Abstract
Cave animals are excellent models to study the general principles of evolution as well as the mechanisms of adaptation to a novel environment: the perpetual darkness of caves. In this article, two of the major model systems used to study the evolution and development (evo–devo) of cave animals are described: the teleost fish Astyanax mexicanus and the isopod crustacean Asellus aquaticus. The ways in which these animals match the major attributes expected of an evo–devo cave animal model system are described. For both species, we enumerate the regressive and constructive troglomorphic traits that have evolved during their adaptation to cave life, the developmental and genetic basis of these traits, the possible evolutionary forces responsible for them, and potential new areas in which these model systems could be used for further exploration of the evolution of cave animals. Furthermore, we compare the two model cave animals to investigate the mechanisms of troglomorphic evolution. Finally, we propose a few other cave animal systems that would be suitable for development as additional models to obtain a more comprehensive understanding of the developmental and genetic mechanisms involved in troglomorphic evolution.
INTRODUCTION
Cave animals are attractive models for evo–devo research because they have evolved an unusual suite of troglomorphic (cave related) traits, including the reduction or loss of eyes and pigmentation, the lengthening of appendages, and the elaboration of nonvisual sensory systems.1 Troglomorphic traits have evolved by convergence in a diverse array of cave-adapted invertebrates and vertebrates. Cave adapted species exhibit several additional attributes making them exceptional models in evo–devo. First, cave animals were originally derived from surface dwelling ancestors, and therefore the direction of troglomorphic changes can be understood. Second, the cue for troglomorphic evolution is the absence of light and its consequences on primary productivity, allowing trait changes to be attributed to a specific environmental factor. Third, animals resembling the surface ancestors of cave animals are sometimes extant, providing a dual system of surface and cave forms in which the starting and ending points of evolution can be compared. In this article, we review two cave species that have been most successfully developed as model systems to understand troglomorphic evolution: the teleost fish Astyanax mexicanus and the isopod crustacean Asellus aquaticus.
REQUIREMENTS OF AN EVO–DEVO CAVE SYSTEM
Certain characteristics can be regarded as integral components for investigating the genetic basis of evolution and development in a cave model system.2,3 Both A. mexicanus and A. aquaticus fulfill the characteristics necessary to study both microevolutionary genetics and comparative developmental biology (see Figure 1). However, many other cave species that do not fulfill all of these characteristics have already provided interesting insights about the evolution and development of cave animals. Still other systems have the potential to fulfill all of these characteristics and some of these will be described as up-and-coming systems in the final section of this review.
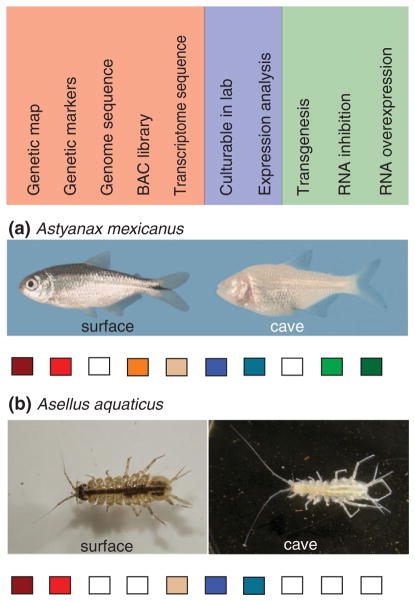
FIGURE 1.
Requirements of an evo–devo cave system. In red are genetic requirements, in blue developmental requirements, and in green requirements for functional analysis. (a) The cavefish Astyanax mexicanus. (b) The isopod crustacean Asellus aquaticus. Boxes filled with the appropriate color indicate that the above tool is present for the particular species. An empty box indicates that the tool has not yet been generated for the species. (Reprinted with permission from Ref 4. Copyright 2011 National Academy of Sciences)
Species with Cave and Surface Morphotypes
To address both genetic and developmental questions, a cave model must harbor both cave and surface forms with different phenotypes that can interbreed to produce fertile offspring. Therefore, all cave animals where the surface counterpart is extinct or unknown must be excluded. In addition to A. mexicanus and A. aquaticus, species with surface and cave forms capable of interbreeding include the amphipod Gammarus minus, the fishes Poecilia mexicana and Garra barreimiae, leeches of the genus Dina, the freshwater limpet Ancylus fluviatilis, the amphipod Synurella ambulans, and planthoppers of the genus Oliarus.5–12
Collecting Animals
Some caves in which A. mexicanus can be found are relatively easy to approach, but others require a rope descent.13 Furthermore, some of these caves have large amounts of bat guano, raising the risk of spore inhalation of the disease-causing fungus, Histoplasma capsulatum. A. aquaticus can be found in the Planina cave, in Slovenia, which has a very easy collecting environment, and in the Movile cave in Romania, which is much less hospitable with high concentrations of hydrogen sulfide and ammonia.14
Another challenge to studying certain species is that unfortunately many cave animals are endangered. Factors contributing to the risk of extinction of cave animals include high endemism of some cave species; human interference with the physical habitat; and water quantity and quality issues resulting from mining, urbanization, deforestation, flooding, storm water drainage, erosion, and dumping.1,15,16 Fortunately, both A. mexicanus and A. aquaticus are found over an extensive geographic range and are relatively abundant within the caves where they are found.13,17 In addition, both A. mexicanus and A. aquaticus are easy to collect; A. mexicanus swims to the surface of disturbed water and A. aquaticus can be collected with a dip net in shallow water.
Practical Considerations: Rearing Animals in the Laboratory, Generation Time, and Number of Offspring
The next challenge is to breed and rear the animals in the laboratory. It is necessary to know what to feed the animals, cues to breeding, and necessary physical characteristics of the environment. Some systems might be too impractical to pursue further. Researchers began breeding A. mexicanus in the laboratory in the 1940s18 and since then, several groups have devoted significant time and effort to raise individuals from different populations in the laboratory environment. A. aquaticus had a promising start as a laboratory organism from some crosses done in the 1940s.5,6 However until recently, raising both cave and surface populations of A. aquaticus (and crosses between them) had not been revisited.4
Cave animals are known to have extremely long generation times. For example, the average lifespan of the cave salamander, Proteus anguinus, is predicted to be 68.5 years.19 Fortunately, the time to sexual maturity of A. mexicanus is relatively short, around 4–6 months,20 and in A. aquaticus is population dependent but can be as fast as 2 months in one particular Italian population.21
In addition, both A. aquaticus and A. mexicanus produce large numbers of easily accessible embryos. A. mexicanus has external fertilization and the embryos can be cultured in a dish until they are free-swimming.22 A. aquaticus females deposit embryos into an external brood pouch and it is possible to remove the embryos without harming the female and culture them in vitro.23
Convergent and Parallel Evolution in Multiple Cave Populations
Multiple cave populations allow for the study of convergent and parallel evolution. At least 29 cave populations of A. mexicanus have been discovered.13 Likewise, there are many cave populations of A. aquaticus.17,24–26
Genomic Information
The genome has not yet been sequenced for either A. aquaticus or A. mexicanus, but many other genomic tools have been generated. For both species, there are linkage maps and hundreds of genetic markers.4,25,27–31 In addition, a BAC library is available for A. mexicanus and there is evidence of synteny with several other fish genomes.32,33 Transcriptome sequencing has been generated for both systems (Protas, Speiser, Patel and Oakley, unpublished; Furterer and Gross, unpublished). All of these molecular tools have aided and will aid in the identification of pathways, genes, and mutations responsible for different phenotypes.
Functional Assays
The final requirement for a cave animal evo–devo system is the ability to determine the relationship of genes to phenotypes using functional assays. Functional assays are well-developed in A. mexicanus and involve gene ‘knock up’ via introduction of synthetic mRNAs or conditionally expressed DNA expression constructs and gene ‘knock down’ using anti-sense oligonucleotides.34 In these functional assays, molecules are injected into the egg or early cleaving embryo and effects can be seen as long as 5–6 days post-fertilization. In some cases, functional analysis of A. mexicanus genes can also be carried out in zebrafish (Danio rerio) or in cultured mouse cell lines. Functional analyses have not yet been attempted in A. aquaticus, although they have been generated for another peracardian crustacean, Parhyale hawaiensis.35,36
ASTYANAX MEXICANUS
The teleost A. mexicanus consists of a surface dwelling form (surface fish) and 29 known cave (cavefish) dwelling populations13 (see Figure 1(a)). The cavefish are native to limestone caves in the Sierra de El Abra region of northeastern Mexico and named according to their caves of origin: for example, Pachón cavefish are from La Cueva de El Pachón, Molino cavefish from El Sotano del Molino, and so forth. The cave-fish populations have arisen independently at least three times when ancestral surface fish entered limestone cave systems and became isolated from their above ground counterparts.20,34,37–42 Estimates for the length of time since the initial cavefish and surface fish divergence vary from about 100,000 to several million years.13,43 After their separation from surface fish, the cave adapted fish dispersed through underground channels and subsequently became isolated in different caves. The repertoire of troglomorphic features that evolved during adaptation to the dark cave environment includes both constructive and regressive traits.44,45 Regressive traits were present in the surface ancestors of cavefish but subsequently were reduced or lost during adaptation to subterranean life. Constructive changes can entail either the enhancement of pre-existing traits or the evolution of novel traits.
Regressive Traits
The most prominent regressive traits in cavefish are the loss of eyes and pigmentation. During the last decade, significant progress has been made in understanding the developmental and genetic basis of eye and pigment regression in A. mexicanus.
Developmental Basis of Eye Regression
Normal development of the lens, retina, photoreceptors, neural connections, retinal pigment epithelium (RPE), and optic processing areas in the brain are required to form a correct visual image. The loss of vision in cavefish is due to developmental defects in these components. Eyes initially develop in the cavefish embryo,46 but optic defects appear later during larval development and the eye subsequently degenerates47–49 (see Figure 2(k)). Although appearing eyeless, adult cavefish actually have small degenerated eye rudiments buried within their orbits. Different cavefish populations show various degrees of eye reduction, and variations in the size of vestigial eyes can also occur within a single cavefish population.44 The developmental and genetic mechanisms of eye degeneration have been most extensively studied in Pachón cavefish; these experiments are described below.
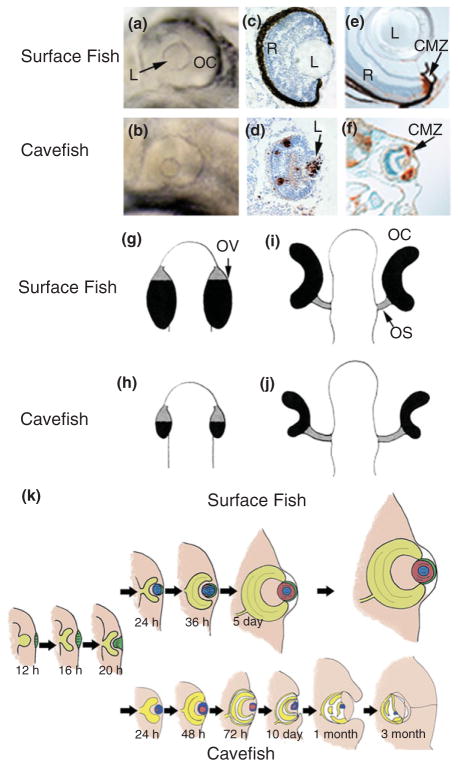
FIGURE 2.
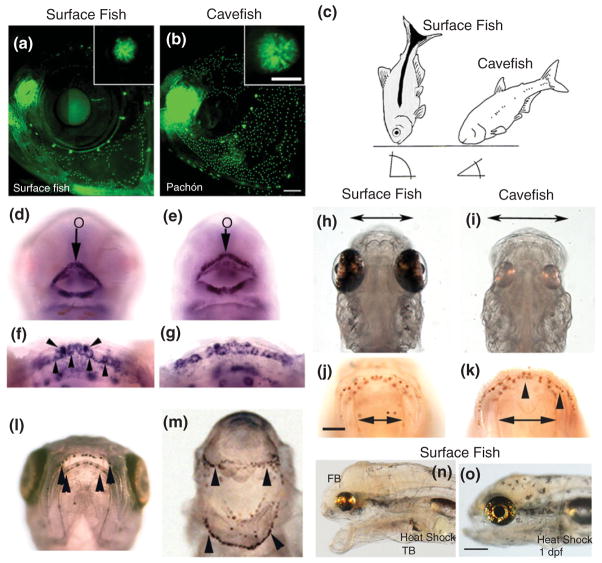
Eye development in Astyanax mexicanus surface fish and cavefish. (a and b) Surface fish (a) and cavefish (b) eye primordia viewed laterally at 1 day post-fertilization (dpf) showing the small lens (L) and reduced ventral optic cup (OC) in the latter. (Reprinted with permission from Ref 50. Copyright 2004 Nature Publishing Group). (c and d) Section through surface fish (c) and cavefish (d) eye primordia at 1.5 dpf showing apoptotic cells (brown color) detected by the terminal deoxynucleotidyl transferase (TUNEL) assay in the lens and retina in cavefish but not surface fish. (Reprinted with permission from Ref 48. Copyright 2007 Elsevier Limited). (e and f) Sections through surface fish (e) and cavefish (f) eyes at 10 dpf showing dividing cells in the ciliary marginal zone (CMZ) stained with anti-proliferating cell nuclear antigen (PCNA) (brown color). (Reprinted with permission from Ref 51. Copyright 2002 University of the Basque Country Press). (g–j) Diagram showing OC morphogenesis in surface fish (g and i) and cavefish (h and j). OV, optic vesicle; OS, optic stalk. Black and gray areas represent OC and OS domains, respectively. (Reprinted with permission from Ref 52. Copyright 2003 Oxford University Press). (k) Diagram comparing eye development and growth in surface fish (top) with eye degeneration in cavefish (bottom). (Reprinted with permission from Ref 34. Copyright 2009 Annual Reviews)
When the eye primordium first appears in cavefish, the lens and optic cup are smaller than those of their surface fish counterparts46 (see Figure 2(a), (b), (i), and (j)). As development proceeds, the lens fails to differentiate, gradually decreases in size, and in some cases may eventually disappear.48,49,53 In contrast, the embryonic retina initiates growth, neuronal, glial and photoreceptor cell layers are formed, and retinal ganglion cells project neural connections to the optic tectum via the optic stalk.46–49,53,54 The photoreceptor cells are transient and disappear later in development.55 Eventually, however, the retinal layers become disorganized and further growth ceases. The cavefish RPE is poorly differentiated and usually lacks black pigment44,48 (see Figure 2(c) and (d)). The cornea, iris, and ciliary body also fail to differentiate, and the sclera, the partially ossified tissue surrounding the eyeball, remains cartilaginous.55,56 During the larval period, the degenerating eye sinks into the orbit and is covered by epidermis and connective tissue (see Figure 2(k)).
Most of the lens and parts of the retina and RPE undergo apoptotic cell death during cavefish eye development48,54,55,57 (see Figure 2(c) and (d)). Cell death begins in the eye just after larval hatching and continues for at least 3 months.48 Apoptosis is detectable in the lens at least a day prior to its initiation in other eye tissues, suggesting that lens death could trigger optic degeneration. Lens transplantation was developed to test this possibility55: a surface fish lens was transplanted into a cavefish optic cup, and a cavefish lens was transplanted into a surface fish optic cup (see Figure 3(a)). The transplanted surface fish lens continued to grow in the cavefish host and mediated the restoration of a large eye (see Figure 3(b) and (c)) with a growing retina, permanent photoreceptors, neural connections to the optic tectum, and an ossified sclera. In contrast, the cavefish lens underwent apoptosis on schedule, and growth and development of the surface fish eye was decreased (see Figure 3(d) and (e)). Sophisticated experiments have not been performed to determine whether the rescued cavefish eye or the retarded surface fish eye are functional and can form visual images. However, the results given above indicate that the lens has an important role in cavefish eye degeneration.
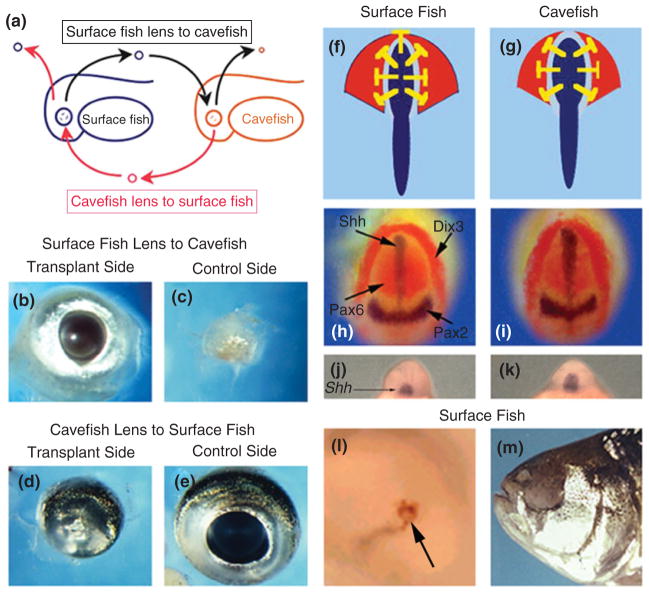
FIGURE 3.
Role of the lens and shh expression in Astyanax mexicanus eye degeneration. Left columns. Rescue of the cavefish eye by lens transplantation. (a) Diagram showing transplantation of a surface fish embryonic lens to a cavefish embryo (top) and a cavefish embryonic lens to a surface fish embryo (bottom). The host lenses were removed prior to transplantation. (b and c) Dissected eyes from an adult cavefish that received a surface fish lens transplant on one side as an embryo. (b) Transplant side. (c) Control side. (d and e) Dissected eyes from an adult surface fish that received a cavefish lens on one side as an embryo. (d) Transplant side. (e) Control side. Photos (b)–(e) courtesy of Yoshiyuki Yamamoto. Right columns. Role of Shh in cavefish eye degeneration. (f and g). Diagram showing expanded shhA gene expression along the embryonic midline (blue) and inhibitory effects (yellow blocked lines) on pax6 expression (red) in the optic domain of the neural plate in surface fish. (h and i) Four-gene in situ hybridization showing expanded shhA expression at the cavefish (h) midline (blue) and corresponding reduction of pax6 expression relative to surface fish (i) at the neural plate stage. No changes were seen in pax2a (blue) or dlx3 (red) gene expression. (j and k) In situ hybridization showing expanded shhA expression in the cavefish (j) compared to surface fish (k) rostrum. (l and m) Overexpression of shhA causes lens apoptosis (l, arrow) and eye degeneration (m) in surface fish. (Reprinted with permission from Ref 50. Copyright 2004 Nature Publishing Group)
How does lens apoptosis affect overall eye development in cavefish? The absence of the cornea, iris, and ciliary body is simple to explain because a functional lens induces these tissues during normal eye development. The role of the lens in controlling retinal growth is more complex. The teleost retina grows continuously throughout life by adding new cells produced in two stem cell niches: the inner nuclear layer, which replenishes the recycling photoreceptor cells, and the ciliary marginal zone, which contributes to the growing retinal layers and RPE.58,59 Thus, the defective lens could possibly curtail retinal differentiation and growth by affecting stem cell proliferation. However, this is not the case: cell proliferation continues for at least 3 months in both stem cell niches after retinal growth is arrested48,51,54 (see Figure 2(e) and (f)). Instead, retinal growth stops because the cells produced by mitosis die before they can differentiate and contribute to the retina.48 Thus, cavefish undergo repeated cycles of retinal cell division followed by death rather than differentiation, a process occurring throughout the period of larval development and into the adult. Transplantation of a surface fish lens into the cavefish optic cup protects the retinal stem cells from apoptosis, allowing retinal differentiation and growth to continue.48
Lens dysfunction and its secondary effects on the retina are not the only defects in the cavefish eye, as revealed by two further observations. First, as mentioned above, the cavefish optic vesicle (the precursor of the optic cup) is smaller than in surface fish, and later in development the ventral sector of the optic cup is reduced (see Figure 2(a) and (b)). It has been demonstrated that premature (heterochronic) fgf8 expression is involved in decreasing the ventral sector of the optic cup.60 Accordingly, treatment of cavefish embryos with a fibroblast growth factor (FGF) inhibitor can restore the optic cup to normal size.60 Second, when a surface fish lens is deleted during embryogenesis, there is no induction of retinal apoptosis, as would be expected if the lens were the sole protector of the retina.48 This result suggests that a part of the eye besides the lens is capable of protecting the retina from apoptosis and this eye part is also defective in cave-fish. A good candidate for this second protector is the RPE, which is known to influence retinal growth and differentiation during vertebrate eye development. The protective factor(s) secreted by the lens and possibly the RPE remains to be identified.
Genetic Basis of Eye Regression
How many genes are involved in cavefish eye regression, what is their identity, and how do they function? Crosses between cavefish and surface fish and their F1 progeny have addressed the question of gene number. The eyes of the F1 progeny are fairly uniform in size, although smaller than normal, but the F2 generation shows a wide range of eye sizes, indicating that eye reduction is a multigenic trait.8,44,61 Genetic linkage mapping has identified 13 quantitative trait loci (QTL) responsible for eye formation: seven affecting eye size and six affecting the lens28 (see Figure 4). In addition, some lens and eye size QTL overlap, suggesting the possibility of single genes with effects on both the lens and retina. Interestingly, some of the genes responsible for eye reduction are unique to different cavefish populations, as demonstrated by genetic complementation experiments:34,61–63 when cavefish from two different populations are interbred, eye size can be significantly larger in the F1 hybrids than their parents, and in some cases vision is restored.63 The loss or alteration of three or four genes in each cavefish population is thought to be enough to abolish vision.63 The genetic complementation results imply that eye regression can be accomplished differently, at least partially, in independently evolved cavefish lineages.
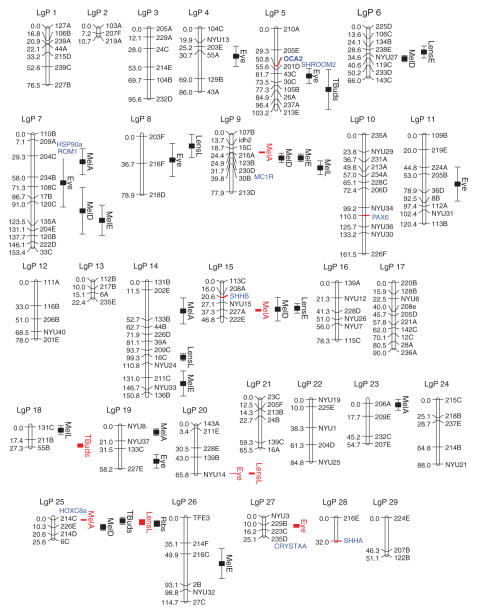
FIGURE 4.
Astyanax mexicanus genetic map showing locations of quantitative trait loci (QTL) and candidate genes. Shown is a linkage map generated from a Pachón F2 cross. 28 QTL are shown for eye size, lens size (Lens E, L), melanophore number (Mel A, D, E, L) in four different places on the body, and number of tastebuds (Tbuds). QTL with more precise locations are shown in red. The location of a rib QTL from Ref 28 is also shown. Candidate genes are shown in blue. Candidate genes with red tick marks were mapped in Ref 28. Candidate genes without red tick marks were mapped in a different analysis64 and are shown next to the most closely linked marker in common between the two analyses. (Reprinted with permission from Ref 28. Copyright 2007 Elsevier Limited)
The identity of genes involved in eye regression has been explored by candidate gene analysis and QTL characterization. In some cases, these studies have also provided clues about gene function. Since many genes are known with roles in vertebrate eye development, candidate gene analysis focused on previously identified eye genes whose expression is modified in cavefish relative to surface fish. Most of these genes showed no changes in expression, but a few were substantially decreased or increased.52 For example, the expression of αA-crystallin, which encodes an anti-apoptotic factor, is greatly reduced in the cavefish lens.65,66 In contrast, the expression of hsp90α, which encodes a pro-apoptotic factor, is activated in the cavefish lens just prior to apoptosis.67 Therefore, it is possible that lens apoptosis is controlled by an antagonistic interaction between αA-crystallin and hsp90α.
Other changes in gene expression are subtler and observed by in situ hybridization as an expansion or contraction of expression domains. For example, the expression domains of pax6 are diminished, whereas those of pax2a and vax1 are expanded in the cave-fish optic vesicle and cup50,68 (see Figure 2(f) and (g)). During vertebrate eye development, the optic vesicle is subdivided into preoptic cup and preoptic stalk domains via antagonistic interactions between the Pax6, Pax2, and Vax1 transcription factors: Pax6 directs optic vesicle development and suppresses optic stalk development, whereas Pax2 and Vax1 promote optic stalk and suppress optic vesicle development.69 Accordingly, expansion and contraction of the three transcription factor domains can account for a smaller optic cup in cavefish.
The anterior embryonic midline is also a location of expanded gene expression in cavefish.50,60,68,70 The midline expression domains of the two teleost sonic hedgehog (shh) genes, shhA (see Figure 3(h)–(k)) and shhB (once called twiggy winkle hedgehog or twhh) are expanded in cavefish relative to surface fish.50 During vertebrate development, Sonic Hedgehog (Shh) signals emanating from this region inhibit pax6 expression, producing an expression gap of the latter gene in the overlying neuroectoderm.71,72 As a result of Shh expansion, the gap in pax6 expression is larger in cavefish relative to surface fish (see Figure 3(f) and (g)). The role of enhanced Shh signaling in cavefish eye development was further investigated by increasing shh expression in surface fish embryos.50 Overexpression of shhA induced lens apoptosis and caused larvae to develop lacking external eyes (see Figure 3(l)). Thus, a blind cavefish phenocopy was produced in surface fish (see Figure 3(m)), demonstrating a key role for increased Shh signaling in eye degeneration.
No known eye QTL are located near the pax6, shhA, or shhB genes on the genetic map,28 suggesting that they are not mutated and may be an indirect rather than a direct cause of eye degeneration. In contrast to candidate analysis, QTL characterization has the potential to identify those genes and their mutations that directly cause eye degeneration. In the current absence of a sequenced A. mexicanus genome (see Figure 1), however, this identification is extremely difficult. Nevertheless, progress has been made by anchoring the existing genetic map to syntenic regions in the sequenced zebrafish genome.33 This approach revealed four promising genes located near eye QTL: αA-crystallin and hsp90α, which were also predicted to be involved in eye degeneration by candidate gene analysis,65–67 shroom2, which has a role in the synthesis and localization of melanin granules in the RPE,73 and rom1, which encodes a structural protein required for differentiation of photoreceptor cells.74 Further work will be necessary to investigate whether mutations in these genes actually cause eye degeneration. In addition, future candidate genes to be considered include genes that upregulate the Shh midline signaling system.
Developmental Basis of Pigmentation Regression
Pigmentation is one of the most commonly studied morphological traits in evo–devo studies because it is easily observed and much is known about the genetic pathways responsible for pigmentation in vertebrate and invertebrate model systems.75–77 Pigmentation in fish involves multiple types of pigment cells;78 most of the focus on pigmentation in A. mexicanus involves melanophores, which are the pigment cells that produce black melanin.
The developmental basis of albinism was investigated in Pachón cavefish. To address whether melanophores were absent in albino animals or whether they were present but did not produce melanin, the experimenters added L-DOPA, a substrate of tyrosinase in the melanin pathway, which resulted in melanin production in melanophores79 (see Figure 5(a)). However, addition of a substrate that is active immediately upstream in the same biochemical pathway, L-tyrosine, did not result in melanin production. Therefore, the investigators identified the location of the defect in the melanin pathway that causes the albino phenotype in Pachón cavefish.
FIGURE 5.
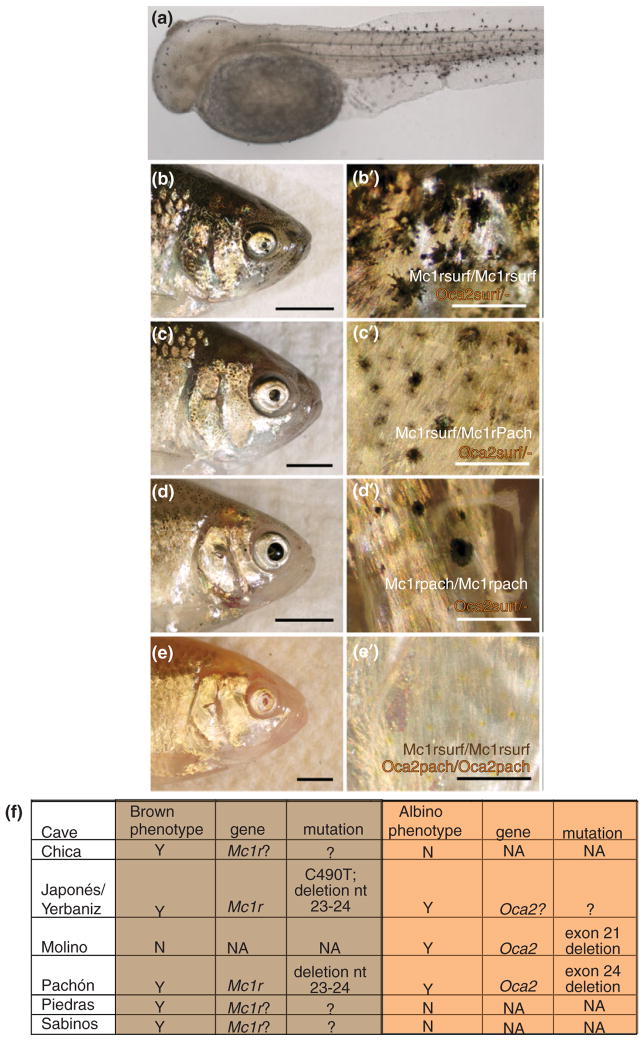
Pigmentation in Astyanax mexicanus. (a) Tyrosinase positive cells in a Pachón cavefish embryo at 72 hpf. (b–e′) F2 animals from a cross between a surface and a Pachón cave individual. (b–e) Side views of each fish head. (b′–e′) Higher magnification of (b–e). Genotypes of each individual are written in (b′–e′) for both mc1r and oca2. (f) Table describing certain cave populations of A. mexicanus: whether the albino and/or brown phenotypes are present, what genes are responsible, and what mutations are responsible. nt, nucleotide. ‘Mc1r?’ or ‘Oca2?’ shows that complementation tests indicate that the particular gene is involved but no coding mutation has been observed. (b–e′) from Gross et al.64 (f) Summarizing results in Gross et al.64 Protas et al.30 and Wilkens and Strecker.80
Genetic Basis of Pigment Regression
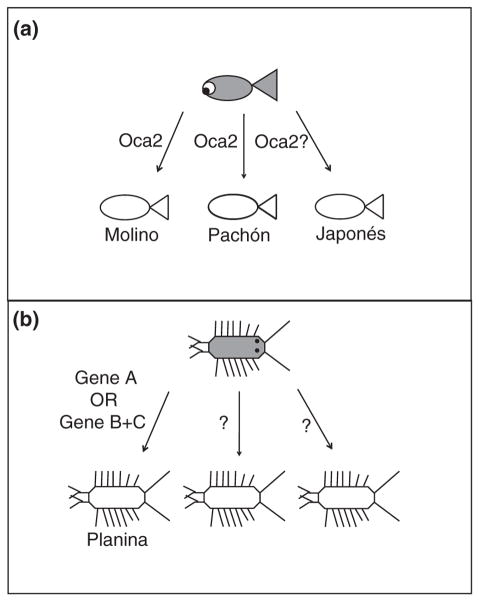
Early experiments documented two recessive monogenic traits, albinism and the ‘brown’ mutation, which generated brown instead of black pigment.81,82 A genetic approach was used to map the location of the monogenic trait responsible for albinism.30 An albinism candidate gene, ocular and cutaneous albinism 2 (oca2), was mapped to the albino locus in crosses from two different cavefish, Pachón and Molino. Two deletions were observed in the coding region of oca2, one in Molino cavefish and the other in Pachón cavefish (see Figure 5(f)). A mouse melanocyte cell line deficient in oca2 was used to demonstrate that these deletions resulted in nonfunctional OCA2 proteins. A third albino cavefish, Japonés, which did not contain either deletion, did not complement when crossed to either Pachón or Molino cavefish. Therefore, albinism has evolved independently in three different cavefish through the same gene, oca2, via two different coding mutations and possibly a regulatory mutation.
The genetic basis of the ‘brown’ mutation82 was investigated using similar genetic methods.64 The melanocortin receptor 1 (mc1r) gene mapped to the brown locus. Two coding mutations were found.64 One was a 2 base pair (bp) deletion resulting in a frameshift, found in Pachón cavefish and at a low frequency in the connected Japonés and Yerbaniz Caves. The other was a point mutation leading to an amino acid change, also found in humans with red hair and pale skin, present only in Japonés and Yerbaniz cavefish.64 When an F2 cross of fish originating from Pachón cavefish was observed, those animals with two copies of the mc1r surface allele had larger and darker melanophores, those fish with one copy of the Pachón mc1r allele had fewer and lighter melanophores, and those fish with two copies of the Pachon mc1r allele had still fewer and lighter melanophores (see Figure 5(b)–(d)). Individuals who had two copies of the Pachón oca2 allele were albino regardless of the genotype at mc1r64 (see Figure 5(e)). Functional analysis in Danio rerio showed that both the amino acid change and the 2bp deletion altered the function of mc1r.64 In addition, other cavefish expressed the ‘brown’ phenotype based on complementation tests,80 but coding changes were not observed in the mc1r open reading frame in representatives from these caves. This may indicate that cis-regulatory changes in cavefish from these populations lead to reduced expression of the MC1R receptor (see Figure 5(f)).
Several cave populations only express the brown mutation, while Molino cavefish harbor albinism in the absence of brown (see Figure 5(f)). Interestingly, Pachón, Yerbaniz, and Japonés cavefish all express both brown and albinism. Furthermore, many other QTL were mapped for the trait of melanophore number28 showing that multiple genes are responsible for pigmentation differences in cave versus surface A. mexicanus.
Other Regressive Traits
Other regressive traits in cavefish are listed in Table 1. Some of these may be a direct consequence of the absence of functional eyes and vision. For example, reduction of the optic tectum is probably due to decreased neuronal targeting from the retina.53 The loss of aggressive and schooling behaviors has also been attributed to the absence of eyes,44 although there is no compelling evidence that this is the case. In fact, Molino cavefish have lost eyesight but continue to show aggressive behavior.83 Other behaviors that are modified or missing in cavefish are the dorsal light reaction,84 which orients the dorsal side of a fish at a constant 90° angle to a light source, and the alarm or fear behavior,85 which is mediated by a substance released from the skin in the presence of a predator or a sustained physical disturbance. The alarm substance triggers rapid swimming, taking cover, or a sudden freeze in motion, presumably to avoid or confuse predators. Interestingly, despite their inability to respond to the alarm substance, cavefish appear to have conserved the ability to produce it.85 Genetic analysis shows that a minimum of 3–6 genes are responsible for changing the dorsal light reaction,84 and two dominant genes may cause loss of the alarm response.85
TABLE 1.
Major Constructive and Regressive Troglomorphic Traits in Astyanax mexicanus Cavefish
| Constructive Traits | Regressive Traits |
|---|---|
| Jaws | Eyes (lens, cornea, iris, ciliary body, retinal photoreceptors, RPE, scleral bones) |
| Forebrain | Melanophores |
| Olfactory bulbs and interneurons | Melanin |
| Hypothalamus | Rib-bearing vertebrae |
| Feeding posture behavior | Sleep |
| Vibration attraction behavior | Aggressive behavior |
| Neuromasts | Dorsal light reaction |
| Fat deposition | Feeding angle behavior |
| Maxillary teeth | Scales |
| Nares | Fin rays |
| Orbit bones | — |
Sleep is another behavioral trait that has regressed in cavefish.86 Cavefish show dramatic reductions in the duration of sleep, as measured by daily activity patterns. Sleep regression has converged in three independently evolved cavefish populations and is controlled by a small number of genes with dominant effects.86
Some cavefish populations show a reduction in anal fin rays,29 smaller body scales,44 and have one or two fewer rib-bearing thoracic vertebrate than surface fish.38 The loss of rib-bearing vertebrae reduces body length in Pachón cavefish (see Figure 1(a)) and is a polygenic trait.29 QTL analysis (see Figure 4) and anchoring to the zebrafish genome show that one of the rib QTL is tightly linked to the hoxc8a gene,33 which patterns rib numbers in other vertebrates.87
Constructive Traits
Most constructive traits fall into one of two categories: those associated with (1) food detection, feeding, or energy storage and (2) changes in brain organization (Table 1). The developmental and genetic basis of constructive changes is just beginning to be studied, and consequently much less is currently known about them compared to regressive traits. Constructive traits are extremely important, however, because in contrast to regressive traits they were probably directly involved in the adaptation of A. mexicanus to life in caves.
Food Detection, Feeding Activities, and Energy Storage
Compensation for the loss of visual feeding is a major challenge for survival in a dark cave. One of the ways in which ancestral surface fish may have adapted to caves is by the evolution of vibration attraction behavior (VAB), which is defined as the movement of cavefish toward oscillating objects in cave pools.88 VAB is advantageous for detecting moving prey in darkness, heritable, and dependent on an enhanced neuromast system. A. mexicanus contains two types of lateral line sensory organs: canal neuromasts located in the lateral line proper, and superficial neuromasts (SN) distributed outside the lateral line. The size of both types of neuromasts is increased in cavefish, but SN specifically show a several fold increase in number compared to surface fish, which is particularly evident in the cranial region88,89 (see Figure 6(a) and (b)). When cranial or trunk SN are ablated, VAB also disappears indicating that SN are the VAB receptors.88 While most surface fish lack VAB, a small proportion of laboratory raised animals exhibit low levels of VAB,88 suggesting that there may be standing genetic variation for this trait in surface fish populations. VAB is not expected to be adaptive in lighted environments because of the risk of predation. In caves, however, where there is a much lower risk of predation, the benefits of selection for increased food detection via VAB may be quite strong. Therefore, continuous selection for SN-mediated VAB enhancement in cavefish may be a major adaptation to life in darkness.
FIGURE 6.
Constructive traits in Astyanax mexicanus cavefish. (a and b) Superficial neuromast number and diameter (insets) is increased in cavefish relative to surface fish (a). (Reprinted with permission from Ref 88. Copyright 2010 Elsevier Limited). (c) Differences in feeding posture behavior in surface fish and cavefish. (Reprinted with permission from Ref 90. Copyright 2005 Oxford University Press). (d–g) Cavefish embryos (e) have a larger mouth (o) than surface fish (d). The mouth is encircled by shhA expression (d and e). Later in development (f and g), shhA expression is attenuated to tooth germs (downward arrowheads) and developing taste buds (upward arrowheads). (h–k) Cavefish larvae (i) show increased jaw span (double headed arrows) and anti-calretinin-stained taste buds (arrowheads) on their lips (k) relative to surface fish (h and j). (l and m) Overexpression of shhA by mRNA injection into cavefish embryos causes the formation of a large gaping mouth with excessive taste bud development. (l) Surface fish control and (m) shhA-injected cavefish. (n and o) Conditional heat shock shhA overexpression at the tailbud stage (n) but not at 1 dpf (o) induces eye degeneration, forebrain enlargement, and a larger mouth with more taste buds (not shown) in surface fish larvae. (Reprinted with permission from Ref 91. Copyright 2009 Elsevier Limited)
A change in feeding posture has evolved in cavefish to facilitate foraging on the substratum of cave pools92 (see Figure 6(c)). Surface fish usually feed in the water column under lighted conditions. When forced to feed from the substratum in darkness, however, they position themselves at a 90° angle with respect to the bottom. Then, by swiveling their body, they pick up pieces of food by a rather awkward and inefficient process. In contrast, under the same conditions, cavefish position their bodies at a 45° angle with respect to the substratum and move steadily forward to more efficiently collect food particles. The feeding posture of cavefish is likely to be adaptive for feeding in the cave environment. When surface fish and cavefish are placed in the same lighted tank and required to compete for limited food, surface fish obtain most of it and thrive, but in darkness cavefish have better food finding ability and flourish.93 Genetic analysis suggests that the regulation of feeding posture behavior is controlled by only a few genes.95
Possibly to accompany their novel feeding posture, cavefish have evolved wide shovel-like jaws with several additional maxillary teeth, more taste buds, and higher sensitivity to dissolved amino acids compared to their surface fish counterparts29,56,91 (see Figure 6(h), (i), (j) and (k)). In surface fish, taste buds are located along the lips and in the pharynx, whereas in cavefish they are increased in these locations but also found in large numbers in the ventral skin of the head.91,94,95 Taste bud formation begins relatively early in A. mexicanus embryos, just after the opening of the larval mouth, and their number increases continuously during subsequent development.91,94 In addition to its role in eye degeneration, the Shh signaling system mediates enhancement of mouth/jaw size and taste bud number.91 The midline expansion of shhA expression continues during later development, encompassing the developing mouth and pharynx (see Figure 6(d) and (e)), and is eventually attenuated to the taste buds (see Figure 6(f) and (g)). The expression of shhA has been shown to be necessary and sufficient (see Figure 6(l) and (m)) for increasing mouth/jaw size and taste bud number.91 Although 11–12 genes were orginally predicted to be involved in determining taste bud numbers,44 this has been a difficult trait to quantify and consequently only three taste bud QTL have been detected so far28 (see Figure 4). Similar to eye QTL, however, none of the taste bud QTL are closely linked to the shh genes.28 Perhaps, upstream regulation of Shh signaling by modulating genes could also be important in oral and taste bud enhancement.
In addition to changes in the abundance and types of food, the availability of food also fluctuates in caves with major inputs probably corresponding to sporadic flooding. Thus, cavefish are faced with the challenge of survival during periods of low food input. To accomplish this feat, they have evolved capacities for increased lipid metabolism and fat deposition.96 The developmental and genetic basis of increased fat formation is unknown.
Brain Organization
The cavefish brain is organized much differently from that of surface fish.97 In addition to a reduced optic tectum,53 it has larger olfactory lobes, telencephalon, and hypothalamus than the surface fish brain.70,97,98 These constructive changes may be in part caused by amplifications in the senses of taste (gustation) and olfaction. The gustatory relay centers, which are located in the three enlarged areas of the cavefish brain, may have increased to accommodate excess inputs from more taste buds. Although the possibility of changes in the olfactory epithelium has not been investigated in cavefish, their nasal volume (nares) is much larger than surface fish,56 suggesting more internal space to accommodate receptors. Moreover, a specific population of GABAergic interneurons that are part of the olfactory bulb neural circuitry is enhanced in cavefish.70 Thus, although further investigation is necessary, it seems likely that the olfactory epithelium has been enhanced in cavefish.
Differences in cavefish brain organization first appear during early development and are controlled by the Shh and FGF pathways.60,70,98 In addition to its expansion along the anterior embryonic mid-line, Shh is also increased in the floor plate of the developing anterior neural tube.70 There are two lines of evidence suggesting that Shh signals have important consequences on cavefish brain organization.70 First, the large hypothalamus is probably due to Shh-induced increase in cell proliferation. Second, Shh is responsible for an increase in the development of GABAergic olfactory interneurons via the downstream activation of NKX and LHX transcription factors. These GABAergic interneurons are first produced in the ventral telencephalon and later migrate into the olfactory bulbs
Pleiotropic Tradeoffs between Constructive and Regressive Traits
On the basis of developmental studies, it was predicted that tradeoffs based on pleiotropy could occur between constructive and regressive changes in cavefish.99 Consistent with this idea, genetic linkage analysis has revealed numerous overlaps both among different regressive trait QTL and between constructive and regressive trait QTL that are hard to reconcile simply by chance.29 For example, some eye QTL are linked to taste bud QTL (see Figure 4). The possibility of an eye/oral/taste bud antagonism was substantiated by developmental methods, and once again expansion of the Shh signaling system was found to be a key factor. Evidence for coupling between oral size/taste bud enhancement and eye degeneration via Shh signaling was obtained by experiments in which shhA was conditionally expressed at specific times during surface fish development.91 Overexpression at early developmental stages resulted in subsequent eye degeneration and mouth and taste bud enhancement in the same surface fish larvae, but had no effects when carried out later in development (see Figure 6(n) and (o)). It was concluded that the critical developmental period for eye degeneration and oral enlargement/taste bud expansion is the same and likely to be controlled by an antagonistic tradeoff based on pleiotropic Shh signaling.
Convergent/Parallel Evolution and Evolutionary Forces
Depending on the identity of the founder surface fish, troglomorphic traits could have evolved by convergent or parallel evolution. Whichever the case, an understanding of the evolutionary forces responsible for troglomorphism is a major goal in the A. mexicanus model system.
The forces responsible for the evolution of constructive traits seem obvious: natural selection is acting on standing genetic variation to produce adaptive phenotypes with the most fitness for cave life. A good example is the evolution of VAB.88 The evolutionary forces responsible for regressive traits are less clear and have been the subject of considerable debate.1,100 Two major hypotheses have been proposed: (1) the accumulation of neutral (loss of function) mutations and their fixation via genetic drift and (2) natural selection acting either directly based on the benefits of energy conservation or indirectly via antagonistic pleiotropy. In the case of eye regression, some of the available evidence supports the first hypothesis, whereas other evidence seems to favor the second hypothesis. Thus, it has been proposed that both neutral mutation and indirect selection may be acting in concert.90 Evidence for the neutral hypothesis is primarily based on the observation that vestigial eye sizes vary within cavefish populations, which seems hard to account for by selection.62 Evidence for selection comes from QTL studies in which the direction of eye size changes is always negative,28 whereas random genetic drift would predict both positive and negative QTL. Other information also supports selection over neutral mutation.101 First, most of the genes involved in eye development do not appear to have mutated to a degree in which they have lost function. Second, the restoration of eyes by lens transplantation suggests that many of the genes acting downstream of lens function are present and potentially active in cavefish. Third, most genes with modified expression patterns, such as those in the Shh signaling pathway and hsp90α, increase rather than decrease activity in cavefish. If natural selection is actually involved, what are the benefits of losing eyes? Direct evidence for a role of energy conservation is lacking, and perhaps this explanation is questionable considering the costly cycles of cell division and death that occur in the cavefish retina.48 Another explanation may be pleiotropy involving the expanded Shh midline signaling system.90,91
In contrast to eye regression, individual QTL governing the number of pigment cells can either increase or decrease,28 supporting the role of neutral mutation and genetic drift in this regressive trait. In contrast, convergent or parallel evolutionary changes in the oca2 gene suggest the possibility that the block in melanin synthesis (albinism) could be adaptive, although its beneficial aspects remain to be discovered. Further progress in understanding the evolutionary forces involved in regressive evolution awaits the identification of the specific genes and mutations involved in this process.
ASELLUS AQUATICUS
A. mexicanus is an excellent species for the study of evolutionary and developmental biology of troglomorphic evolution. However, to understand if there are common themes in the evolution of cave related characteristics, it is critical that multiple species be explored. Ideally one would investigate several cave species, both similar (perhaps other fish) and dissimilar (invertebrates). Because of the significant time and resources needed for genetic and developmental studies, one invertebrate system was first chosen.
Arthropods are very common cave dwellers (see Figure 7) and, in particular, crustaceans have been successful in adapting to the cave environment with greater than 2900 known obligate species.102 The isopod crustacean, A. aquaticus, has recently been developed as an emerging invertebrate cave model.
FIGURE 7.
Cave arthropods. (a) The crustacean, Monolistra monstruosa, photo courtesy of Helena Bilandžija. (b) The spider, Travunia jandai and the isopod crustacean, Aegonethes cervinus, photo courtesy of Helena Bilandžija. (c) The beetle, Ptomaphagus hirtus, photo courtesy of Markus Friedrich. (d) The crustacean, Gammarus minus, photo courtesy of Dan Fong. (e) The beetle, Leptodirus hochenwartii, photo courtesy of Helena Bilandžija. (f) The collembolan, Verhoeffiella longicornis, photo courtesy of Marko Lukić. (g) The spider, Sulcia nocturna, photo courtesy of Martina Pavlek. (h) The millipede, Brachidesmus sp., photo courtesy of Helena Bilandžija.
A. aquaticus is commonly found throughout much of Europe, in freshwater habitats including ponds, rivers, lakes, caves, and wells. This species is a good model for studying evolutionary and developmental biology because cave and surface populations are inter-fertile,4,5 the surface form is easy to breed in the laboratory and is widely used for eco-toxicological studies, the life cycle is relatively short, and there are many morphological differences between cave and surface populations.103,104 Furthermore, the phylogenetic history of A. aquaticus has been well studied, despite its remarkable complexity, describing multiple different invasions of the cave environment both within a relatively small geographic location in Slovenia as well as in Italy and Romania17,24–26 (Figure 8(a)).
FIGURE 8.
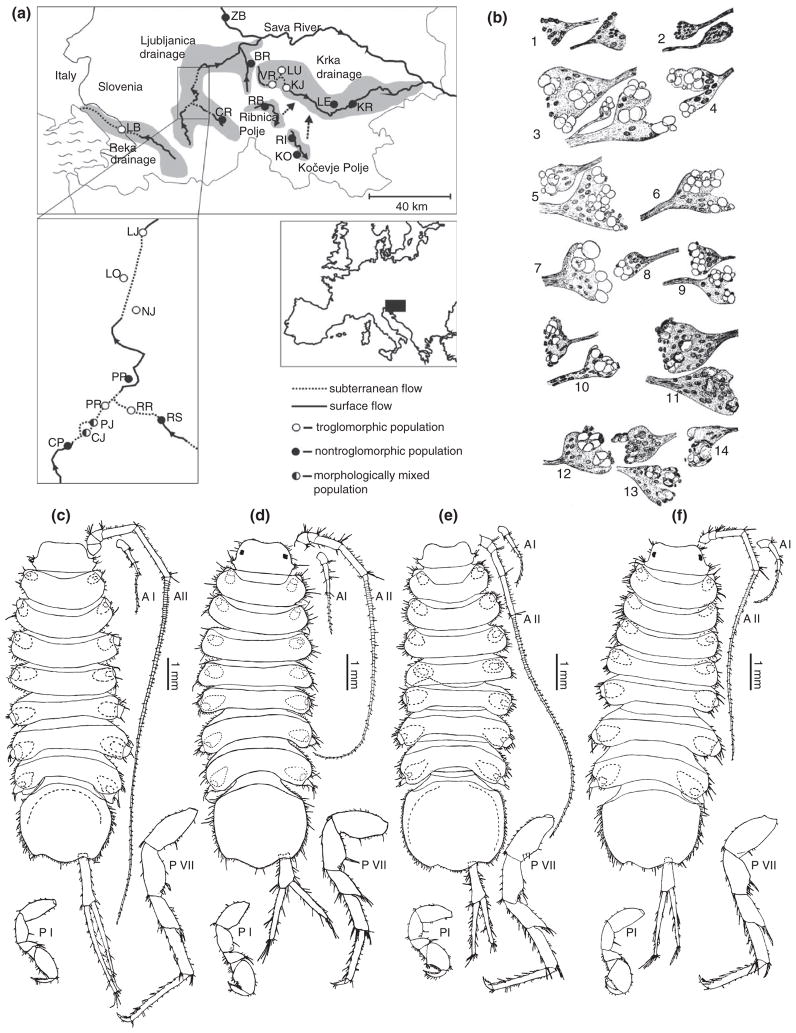
Different cave and surface populations of Asellus aquaticus. (a) Map of cave and surface populations of A. aquaticus. (Reprinted with permission from Ref 17. Copyright 2010 John Wiley and Sons). (b) Drawings of eye rudiments from individuals from the Planina cave population. Pairs of eye rudiments depict right and left sides from the same individual. (Reprinted with permission from Ref 105. Copyright 1965 Guy Demortier). (c–f) Drawings of individuals from (c) Planina cave in Slovenia, (d) Planina polje (surface water) in Slovenia, (e) Mangalia well in Romania, and (f) Bucharest, Romania surface waters. Note differences in length of appendages and numbers of setae. P, pereopod (thoracic leg) and A, antennae. (Reprinted with permission from Ref 104. Copyright 1996 Springer)
Regressive Traits
Eye Regression
The surface form of A. aquaticus has four ommatidia, each having between 1 and 4 crystalline cone cells.106 Different cave populations of A. aquaticus indicate a high diversity of phenotypes within caves, between caves, and even within individuals6,107 (see Figure 8(b)). Some animals have fragmented ommatidia with disassociated crystalline cone cells of different sizes, shapes, and different numbers of fragments. Other individuals have no visible ommatidial remnants.
Genetic Basis of Eye Regression
Genetic mapping analyses were performed in this species to identify regions of the genome responsible for the morphological differences. Genetic markers were isolated, including anonymous genomic markers and those associated with candidate genes in eye, pigment, and appendage development.4 Backcrosses were generated using individuals from the Planina cave and individuals from the surrounding surface waters. Based on a molecular clock approach, these populations are thought to have been separated for less than 100,000 years.17 A linkage map was generated with eight linkage groups, which is the karyotypic number in A. aquaticus.108
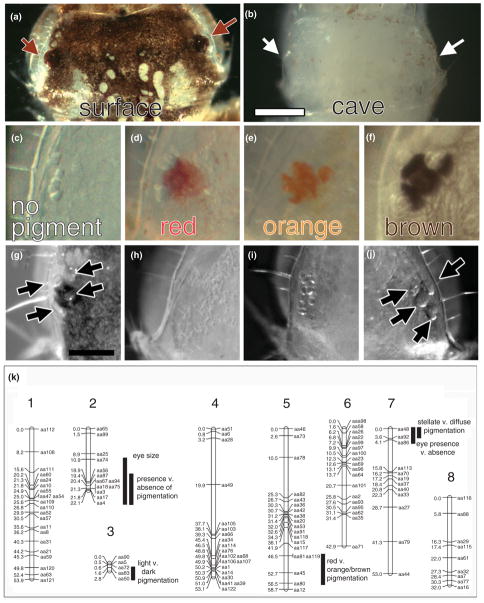
The Planina cave population has great variability in the eye phenotype (see Figure 8(b)). Eye size was measured as a quantitative trait and a QTL was observed accounting for 30% of the variance (see Figure 9(k)). As the analysis was fairly small, it is expected that with a larger cross size, more regions of the genome would be discovered accounting for the rest of the variation. A qualitative trait was also measured, presence of ommatidia or ommatidial remnants versus absence of ommatidial remnants. A single region of the genome, potentially even a single gene, is responsible for the observed variation (see Figure 9(k)). Therefore, as was predicted from earlier genetic crosses,6 multiple genes are responsible for the variation in eye phenotypes in the Planina cave population.
FIGURE 9.
Quantitative trait loci (QTL) map, phenotypes, and mapped loci in a backcross of cave and surface Asellus aquaticus. (a) Surface head, arrows pointing to the eyes. Scale bar = 0.25 mm. (b) Cave head, arrows pointing to the eyes. (c–f) Four different eye colors present in the backcross offspring. (g) Surface eye. Scale bar = 0.125 mm. (h–j) Representative eye phenotypes in the backcross offspring. (k) Linkage map with eight linkage groups. Distance in centimorgans is on the left side of the linkage group and marker name is on the right side of the linkage group. Mapped locations of various QTL and loci for eye and pigment traits. The length of the black bar represents the 1.5 logarithm (base 10) of odds (LOD) interval of the QTL or locus. (Reprinted with permission from Ref 4. Copyright 2011 National Academy of Sciences)
Pigment Regression
Researchers first described the body pigment in A. aquaticus as a melanin5 but more recent evidence indicates that the pigments could be ommochromes (one of the pigments present in Drosophila melanogaster eyes).109
Early work on pigmentation variation in surface and cave populations of A. aquaticus described six levels of body pigmentation where the faintest levels were only present in cave populations.6 Furthermore, in certain cave populations, different colors of eyes were observed including unpigmented, red, light purple, purple, and black. Genetic crosses were performed with individuals from caves that had great variation in pigmentation phenotypes and the experimenters concluded that multiple genes were responsible for the variation. However, not all cave populations demonstrated variability in pigmentation; the Planina cave was documented to contain only unpigmented individuals.105
Genetic Basis of Pigment Regression
The same methods and crosses used to analyze the eye phenotype were also utilized for pigmentation.4 Backcross offspring fell into five different color categories: no pigment, light red eye pigment and no head pigment, red eye and head pigment, orange eye and head pigment, and brown eye, head and body pigment (see Figure 9(c)–(f)). The genetic bases of several qualitative traits were examined: presence versus absence of pigment, light red and red versus orange and brown pigment, light red and orange versus red and brown pigment, and stellate versus diffuse pigment. For each of these qualitative traits, a different single region of the genome was discovered to be responsible for the majority of the variation (see Figure 9(k)). Further work investigating genotypes of these different regions of the genome has generated a model where it appears that there are two ways to generate the unpigmented phenotype—either by a single region of the genome or the combined effect of two other regions of the genome. Although, the Planina population is described as having a uniform loss of pigment, it is likely that multiple genes are involved in pigmentation. The fact that there are multiple ways to achieve albinism in the same population might support the neutral mutation theory. However, it is also possible that pleiotropy is responsible for some or all of the pigmentation mutations. Further experiments will examine additional constructive traits and whether the genetic basis of these traits is linked to the pigmentation loci.
Constructive Traits: Appendage and Sensory Phenotypes
Other traits compared between surface and cave individuals of different populations include the common constructive traits of appendage length and enhancement of sensory systems.24,103,104 In these studies, multiple cave populations from Slovenia were examined in addition to the Romanian cave population (see Figure 8(a)). Comparisons between the Planina cave population and the neighboring surface population showed that the relative length of antennae II, the relative length of the seventh thoracic leg, and the number of aesthetascs (thought to be chemosensory cells) were increased in the cave population104 (see Figure 8(c)–(f)).
Future Directions
Genetic Analyses
It will be very interesting to investigate if QTL for the constructive traits are located near QTL for the eye and pigmentation traits. By investigating these additional traits it will be possible to address the question of pleiotropy in the evolution of cave characteristics in A. aquaticus.
Furthermore, many behavioral traits have been pursued in A. mexicanus and likewise it would be fascinating to expand the analysis to the genetic basis of behavioral evolution in A. aquaticus. For example, photobic/photophilic behavior has already been examined in differentially pigmented individuals of A. aquaticus.110
Developmental Analyses
The embryonic development of A. aquaticus has also been investigated111 (see Figure 10). The method of in situ hybridization has been performed in this species111,112 and, therefore, it should be possible to compare expression patterns of promising candidate genes between cave and surface embryos.
FIGURE 10.
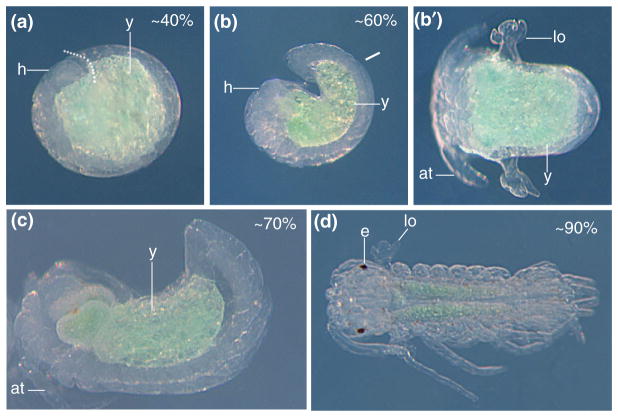
Embryonic development in the surface form of Asellus aquaticus. (a) Lateral view of an embryo at 40% of development. h, head; y, yolk. The dotted line shows the separation between the head and the posterior. (b) Lateral view of an embryo at 60% of development. (b′). Dorsal view of the same embryo. lo, lateral organs; at, antenna. (c) Lateral view of an embryo at 70% of development. (d) Dorsal view of an embryo at 90% of development, just before hatching. e, eye. (Reprinted with permission from Ref 111. Copyright 2010 Staatliches Museum für Naturkunde Stuttgart)
A. aquaticus demonstrates many of the same advantages present in A. mexicanus. Through continued development of A. aquaticus as a system for evolutionary and developmental biology research, more discoveries will be enabled, allowing a much better understanding of the evolution of cave characteristics in these isopods. Furthermore, comparing A. mexicanus and A. aquaticus will allow for a broader view of how and why cave-associated traits evolve.
Similarities and Differences between Astyanax and Asellus
Cave animals represent a unique system in which it is possible to investigate the evolution of the same characteristics in very different species from vertebrates to invertebrates. We will compare eye and pigmentation in A. mexicanus and A. aquaticus.
Eye and Pigment Reduction—Monogenic or Polygenic?
Eye reduction appears to be a polygenic trait in both A. aquaticus and A. mexicanus.4,27,44 On the other hand, the phenotype of ommatidial absence in A. aquaticus appears to be encoded by a single locus of large effect.
Pigmentation seems to involve multiple monogenic traits in both A. mexicanus and A. aquaticus. In A. mexicanus, there appears to be three monogenic recessive traits involved in pigmentation phenotypes: the yellow, albino, and brown phenotypes.81,82,100 Similarly, in A. aquaticus, four qualitative pigmentation traits are each determined primarily by a single region of the genome. However, melanophore number is a polygenic trait in A. mexicanus and has not yet been described for A. aquaticus. Therefore, it seems that both pigmentation and eye reduction can be either monogenic or polygenic traits.
Eye Reduction and Pigment Reduction—Multiple Mechanisms or One Favored Method?
Within the single A. aquaticus population examined, there appears to be multiple genetic modes of eye reduction—a potentially monogenic mode of ommatidial absence and a polygenic mode of fragmented ommatidia.4 Multiple methods also are apparent in A. mexicanus where complementation studies between different populations result in individuals with eyes more developed than either parent.34,62,63,80 This might indicate that there are multiple possible ways to cause eye reduction and not a single favored genetic path in both species.
If one looks at pigmentation as a whole, it seems as though there are multiple methods in both systems to degenerate the amount and types of pigments. For example, in A. mexicanus, there are the brown, albino, yellow, and iridophore mutations81,82,100 and in A. aquaticus there are mutations that cause albino, orange, red, or stellate chromatophores.4,6
However, in A. mexicanus, the most extreme phenotype in both species, albinism, the same gene, oca2, appears to be responsible in three different cave populations with two discovered coding mutations and potentially an unknown regulatory mutation30 (see Figure 11(a)). Contrastingly, in A. aquaticus, within a single cave population there appears to be two different ways to achieve albinism4 (see Figure 11(b)). Within the A. mexicanus populations examined, there seems to be a bias in the method of evolving albinism but it does not appear that there is such a bias in A. aquaticus. Perhaps, loss of oca2 could confer some kind of advantageous pleiotropic effect in the fish populations, or other genes in the pigmentation pathway when mutated have adverse effects. Investigation of additional populations and species will be needed to further conclude whether there are more constraints in complete pigment loss in vertebrates versus invertebrates.
FIGURE 11.
Differences in albinism between Astyanax mexicanus and Asellus aquaticus. (a) Three different cavefish populations: Molino, Pachón, and Japones have an albino phenotype and the causative gene appears to be oca2. (In Japonés, the actual mutation has not been mapped—the evidence for oca2 as the responsible gene is by complementation test).30 (b) In A. aquaticus, only one cave population has been examined but there are two different methods observed to cause albinism; either a single locus or mutations at two different loci.4
Similar Degree of Reduction?
The degree of reduction in pigment seems similar in A. mexicanus and A. aquaticus; in both there are either populations or individuals that exhibit complete loss of melanin (A. mexicanus) or melanin/ommochrome (A. aquaticus).6,81 However, the degree of eye reduction can be different in A. aquaticus and A. mexicanus. In all of the populations of A. mexicanus investigated, a rudimentary eye is present. Although some vertebrate cave animals have no exterior evidence of an eye, when the face is dissected or sectioned, there is evidence of a rudimentary eye. It has been suggested that complete eye loss in vertebrates is not possible due to developmental constraints.80 In A. aquaticus some individuals are missing all ommatidia. These animals were described as blind with undifferentiated photoreceptors and an optic nerve.6 However, other cave arthropod species have been described as eyeless (reviewed in113). Therefore, the same developmental constraints present in vertebrates may not be present in arthropods. Another method of addressing the degree of reduction in cave animals is to sequence the transcriptome of putatively eyeless animals and ask whether any transcripts involved in phototransduction are present. Recently such an analysis found many transcripts involved in phototransduction in the cave beetle Ptomaphagus hirtus, which had been previously described as blind.114 In terms of structures, it seems that invertebrates can have a greater degree of eye reduction than vertebrates. However, whether this is a functional reduction (as evidenced by transcriptome sequencing), as well as a structural reduction, remains to be seen.
Unresolved Questions and Future Comparisons
Many questions and comparisons remain to be addressed either because of the complexity of the question or the recent development of A. aquaticus as a model for evolutionary and developmental biology.
First, are the same genes affected in both A. mexicanus and A. aquaticus? For pigmentation it is unlikely because many genes in the melanin pathway are not conserved between vertebrates and arthropods and the type of pigment present in the isopods could be ommochromes instead of melanin.105 However, there are many conserved genes in eye development in both vertebrates and invertebrates. Therefore, it could be possible to see the same genes affected in eye reduction in A. mexicanus and A. aquaticus.
Second, what are the evolutionary mechanisms leading to eye and pigment loss in both systems? In A. aquaticus, additional phenotypic traits must be investigated to determine whether phenotypes such as antennal length and sensory cell elongation are genetically or developmentally linked to the eye and pigment loss characteristics. Furthermore, once the actual mutations responsible for eye and pigment loss are found in both systems, researchers can investigate whether or not there are signatures of selection around causal mutations. With this information, it should be possible to understand if there is a ‘universal answer’ in cave evolution of pigment and eye loss, whether a single loss phenotype within a population can be governed by multiple varied mechanisms, or whether the answer lies between these two extremes.
Overall Comparisons between A. mexicanus and A. aquaticus
There are many similarities and differences in the evolution of cave-associated characteristics of A. mexicanus and A. aquaticus. To truly understand the evolution of cave-related characteristics in cave animals it will be necessary to examine many other species. Only through examination of additional systems will it be possible to identify trends unique to certain types of cave animals or to a particular trait.
UP AND COMING SYSTEMS: OTHER CAVE ANIMALS AS EVO–DEVO MODELS
The following are just a few examples of cave animals that might be appropriate as additional model systems. Different cave adapted species have different advantages. For example, the cave beetle P. hirtus profits by its phylogenetic relationship to the genetic model organism Tribolium castaneum.114 The primary consideration in selecting the animals described below, however, is the existence of closely related surface and cave-dwelling forms.
Planarians have surface dwelling and cave-adapted representatives, the latter lacking eyes and pigmentation.115 Although constructive traits are not obvious, cave-adapted planaria are an attractive system because of their basal position in metazoan phylogeny and their intense current use as a developmental model. The Hawaiian planthopper genus Oliarus has closely related surface and numerous cave dwelling forms, and the latter have lost eyes and pigmentation, reduced their wings, and increased the size of their antennae.12 Albino cave planthoppers, like Astyanax cavefish, have interupted melanin synthesis at its first step (the conversion of L-tyrosine to L-DOPA), suggesting convergence in the evolution of albinism between a cave adapted insect and a vertebrate.116 The planthopper system is also attractive because of its ecology. Hawaiian cave-adapted planthoppers inhabit lava caves, a different type of habitat than the limestone caves typical of most other troglomorphic cave animals. In addition, they feed on the roots of surface foliage that extend into the relatively shallow lava caves and thus may not be food limited, making it possible to address the role of energy conservation in troglomorphic evolution.
Two other systems would be attractive for further development because of their close phylogenetic affinities to A. aquaticus and A. mexicanus. The amphipod Gammarus minus has already provided insights as a model system in understanding cave animal evolution.3,7 Side by side comparisons of A. aquaticus and G. minus would give useful insights into the convergent evolution of troglomorphic features in cave crustaceans. Likewise, the teleost Poecilia mexicanus is being developed to examine the evolution of cave adapted traits, particularly in the extreme environment of a sulfurous cave system.117 P. mexicanus has small eyes and reduced pigmentation and is interesting to compare with A. mexicanus due to its less pronounced troglomorphic characters. Further progress in developing these and other systems could significantly increase our overall understanding of the comparative evolutionary biology of cave animals and take maximum advantage of the cave environment as a true evolutionary laboratory.
Acknowledgments
We thank H. Bilandžija, D. Fong, M. Friedrich, J. Gross, and P. Trontelj for critical reading of the manuscript; to R. Borowsky, J. Gross, S. Prevorčnik, R. Verovnik, P. Vick, and Y. Yamamoto for figures and drawings that were included and to H. Bilandžija, D. Fong, M. Friedrich, M. Pavlek, and M. Lukić for the use of photos of cave animals.
References
- 1.Culver DC, Pipan T. The Biology of Caves and Other Subterranean Habitats. New York: Oxford University Press; 2009. p. 254. [Google Scholar]
- 2.Abzhanov A, Extavour CG, Groover A, Hodges SA, Hoekstra HE, Kramer EM, Monteiro A. Are we there yet? Tracking the development of new model systems. Trends Genet. 2008;24:353–360. doi: 10.1016/j.tig.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Fong DW. Gammarus minus: a model system for the study of adaptation to the cave environment. In: Culver DC, White WB, editors. Encyclopedia of Caves. 2. Amsterdam: Elsevier; 2011. [Google Scholar]
- 4.Protas ME, Trontelj P, Patel NH. Genetic basis of eye and pigment loss in the cave crustacean, Asellus aquaticus. Proc Natl Acad Sci U S A. 2011;14:5702–5707. doi: 10.1073/pnas.1013850108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin E, Beatty RA. The pigmentation of cavernicolus animals. J Exp Biol. 1941;18:136–143. [Google Scholar]
- 6.Kosswig VC, Kosswig L. Die Variabilitat bei Asellus aquaticus, unter besonderer Berucksichtigung der Variabilitat in isolierten unter-und oberirdischen Populationen. R Facultie Sci. 1940;B5:1–55. [Google Scholar]
- 7.Fong DW. Morphological evolution of the amphipod Gammarus minus in caves: quantitative genetic analysis. Am Midland Natural. 1989;121:361–378. [Google Scholar]
- 8.Sadoglu P. Mendelian inheritance in hybrids between the Mexican blind fish and their overground ancestors. Verhandlungen Deutschen Zool Gesellschaft, Graz. 1957;1957:432–439. [Google Scholar]
- 9.Parzefall J. Zur genetik und biologischen bedeutung des aggressionsverhaltens von Poecilia sphenops (Pisces, Poeciliidae). Untersuchungen an Bastarden ober-und unterirdisch lebender Populationen. Zeitschrift Tierpsychol. 1979;50:399–422. [Google Scholar]
- 10.Sket B. Postojna-Planina cave system: biospeleology. In: Gunn J, editor. Encyclopedia of Cave and Karst Science. New York/London: Fitzroy Dearborn; 2004. pp. 603–604. [Google Scholar]
- 11.Kruckenhauser L, Haring E, Seemann R, Sattmann H. Genetic differentiation between cave and surface dwelling populations of Garra barreimiae (Cyprinidae) in Oman. BMC Evolution Biol. 2011;11:172. doi: 10.1186/1471-2148-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howarth FG. Cavernicoles in lava tubes on the island of Hawaii. Science. 1972;175:325–326. doi: 10.1126/science.175.4019.325. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell RW, Russell WH, Elliott WR. Mexican eyeless Characin fishes, genus Astyanax: environment, distribution, and Evolution. Special Publ Museum Texas Tech Univ. 1977;12:1–89. [Google Scholar]
- 14.Sarbu SM. Movile Cave: a chemoautotrophically based groundwater ecosystem. Subterranean Ecosyst. 2000;30:319–343. [Google Scholar]
- 15.Sket B. The nature of biodiversity in hypogean waters and how it is endangered. Biodiv Conserv. 1999;8:1319–1338. [Google Scholar]
- 16.Romero A. Cave Biology Life in Darkness. Cambridge: Cambridge University Press; 2009. [Google Scholar]
- 17.Verovnik R, Sket B, Trontelj P. Phylogeography of subterranean and surface populations of water lice Asellus aquaticus (Crustacea: Isopoda) Mol Ecol. 2004;13:1519–1532. doi: 10.1111/j.1365-294X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 18.Breder CM, Rasquin P. Comparative studies in the light sensitivity of blind characins from a series of Mexican caves. Bull Am Museum Nat Hist. 1947;89:323–351. [Google Scholar]
- 19.Voituron Y, de Fraipont M, Issartel J, Guillaume O, Clobert J. Extreme lifespan of the human fish (Proteus anguinus): a challenge for ageing mechanisms. Biol Lett. 2011;7:105–107. doi: 10.1098/rsbl.2010.0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffery WR. Evolution and development in the cavefish Astyanax. Curr Topics Dev Biol. 2009;86:191–221. doi: 10.1016/S0070-2153(09)01008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vitagliano Tadini G, Valentino F. Reproductive cycle in various stocks of Asellus aquaticus of different geographical origin. Rivista Biol. 1964;57:327–346. [PubMed] [Google Scholar]
- 22.Borowsky R. Astyanax mexicanus, the blind Mexican cave fish: a model for studies in development and morphology. Emerging Model Organsims: A Laboratory Manual. 2008;1:469–480. doi: 10.1101/pdb.emo107. [DOI] [PubMed] [Google Scholar]
- 23.Holditch DM, Tolba MR. The effect of temperature and water quality on the in vitro development and survival of A. aquaticus (Crustacea; Isopoda) eggs. Hydrobiologia. 1981;78:227–236. [Google Scholar]
- 24.Turk-Prevorcnik S, Blejec A. Asellus aquaticus infernus, new subspecies (isopoda:asellota:asellidae), from Romanian hypogean waters. J Crustacean Biol. 1998;18:763–773. [Google Scholar]
- 25.Verovnik R, Sket B, Prevorcnik S, Trontelj P. Random amplified polymorphic DNA diversity among surface and subterranean populations of Asellus aquaticus (Crustacea: Isopoda) Genetica. 2003;119:155–165. doi: 10.1023/a:1026085716777. [DOI] [PubMed] [Google Scholar]
- 26.Verovnik R, Sket B, Trontelj P. The colonization of Europe by the freshwater crustacean Asellus aquaticus (Crustacea: Isopoda) proceeded from ancient refugia and was directed by habitat connectivity. Mol Ecol. 2005;14:4355–4369. doi: 10.1111/j.1365-294X.2005.02745.x. [DOI] [PubMed] [Google Scholar]
- 27.Borowsky R, Wilkens H. Mapping a cave fish genome: polygenic systems and regressive evolution. J Hered. 2002;93:19–21. doi: 10.1093/jhered/93.1.19. [DOI] [PubMed] [Google Scholar]
- 28.Protas ME, Conrad M, Gross JB, Tabin C, Borowsky R. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr Biol. 2007;17:452–454. doi: 10.1016/j.cub.2007.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Protas ME, Tabansky I, Conrad M, Gross JB, Vidal O, Tabin CJ, Borowsky R. Multi-trait evolution in a cave fish, Astyanax mexicanus. Evol Dev. 2008;10:196–209. doi: 10.1111/j.1525-142X.2008.00227.x. [DOI] [PubMed] [Google Scholar]
- 30.Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nature Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- 31.Austin JD, Bertin A, Bórquez JP, Cárdenas L, Cardoza TB, Chapman F, De Sousa AC, De Souza AP, Douglas KC, Ellwood SR, et al. Molecular Ecology Resources Primer Development Consortium. Permanent genetic resources added to Molecular Ecology Resources Database 1 February 2011–31 March 2011. Molecular Ecology Resources. 2011;11:757–758. doi: 10.1111/j.1755-0998.2011.03028.x. [DOI] [PubMed] [Google Scholar]
- 32.Di Palma F, Kidd C, Borowsky R, Kocher TD. Construction of bacterial artificial chromosome libraries for the Lake Malawi cichlid (Metriaclima zebra), and the blind cavefish (Astyanax mexicanus) Zebrafish. 2007;4:41–47. doi: 10.1089/zeb.2006.9996. [DOI] [PubMed] [Google Scholar]
- 33.Gross JB, Protas M, Conrad M, Scheid PE, Vidal O, Jeffery WR, Borowsky R, Tabin CJ. Synteny and candidate gene prediction using an anchored linkage map of Astyanax mexicanus. Proc Natl Acad Sci U S A. 2008;105:20106–20111. doi: 10.1073/pnas.0806238105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeffery WR. Regressive evolution in Astyanax cavefish. Ann Rev Genet. 2009;43:25–47. doi: 10.1146/annurev-genet-102108-134216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liubicich DM, Serano JM, Pavlopoulos A, Kontarakis Z, Protas ME, Kwan E, Chatterjee S, Tran KD, Averof M, Patel NH. Knockdown of Parhyale Ultrabithorax recapitulates evolutionary changes in crustacean appendage morphology. Proc Natl Acad Sci U S A. 2009;106:13892–13896. doi: 10.1073/pnas.0903105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pavlopoulos A, Kontarakis Z, Liubicich DM, Serano JM, Akam M, Patel NH, Averof M. Probing the evolution of appendage specialization by Hox gene misexpression in an emerging model crustacean. Proc Natl Acad Sci U S A. 2009;106:13897–13902. doi: 10.1073/pnas.0902804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Espinasa L, Borowsky RB. Origin and relationships of cave populations of the blind Mexican tetra, Astyanax fasciatus, in the Sierra de El Abra. Environ Biol Fishes. 2001;62:233–227. [Google Scholar]
- 38.Dowling TE, Martasian DP, Jeffery WR. Evidence for multiple genetic lineages with similar eyeless phenotypes in the blind cavefish, Astyanax mexicanus. Mol Biol Evol. 2002;19:446–555. doi: 10.1093/oxfordjournals.molbev.a004100. [DOI] [PubMed] [Google Scholar]
- 39.Strecker U, Bernachez L, Wilkens H. Genetic divergence between cave and surface populations of Astyanax in Mexico (Characidae, Teleostei) Mol Ecol. 2003;12:699–710. doi: 10.1046/j.1365-294x.2003.01753.x. [DOI] [PubMed] [Google Scholar]
- 40.Strecker U, Faúndez VH, Wilkens H. Phylogeography of surface and cave Astyanax (Teleostei) from Central and North America based on cytochrome b sequence data. Mol Phylogenet Evol. 2004;33:469–481. doi: 10.1016/j.ympev.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Ornelas-García CP, Domínguez-Domínguez O, Doadrio I. Evolutionary history of the fish genus Astyanax Baird Girard (1854) (Actinopterygii, Characidae) in Mesoamerica reveals multiple morphological homoplasies. BMC Evol Biol. 2008;8:340–357. doi: 10.1186/1471-2148-8-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strecker U, Hausdorf B, Wilkens H. Parallel speciation in Astyanax cave fish (Teleostei) in Northern Mexico. Mol Phylogenet Evol. 2012;62:62–70. doi: 10.1016/j.ympev.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Porter ML, Dittmar K, Perez-Losada M. How long does the evolution of the troglomorphic form take? Estimating divergence times in Astyanax mexicanus. Acta Carsol. 2007;36:173–182. [Google Scholar]
- 44.Wilkens H. Evolution and genetics of epigean and cave Astyanax fasciatus (Characidae, Pisces) Evol Biol. 1988;23:271–367. [Google Scholar]
- 45.Jeffery WR. Cavefish as a model system in evolutionary developmental biology. Dev Biol. 2001;231:1–12. doi: 10.1006/dbio.2000.0121. [DOI] [PubMed] [Google Scholar]
- 46.Cahn PH. Comparative optic development in Astyanax mexicanus and two of its blind cave derivatives. Bull Am Museum Nat Hist. 1958;115:75–112. [Google Scholar]
- 47.Jeffery WR, Strickler AG, Guiney S, Heyser D, Tomarev SI. Prox1 in eye degeneration and sensory organ compensation during development and evolution of the cavefish Astyanax. Dev Genes Evol. 2000;210:223–230. doi: 10.1007/s004270050308. [DOI] [PubMed] [Google Scholar]
- 48.Strickler AG, Yamamoto Y, Jeffery WR. The lens controls cell survival in the retina: evidence from the blind cavefish Astyanax. Dev Biol. 2007;311:512–523. doi: 10.1016/j.ydbio.2007.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilkens H. Regressive evolution: ontogeny and genetics of cavefish eye rudimentation. Biol J Linnean Soc. 2007;92:287–296. [Google Scholar]
- 50.Yamamoto Y, Stock DW, Jeffery WR. Hedgehog signalling controls eye degeneration in blind cavefish. Nature. 2004;431:844–847. doi: 10.1038/nature02864. [DOI] [PubMed] [Google Scholar]
- 51.Strickler AG, Famuditimi K, Jeffery WR. Retinal homeobox genes and the role of cell proliferation in cavefish eye degeneration. Int J Dev Biol. 2002;46:285–294. [PubMed] [Google Scholar]
- 52.Jeffery WR, Strickler AG, Yamamoto Y. To see or not to see: evolution of eye degeneration in Mexican blind cavefish. Comparat Integrat Biol. 2003;43:531–541. doi: 10.1093/icb/43.4.531. [DOI] [PubMed] [Google Scholar]
- 53.Soares D, Yamamoto Y, Strickler AG, Jeffery WR. The lens has a specific influence on optic nerve and tectum development in the blind cavefish Astyanax. Dev Neurosci. 2004;26:308–317. doi: 10.1159/000082272. [DOI] [PubMed] [Google Scholar]
- 54.Alunni A, Menuet A, Candal E, Pénigault J-B, Jeffery WR, Rétaux S. Developmental mechanisms for retinal degeneration in the blind cavefish Astyanax mexicanus. J Compara Neurol. 2007;505:221–233. doi: 10.1002/cne.21488. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto Y, Jeffery WR. Central role for the lens in cavefish eye degeneration. Science. 2000;289:631–633. doi: 10.1126/science.289.5479.631. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto Y, Espinasa L, Stock DW, Jeffery WR. Development and evolution of craniofacial patterning is mediated by eye-dependent and—independent processes in the cavefish Astyanax. Evol Dev. 2003;5:435–446. doi: 10.1046/j.1525-142x.2003.03050.x. [DOI] [PubMed] [Google Scholar]
- 57.Jeffery WR, Martasian DP. Evolution of eye regression in the cavefish Astyanax: apoptosis and the Pax-6 gene. Am Zool. 1998;38:685–696. [Google Scholar]
- 58.Johns P. Growth of the adult goldfish eye III. Source of the new retinal cells. J Comparat Neurol. 1997;176:348–358. doi: 10.1002/cne.901760304. [DOI] [PubMed] [Google Scholar]
- 59.Otteson DC, D’ Costa AR, Hitchcock RF. Putative stem cells and the lineage of rod photoreceptors in the mature retina of the goldfish. Dev Biol. 2001;232:62–76. doi: 10.1006/dbio.2001.0163. [DOI] [PubMed] [Google Scholar]
- 60.Pottin K, Hinaux H, Rétaux S. Restoring eye size in Astyanax mexicanus embryos through modulation of the Shh and fgf8 forebrain organizing centres. Development. 2011;138:2467–2476. doi: 10.1242/dev.054106. [DOI] [PubMed] [Google Scholar]
- 61.Wilkens H. Genetic interpretation of regressive evolutionary processes: studies of hybrid eyes of two Astyanax cave populations (Characidae, Pisces) Evolution. 1971;25:530–544. doi: 10.1111/j.1558-5646.1971.tb01913.x. [DOI] [PubMed] [Google Scholar]
- 62.Wilkens H. Genes, modules, and the evolution of cave fish. Heredity. 2010;105:413–422. doi: 10.1038/hdy.2009.184. [DOI] [PubMed] [Google Scholar]
- 63.Borowsky R. Restoring sight in blind cavefish. Curr Biol. 2008;18:R23–R24. doi: 10.1016/j.cub.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 64.Gross JB, Borowsky R, Tabin CJ. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. Public Lib Sci Genet. 2009;5:e1000326. doi: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Behrens M, Wilkens H, Schmale H. Cloning of the αA-crystallin genes of the blind cave form and the epigean form of Astyanax fasciatus: a comparative analysis of structure, expression and evolutionary conservation. Gene. 1998;216:319–326. doi: 10.1016/s0378-1119(98)00346-1. [DOI] [PubMed] [Google Scholar]
- 66.Strickler AG, Byerly MS, Jeffery WR. Lens gene expression analysis reveals downregulation of the anti-apoptotic chaperone αA-crystallin during cavefish eye degeneration. Dev Genes Evol. 2007;217:771–782. doi: 10.1007/s00427-007-0190-z. [DOI] [PubMed] [Google Scholar]
- 67.Hooven TA, Yamamoto Y, Jeffery WR. Blind cave-fish and heat shock protein chaperones: a novel role for hsp90α in lens apoptosis. Int J Dev Biol. 2004;48:731–738. doi: 10.1387/ijdb.041874th. [DOI] [PubMed] [Google Scholar]
- 68.Strickler AG, Yamamoto Y, Jeffery WR. Early and late changes in Pax6 expression accompany eye degeneration during cavefish development. Dev Genes Evol. 2001;211:138–144. doi: 10.1007/s004270000123. [DOI] [PubMed] [Google Scholar]
- 69.Schwarz M, Cecconi F, Berneir G, Andrejewski N, Kammandel B, Wagner M, Gruss P. Spatial specification of mammalian eye territories by reciprocal transcriptional repression of Pax2 and Pax6. Development. 2000;127:4325–4334. doi: 10.1242/dev.127.20.4325. [DOI] [PubMed] [Google Scholar]
- 70.Menuet A, Alunni A, Joly J-S, Jeffery WR, Rétaux S. Shh overexpression in Astyanax cavefish: multiple consequences on forebrain development and evolution. Development. 2006;134:845–855. doi: 10.1242/dev.02780. [DOI] [PubMed] [Google Scholar]
- 71.Ekker SC, Ungar AR, von Greenstein P, Porter JA, Moon RT, Beachy P. Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Curr Biol. 1995;5:944–955. doi: 10.1016/s0960-9822(95)00185-0. [DOI] [PubMed] [Google Scholar]
- 72.Macdonald R, Anukampa Barth K, Xu Q, Holder N, Mikkola I, Wilson S. Midline signaling is required for Pax6 gene regulation and patterning of the eyes. Development. 1995;121:3267–3278. doi: 10.1242/dev.121.10.3267. [DOI] [PubMed] [Google Scholar]
- 73.Fairbank PD, Lee C, Ellis A, Hildebrand JD, Gross JM, Wallingford JB. Shroom2 (APXL) regulates melanosome biogenesis and localization in the retinal epithelium. Development. 2006;133:4109–4118. doi: 10.1242/dev.02563. [DOI] [PubMed] [Google Scholar]
- 74.Moritz OL, Molday RS. Molecular cloning, membrane topology, and localization of bovine rom-1 in rod and cone photoreceptor cells. Invest Ophthalmol Visual Sci. 1996;37:352–362. [PubMed] [Google Scholar]
- 75.Hubbard JK, Uy JA, Hauber ME, Hoekstra HE, Safran RJ. Vertebrate pigmentation: from underlying genes to adaptive function. Trends Genet. 2010;26:231–239. doi: 10.1016/j.tig.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 76.Protas ME, Patel NH. Evolution of coloration patterns. Annu Rev Cell Dev Biol. 2008;24:425–446. doi: 10.1146/annurev.cellbio.24.110707.175302. [DOI] [PubMed] [Google Scholar]
- 77.McKinnon JS, Pierotti ME. Colour polymorphism and correlated characters: genetic mechanisms and evolution. Mol Ecol. 2010;19:5101–5125. doi: 10.1111/j.1365-294X.2010.04846.x. [DOI] [PubMed] [Google Scholar]
- 78.Kelsh RN. Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 2004;17:326–336. doi: 10.1111/j.1600-0749.2004.00174.x. [DOI] [PubMed] [Google Scholar]
- 79.McCauley DW, Hixon E, Jeffery WR. Evolution of pigment cell regression in the cavefish Astyanax: a late step in melanogenesis. Evol Dev. 2004;6:209–218. doi: 10.1111/j.1525-142X.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 80.Wilkens H, Strecker U. Convergent evolution of the cavefish Astyanax (Characidae, Teleostei): genetic evidence from reduced eye-size and pigmentation. Biol J Linnean Soc. 2003;80:545–554. [Google Scholar]
- 81.Sadoglu P. A Mendelian gene for albinism in natural cave fish. Experientia. 1957;13:394. [Google Scholar]
- 82.Sadoglu P, McKee A. A second gene that affects eye and body color in Mexican blind cave fish. J Heredity. 1969;60:10–11. doi: 10.1093/oxfordjournals.jhered.a107917. [DOI] [PubMed] [Google Scholar]
- 83.Espinasa L, Yamamoto Y, Jeffery WR. Non-optical releasers of regressive behavior in blind and blinded Astyanax. Behav Process. 2005;70:144–148. doi: 10.1016/j.beproc.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 84.Langecker TG. Genetic analysis of the dorsal light reaction in epigean and cave dwelling Astyanax fasciatus (Telelostei, Characidae) Ethol Ecol Evol. 1993;5:357–364. [Google Scholar]
- 85.Parzefall J, Fricke D. Alarm reaction and schooling in cave and river populations of Astyanax fasciatus (Pisces, Characidae) and their hybrids. Memiors Biospeleol. 1989;16:177–182. [Google Scholar]
- 86.Duboué ER, Keene AC, Borowsky RL. Evolutionary convergence of sleep loss in cavefish. Curr Biol. 2011;21:671–676. doi: 10.1016/j.cub.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 87.Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: genomic organization and modified collinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- 88.Yoshizawa M, Gorički Š, Soares D, Jeffery WR. Evolution of a behavioral shift mediated by superficial neuromasts helps cavefish find food in darkness. Curr Biol. 2010;20:1631–1636. doi: 10.1016/j.cub.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teyke T. Morphological differences in neuromasts of the blind cavefish Astyanax hubbsi and the sighted river fish Astyanax mexicanus. Brain Behav Evol. 1990;35:23–30. doi: 10.1159/000115853. [DOI] [PubMed] [Google Scholar]
- 90.Jeffery WR. Pleiotropy and eye degeneration in blind cavefish. Heredity. 2010;105:495–496. doi: 10.1038/hdy.2010.7. [DOI] [PubMed] [Google Scholar]
- 91.Yamamoto Y, Byerly MS, Jackman WR, Jeffery WR. Pleiotropic functions of embryonic sonic hedgehog expression link jaw and taste bud amplification with eye loss during cavefish evolution. Dev Biol. 2009;330:200–211. doi: 10.1016/j.ydbio.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schemmel C. Studies on the genetics of feeding behaviour in the cave fish Astyanax mexicanus f. Anoptichthys An example of apparent monofactorial inheritance by polygenes. Zeitschrift Teirpsychol. 1980;53:9–22. doi: 10.1111/j.1439-0310.1980.tb00730.x. [DOI] [PubMed] [Google Scholar]
- 93.Hüppop K. Food finding ability in cave fish (Astyanax fasciatus) Int J Speleol. 1987;18:59–66. [Google Scholar]
- 94.Varatharasan N, Croll RP, Franz-Odendaal T. Taste bud development and patterning in sighted and blind morphs of Astyanax mexicanus. Dev Dyn. 2009;238:3056–3064. doi: 10.1002/dvdy.22144. [DOI] [PubMed] [Google Scholar]
- 95.Schemmel C. Vergleichende Untersuchungen an den Hautsinnesorgagen over und unterirdischlebender Astyanax-Formen. Zeitschrift Morphol Tiere. 1967;61:255–316. [Google Scholar]
- 96.Rose FL, Mitchell RW. Comparative lipid values of epigean and cave-adapted Astyanax. Southwestern Natural. 1980;27:357–358. [Google Scholar]
- 97.Peters N, Schacht V, Schmidt W, Wilkens H. Gehirn-proportionen und Auspra gungsgrad der Sinnesorgane von Astyanax mexicanus (Pisces, Characinidae) Zeitschrift Zoologische SystemEvolutionsforschung. 1993;31:144–159. [Google Scholar]
- 98.Rétaux S, Pottin K, Alunni A. Shh and forebrain evolution in the blind cavefish Astyanax mexicanus. Biol Cell. 2008;100:139–147. doi: 10.1042/BC20070084. [DOI] [PubMed] [Google Scholar]
- 99.Franz-Odendaal TA, Hall BK. Modularity and sense organs in the blind cavefish, Astyanax mexicanus. Evol Dev. 2006;8:94–100. doi: 10.1111/j.1525-142X.2006.05078.x. [DOI] [PubMed] [Google Scholar]
- 100.Culver DC, Wilkens H. Critical review of the relevant theories of the evolution of subterranean animals. In: Wilkens H, Culver DC, Humphreys WF, editors. Ecosystems of the World. Amsterdam: Elsevier; 2000. [Google Scholar]
- 101.Jeffery WR. Adaptive evolution of eye degeneration in the Mexican blind cavefish. J Heredity. 2005;96:185–196. doi: 10.1093/jhered/esi028. [DOI] [PubMed] [Google Scholar]
- 102.Gibert J, Deharveng L. Subterranean ecosystems: a truncated functional biodiversity. BioScience. 2002;52:473–481. [Google Scholar]
- 103.Prevorcnik S, Blejec A, Sket B. Racial differentiation in Asellus aquaticus (L) (Crustacea: Isopoda: Asellidae) Archiv Hydrobiol. 2004;160:193–214. [Google Scholar]
- 104.Turk S, Sket B, Sarbu S. Comparison between some epigean and hypogean populations of Asellus aquaticus. Hydrobiologia. 1996;337:161–170. [Google Scholar]
- 105.Kosswig C. Génétique et évolution regressive. Revue Questions Sci. 1965;136:227–257. [Google Scholar]
- 106.Nilsson HL. The fine structure of the compound eyes of shallow-water asellotes, Jaera albifrons Leach and Asellus aquaticus L (Crustacean: Isopoda) Acta Zool. 1978;59:69–84. [Google Scholar]
- 107.de Lattin G. Untersuchungen an Isopodenaugen. Zool Jrb Anat. 1939;65:417–468. [Google Scholar]
- 108.Salema H. The chromosomes of Asellus aquaticus (L)—a technique for isopod karyology. Crustaceana. 1979;36:316–318. [Google Scholar]
- 109.Needham AE, Brunet PCJ. The integumental pigment of Asellus. Cell Mol Life Sci. 1957;13:207–209. [Google Scholar]
- 110.Janzer W, Ludwig W. Versuche zur evolutorischen entsteung der hohlentiermerkmale. Zeitschrift Inductive Abstammungs- Vererbungslehre. 1952;84:462–479. [PubMed] [Google Scholar]
- 111.Vick P, Blum M. The isopod Asellus aquaticus: a novel arthropod model organism to study evolution of segment identity and patterning. Paleodiversity. 2010;3:89–97. [Google Scholar]
- 112.Vick P, Schweickert A, Blum M. Cloning and expression analysis of the homeobox gene abdominal-A in the isopod Asellus aquaticus. In: Kurzfield NC, editor. Development Gene Expression Regulation. Hauppauge: Nova Science Publishers; 2009. pp. 285–295. [Google Scholar]
- 113.Friedrich M. Drosophila as developmental paradigm of regressive brain evolution: proof of principle in the visual system. Brain Behav Evol. 2011;78:199–215. doi: 10.1159/000329850. [DOI] [PubMed] [Google Scholar]
- 114.Friedrich M, Chen R, Daines B, Bao R, Caravas J, Rai PK, Zagmajster M, Peck SB. Phototransduction and clock gene expression in the troglobiont beetle Ptomaphagus hirtus of Mammoth cave. J Exp Biol. 2011;214:3532–3541. doi: 10.1242/jeb.060368. [DOI] [PubMed] [Google Scholar]
- 115.Buchanan JW. Notes on an American cave flat-worm, Sphalloplana percaeca (Packard) Ecology. 1936;17:194–211. [Google Scholar]
- 116.Bilandžija H, Ćetković H, Jeffery WR. Evolution of albinism in cave planthoppers by a convergent defect in the first step of melanin biosynthesis. Evol Dev. 2012;14:196–203. doi: 10.1111/j.1525-142X.2012.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Riesch R, Plath M, Schlupp I. Toxic hydrogen sulfide and dark caves: life-history adaptations in a livebearing fish (Poecilia mexicana, Poeciliidae) Ecology. 2010;91:1494–1505. doi: 10.1890/09-1008.1. [DOI] [PubMed] [Google Scholar]
FURTHER READING
- Carlos J, Guzik MT, Jaume D, Cooper SJB. Evolution in caves: Darwin’s ‘wrecks of ancient life’ in the molecular era. Mol Ecol. 2010;19:3865–3880. doi: 10.1111/j.1365-294X.2010.04759.x. [DOI] [PubMed] [Google Scholar]
- Culver D. Cave Life: Ecology and Evolution. Cambridge: Harvard University Press; 1982. p. 189. [Google Scholar]
- Porter ML, Crandall KA. Lost along the way: the significance of evolution in reverse. Trends Ecol Evol. 2003;18:541–547. [Google Scholar]
- Poulson TL, White WB. The cave environment. Science. 1969;165:971–981. doi: 10.1126/science.165.3897.971. [DOI] [PubMed] [Google Scholar]