SENP1 showed no such phenotype, indicating that the critical involvement of SENP1 in apoptosis is through inactivating SIRT1.53
A nuclear protein, formerly identified as a ribosomal S19-binding protein with no known function,54 was shown to promote SIRT1 activity and thereby named as active regulator of SIRT1 (AROS).55 The expression profile of AROS was remarkably similar to that of SIRT1 in multiple cell lines. AROS specifically interacts with SIRT1 but not other sirtuins and its binding promoted SIRT1 to deacetylate p53. As a result, knockdown of AROS by shRNA increased p53-dependant apoptosis in response to DNA damage while AROS overexpression improved cell survival. Those effects were dependent on the presence of SIRT1.55
The identification of those SIRT1 endogenous activators or inhibitors indicates that diverse regulatory mechanisms likely exist to fine tune the activity of SIRT1, ensuring it is only active when in contact with its relevant substrate under specific conditions. In all those modulator studies, only the deacetylation level of p53 and the corresponding functional consequences (mainly on apoptosis) were explored. It would be reasonable to suspect that the known and yet-to-be-identified SIRT1 modulators also have an impact on the deacetylation level of other SIRT1 substrates, such as PGC-1α, FOXO1, etc, which could affect metabolism.
Emerging metabolic functions of other sirtuins
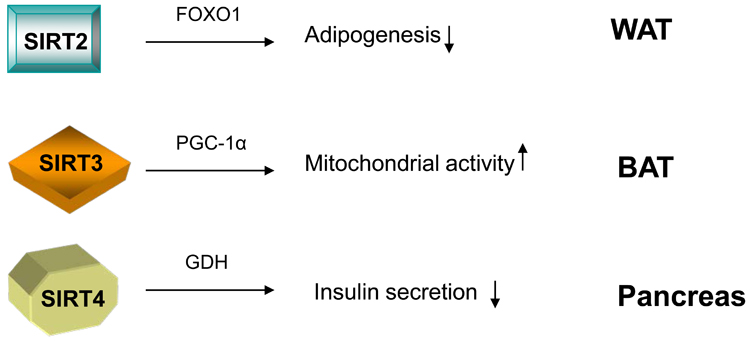
Among other members of sirtuin family, at least three proteins SIRT2, SIRT3, and SIRT4 have been linked with the control of metabolism (Figure 2). SIRT2 is a cytoplasmic sirtuin, but it binds to chromatin in the nucleus during the G2/M phase of the cell cycle.56 Recently, it was shown that SIRT2 inhibits the preadipocyte differentiation.57 SIRT2 protein is abundant in cultured preadipocytes and its expression levels decrease during differentiation. Overexpression of SIRT2 blocked adipogenesis whereas reduction of SIRT2 protein had the opposite effect. Interestingly, SIRT2 interacts with FOXO1. Knockdown of SIRT2 remarkably enhances the acetylation levels of FOXO1, which in turn facilitates its phosphorylation and export to the cytoplasm.58 This allows differentiation to progress, likely by reducing the ability of FOXO1 to associate with the PPARγ promoter and inhibit PPARγ transcription.59 The in vivo evidence of these observations awaits confirmation in gain and /or loss of function experiments in mice.
Figure 2.
The involvement of other sirtuins in the metabolic homeostasis. SIRT2 inhibits the differentiation of white preadipocytes. It achieves this effect likely through deacetylating FOXO1. Thus FOXO1 is retained on the target promoters of PPARγ and inhibits PPARγ-mediated transcription of genes that drive adipogenesis. In brown adipocytes, SIRT3 promotes the expression of mitochondrial-related genes in a PGC-1α-dependent manner and results in enhanced mitochondrial activity. SIRT4 ADP-ribosylates the mitochondrial enzyme GDH, which positively regulates ATP generation and insulin secretion in the pancreas. The resulting ADP-ribosylation inhibits the activity of GDH. Therefore, SIRT4 serves as a negative regulator for insulin secretion in the pancreas.
SIRT3 is generally thought to be located in the mitochondria.60-62 Its expression levels are decreased in muscle from diabetic animals63 and in BAT from obese mice.61 Calorie restriction induces SIRT3 expression in both WAT and BAT and cold exposure increases SIRT3 levels in BAT.61 Constitutive expression of SIRT3 in brown adipocytes promoted the expression of PGC-1α, UCP-1, and other mitochondrial genes in a PGC-1α-dependent manner. As a result, the mitochondrial electron transport activity was enhanced whereas the production of reactive oxygen species was reduced in the SIRT3-overexpressing cells. The mitochondrial enzyme acetyl-CoA synthetase 2 (AceCS2) was identified as the substrate of SIRT3 deacetylase activity.64, 65 The deacetylation of AceCS2 by SIRT3 activates AceCS2, enabling it to convert acetate to acetyl-CoA. It is worthwhile to mention in this context that SIRT1 deacetylates and activates the cytoplasmic acetyl-CoA synthetase, AceCS1.64 Considering that AceCS1 and AceCS2 are the only two known mammalian AceCSs to scavenge the excessive acetate under ketogenic conditions such as prolonged fasting or diabetes,66, 67 it is tempting to speculate that SIRT1 and SIRT3 may serve as important mediators for the acetate metabolism under stress. Somehow, questioning the functional relevance of SIRT3 in metabolic control is the recent report that SIRT3 knockout mice exhibit normal short-term cold resistance and show no metabolic defect on a chow diet, although at the molecular level many mitochondrial proteins are hyperacetylated.68 Before concluding on a metabolic function of SIRT3, it would, however, be interesting to see how those SIRT3 −/− mice react to metabolic stresses such as a high calorie diet, calorie restriction, or a long-term cold exposure.
SIRT4, as mentioned previously, functions as an efficient mitochondrial ADP-ribosyl transferase to ADP-ribosylate the metabolic enzyme glutamate dehydrogenase GDH, inhibiting its activity.11 GDH is known to convert glutamate to α-ketoglutarate, which promotes ATP generation and insulin secretion in pancreatic β cells.11, 69 Consistently, pancreatic mitochondrial lysates from SIRT4 deficient mice displayed higher GDH activity than those of wild type littermate controls. This translated in vivo in a significant increase in insulin secretion from SIRT4 deficient β cells in response to glucose and amino acids. More recently, SIRT4 has also been shown to interact with the insulin-degrading enzyme (IDE),70 which modulates insulin secretion as well,71 thus providing an alternative possible mechanism how SIRT4 could impact on insulin secretion.
Perspectives
The activity of SIRT1, which is regulated by nutrient availability, modulates many aspects of glucose and lipid homeostasis in almost all major metabolic tissues. This points to a complex regulatory role of SIRT1 in the whole body metabolism. Because of the pleiotropic effect of germline SIRT1 deficiency on the development of animals,72 conditional and tissue-specific deletion of the SIRT1 gene will definitely provide novel insights into the physiological actions of SIRT1. Meanwhile, identification of novel SIRT1 targets, modulators, and pathways will help to untangle the comprehensive regulatory SIRT1 circuits. Definition of the metabolic functions of other sirtuin members and their possible cross-regulation with SIRT1 are also needed as this will further enhance our knowledge of sirtuin biology. This progress will pave the way for a better understanding of metabolic diseases and possible development of novel therapies that target members of the sirtuin gene family.
References
- 1.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007;21:1745–55. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- 3.Dali-Youcef N, et al. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med. 2007;39:335–45. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 4.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–12. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Rodgers JT, et al. Metabolic adaptations through the PGC-1alpha and SIRT1 pathways. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JS, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–63. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landry J, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–11. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson RM, et al. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–5. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–63. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 10.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–8. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 11.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 12.Yang H, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liszt G, et al. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–20. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 14.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–29. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 15.Michishita EM, A R, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–6. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 17.Liou GG, et al. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–27. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 18.Hobbs HH, et al. The LDL receptor locus in familial hypercholesterolemia: mutational analysis of a membrane protein. Annu Rev Genet. 1990;24:133–70. doi: 10.1146/annurev.ge.24.120190.001025. [DOI] [PubMed] [Google Scholar]
- 19.Conkright MD, et al. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–23. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell. 2005;120:261–73. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 21.Herzig S, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–83. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 22.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 23.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–95. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 24.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–6. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, et al. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 26.Fontana L, et al. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci U S A. 2004;101:6659–63. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Koo SH, Montminy M. In vino veritas: a tale of two sirt1s? Cell. 2006;127:1091–3. doi: 10.1016/j.cell.2006.11.034. [DOI] [PubMed] [Google Scholar]
- 30.Picard F, Auwerx J. PPAR(gamma) and glucose homeostasis. Annu Rev Nutr. 2002;22:167–97. doi: 10.1146/annurev.nutr.22.010402.102808. [DOI] [PubMed] [Google Scholar]
- 31.Lehrke M, Lazar MA. The many faces of PPARgamma. Cell. 2005;123:993–9. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 32.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun C, et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–19. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Bordone L, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–67. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 35.Lean ME. Brown adipose tissue in humans. Proc Nutr Soc. 1989;48:243–56. doi: 10.1079/pns19890036. [DOI] [PubMed] [Google Scholar]
- 36.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–52. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 37.Timmons JA, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007;104:4401–6. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fulco M, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 39.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kajimura S, et al. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moynihan KA, et al. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–17. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Bordone L, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan CB, et al. Uncoupling protein 2 and islet function. Diabetes. 2004;53(Suppl 1):S136–42. doi: 10.2337/diabetes.53.2007.s136. [DOI] [PubMed] [Google Scholar]
- 44.Zhang CY, et al. Uncoupling protein-2 negatively regulates insulin secretion and is a major link between obesity, beta cell dysfunction, and type 2 diabetes. Cell. 2001;105:745–55. doi: 10.1016/s0092-8674(01)00378-6. [DOI] [PubMed] [Google Scholar]
- 45.Chan CB, et al. Overexpression of uncoupling protein 2 inhibits glucose-stimulated insulin secretion from rat islets. Diabetes. 1999;48:1482–6. doi: 10.2337/diabetes.48.7.1482. [DOI] [PubMed] [Google Scholar]
- 46.Gerhart-Hines Z, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. Embo J. 2007;26:1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elchebly M, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283:1544–8. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 48.Fulco M, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–73. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 50.Zhao W, et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abraham J, et al. Post-translational modification of p53 protein in response to ionizing radiation analyzed by mass spectrometry. J Mol Biol. 2000;295:853–64. doi: 10.1006/jmbi.1999.3415. [DOI] [PubMed] [Google Scholar]
- 52.Sakaguchi K, et al. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–41. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–62. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maeda N, et al. A novel nucleolar protein interacts with ribosomal protein S19. Biochem Biophys Res Commun. 2006;339:41–6. doi: 10.1016/j.bbrc.2005.10.184. [DOI] [PubMed] [Google Scholar]
- 55.Kim EJ, et al. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–90. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 56.Vaquero A, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–61. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105–14. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105–14. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Armoni M, et al. FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and -gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J Biol Chem. 2006;281:19881–91. doi: 10.1074/jbc.M600320200. [DOI] [PubMed] [Google Scholar]
- 60.Onyango P, et al. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99:13653–8. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi T, et al. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–7. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 62.Schwer B, et al. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–57. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yechoor VK, et al. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci U S A. 2004;101:16525–30. doi: 10.1073/pnas.0407574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwer B, et al. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–9. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujino T, et al. Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem. 2001;276:11420–6. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- 67.Yamashita H, Kaneyuki T, Tagawa K. Production of acetate in the liver and its utilization in peripheral tissues. Biochim Biophys Acta. 2001;1532:79–87. doi: 10.1016/s1388-1981(01)00117-2. [DOI] [PubMed] [Google Scholar]
- 68.Lombard DB, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–14. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelly A, Stanley CA. Disorders of glutamate metabolism. Ment Retard Dev Disabil Res Rev. 2001;7:287–95. doi: 10.1002/mrdd.1040. [DOI] [PubMed] [Google Scholar]
- 70.Ahuja N, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–92. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 71.Farris W, et al. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A. 2003;100:4162–7. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McBurney MW, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]