Abstract
Objective
To determine the relationship between allelic variations in genes involved in fluticasone propionate (FP) metabolism and asthma control among children with asthma managed with inhaled FP.
Study design
The relationship between variability in asthma control scores and genetic variation in drug metabolism was assessed by genotyping nine single nucleotide polymorphisms (SNPs) in CYP3A4, CYP3A5, and CYP3A7. Genotype information was compared with asthma control scores (0 = well-controlled to 15 = poorly-controlled), determined by using a questionnaire modified from the National Heart Lung and Blood Institute Expert Panel 3 guidelines.
Results
Our study cohort was comprised of 734 children with asthma (mean age 8.8 ± 4.3 years), who were predominantly male (61%) and non-Hispanic Whites (53%); 413 children (56%) were receiving inhaled glucocorticoids daily, of which FP was prescribed most frequently (65%). Among the children receiving daily FP, SNPs in the genes CYP3A5 and CYP3A7 were not associated with asthma control scores. In contrast, asthma control scores were significantly improved among 20 (7%) children with the CYP3A4*22 allele (median 3, range 0-6), as compared with the 201 patients without the CYP3A4*22 allele (median 4, range 0-15) (P=0.02). The presence of CYP3A4*22 was associated with improved asthma control scores by 2.1 points (95% CI: 0.5-3.8).
Conclusions
The presence of CYP3A4*22, which is associated with decreased hepatic CYP3A4 expression and activity, was accompanied by improved asthma control among FP treated children. Decreased CYP3A4 activity may improve asthma control with inhaled FP.
Keywords: asthma, fluticasone, inhaled glucocorticoids, corticosteroids, children, pharmacogenetics
Inhaled glucocorticoids are the most effective treatment for controlling chronic asthma symptoms and are commonly prescribed to reduce airway inflammation and bronchial hyper-responsiveness.1, 2, 3 Despite broad use of inhaled glucocorticoids, many patients have poor control of asthma symptoms.4, 5 These pharmacodynamic differences may be explained, at least in part, by individual variation in pharmacokinetics or pharmacogenetics.
Fluticasone propionate (FP), a trifluorinated synthetic glucocorticoid, is one of the most widely used inhaled glucocorticoids for the treatment of asthma. This likely relates to its high immunosuppressive potency when measured with a whole-blood lymphocyte proliferation assay.6 The biological activity of FP is terminated by metabolic biotransformation, attributed primarily to cytochrome P450 3A enzymes CYP3A4, CYP3A5, and CYP3A7.7 CYP3A5 is the predominant CYP3A isoform expressed in human lung; however, the most important hepatic 3A enzyme, CYP3A4, is not usually expressed in human lung.8 We have shown that CYP3A4 expression in human lung is repressed by factors that bind to upstream transcriptional regulation sites of the CYP3A4 gene.9 CYP3A7 is the predominant form of CYP3A expressed in human fetal liver,10 and our preliminary studies have shown that it is expressed at low, but detectable levels in lung samples in patients up to 35 years old. These data suggest that CYP3A5 and/or CYP3A7 could be responsible for local biotransformation of FP in the lung due to their higher expression in bronchial mucosa and pulmonary parenchyma.11 Interindividual variability in the response to FP may be explained, in part, by variation in these CYP3A enzymes that mediate FP’s biotransformation in vivo. Furthermore, FP is a highly potent mechanism-based inactivator of CYP3A5, a less potent inactivator of CYP3A4, and has no action on CYP3A7.12
A single nucleotide polymorphism (SNP) in CYP3A5 (CYP3A5*3) was shown recently to reduce the biotransformation of lovastatin, prolonging its half-life and exaggerating its effect on serum cholesterol.13 We hypothesized that CYP3A5*3 would be associated with reduced catalytic activity in the lung, thereby increasing local pulmonary concentrations of FP that would improve asthma control in children treated with inhaled FP. We also speculated that functionally reduced activity, caused by SNPs in CYP3A4 and CYP3A7, might be associated with improved asthma control with inhaled FP. Furthermore, we speculate that mechanism-based inhibition of CYP3A5, and to a lesser degree CYP3A4, by FP may reduce the rate of FP metabolism and improve asthma control.
Methods
Saliva samples were obtained prospectively from children 2-17 years of age with a physician-confirmed diagnosis of asthma. Subjects were randomly recruited as a convenience sample from the emergency department and inpatient wards of Primary Children’s Medical Center in Salt Lake City, Utah according to the availability of the study coordinator. Information about chronic medical conditions, concomitant medication use and chief complaint was collected through a combination of structured interviews and medical chart abstraction. Data were de-identified before genotypic analyses.
Patient levels of chronic asthma control were determined by questionnaire, based upon guidelines, modified from the National Heart Lung and Blood Institute Expert Panel Report 3, which avoided variations in the child’s ability to cooperate with pulmonary function testing.3 The definition of chronic asthma control by Juniper et al as the “full range of clinical impairment that patients with asthma may experience as a result of their disease” was utilized in the development of the questionnaire to assess chronic asthma control among children and young adults.14 Five questions were posed to the child or parent pertaining to the child’s level of chronic asthma control and were quantified using a 4-point scale (Appendix; available at www.jpeds.com). The items were equally weighted, and the subject’s asthma control score was calculated as the sum of all five items. The American Thoracic Society (ATS) and the European Respiratory Society recommend the use of numeric measures of asthma control over categorical variables.15 ATS guidelines support the classification of patients whose asthma control scores fall between poorly-controlled and well-controlled.16 In this study, asthma control scores were analyzed as a numeric variable that could range from 0 (well-controlled) to 15 (poorly-controlled).
Oragene DNA kits (DNA Genotek Inc, Ottawa, ON, Canada) were used to collect 2 mL of saliva per patient. The DNA was extracted from 400 μL of saliva/Oragene buffer mix with a silica based column preparation (GeneElute Mammalian Genomic DNA Miniprep Kit, Sigma, St Louis, MO, USA) according to the manufacturer’s provided protocol. The genomic DNA was eluted from the column with 10 mM Tris-HCl, pH 8.0, and the DNA concentrations were determined by spectrophotometry (NanoDrop, Thermo Scientific, Wilmington, DE, USA). Samples that had concentrations of >1 ng/μL after the column elution were concentrated by lyophilization and resuspended in 1/10 of the original volume.
All genotype analyses were conducted in the Developmental Pharmacology and Experimental Therapeutics Laboratory within the Division of Clinical Pharmacology and Medical Toxicology at Children’s Mercy Hospitals and Clinics (Kansas City, MO). Genotype analysis of nine different SNPs in the CYP3A gene locus on chromosome 7, encompassing the genes CYP3A4, CYP3A5, and CYP3A7, were performed (Table I; available at www.jpeds.com).13, 17-22 The relative location of these SNPs to each other is shown in Figure 1 (available at www.jpeds.com). Eight of the assays are available from Applied Biosystems (Grand Island, NY, USA). The primers and probes for rs2740565 were custom made by Biosearch Technologies, Inc. (Novato, CA, USA; Table I). Approximately 10 ng of genomic DNA was used for each genotyping assay. After denaturing the DNA by heating for 2 min at 95°C, the TaqMan reactions were cycled 50 times (denaturation for 10 seconds at 95°C, annealed/elongated at 60°C for 1 min) on an Applied Biosystems 7900HT Fast Real-Time PCR System instrument. Data clustering analysis was done with the TaqMan Genotyper software (Applied Biosystems).
Table 1.
Candidate genes for genetic analysis of pediatric asthma control with inhaled fluticasone propionate
| SNP | Applied Biosystems Assay # | Reference SNP (rs #) |
|---|---|---|
| CYP3A4*22 | C__59013445_10 | rs35599367 |
| Reported effect: carriers of the variant T allele had lower hepatic CYP3A4 mRNA level and enzyme activity when compared to those with the wild-type C allele.[17] | ||
| CYP3A4 int 7 | C___1845287_10 | rs2246709 |
| Reported effect: carriers of the variant T allele have improved blood pressure in response to amlodipine treatment among high-risk African Americans.[18] | ||
| CYP3A4 int 7 | C__32306227_10 | rs4646437 |
| Reported effect: female carriers of the variant T allele had higher CYP3A4 expression and activity than male carriers and even higher expression and activity than males with the wild-type C allele.[19] | ||
| CYP3A5*3 | C__26201809_30 | rs776746 |
| Reported effect: intronic polymorphism leads to retention of exon 3B, which harbors a premature stop codon.[13,20] | ||
| CYP3A5*6 | C__30203950_10 | rs10264272 |
| Reported effect: causes the skipping of exon 7 and results in a non-functional protein.[13] | ||
| CYP3A5*1D | C__8303531_40 | rs15524 |
| Reported effect: located in the 3′ UTR, 14 nucleotides downstream of a stop codon. Effects upon activity remain unknown.[21] | ||
| CYP3A7 | C__26201872_20 | rs2687133 |
| No published reports. | ||
| CYP3A7*2 | C__25474551_10 | rs2257401 |
| Reported effect: an amino acid change (Thr>Arg) in exon 11 results in a more active form of CYP3A7.[22] | ||
| CYP3A7 6 nt 5′ of ex 14 | custom | rs2740565 |
| Reported effect: variant A allele results in an alternatively spliced product which is translated into a functional protein which may exhibit different stability and catalytic activity.[22] | ||
| rs2740565 custom assay: | ||
| Forward Primer: | 5′ CCCCCATATCTATAAAGTCACAATCC | |
| Reverse Primer: | 5′ TGAATGGGCTCCATATCTACAAAC | |
| Variant Probe: | 5′ TGAGACCTGATTTCTGATT (BHQ plus, FAM) | |
| Wild-Type Probe: | 5′ TGAGACCTGATTTCTGTTT (BHQ plus, TET) | |
Figure 1.
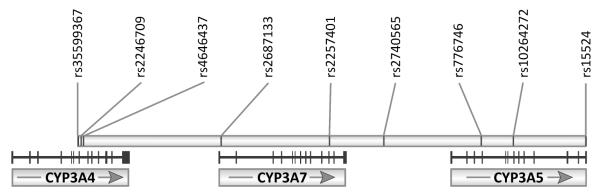
CYP3A gene locus on chromosome 7. The positions of the genotyped SNPs are indicated relative to the genes CYP3A4, CYP3A5, and CYP3A7. Vertical bars identify the individual exons, and arrows indicate the direction of transcription..
Only two samples did not amplify even after several repeat attempts in two of the assays. For all assays, known homozygous wild-type, heterozygous, and homozygous variant DNA samples (the latter genotype was not available for CYP3A4*22) were included on all the plates as positive controls. In addition, as a quality control for genotype calling we randomly selected 10% from all of the samples for each of the nine TaqMan assays and reanalyzed them under the same conditions. All original calls were confirmed. Genetic testing was blinded to both treatment and asthma control score.
Study data were collected and managed using REDCap® electronic data capture tools, hosted at the University of Utah. REDCap (Research Electronic Data Capture) is a secure, web-based application, designed to support data capture for research studies, providing: 1) a graphical user interface for validated data entry; 2) audit trails for tracking data manipulation and export procedures; 3) automated export procedures for seamless data downloads to common statistical packages; and 4) procedures for importing data from external sources.
This study was approved by the University of Utah and Primary Children’s Medical Center’s Institutional Review Boards. Prior to the performance of any study-related procedures, informed parental permission was provided by the legally-authorized representative of all study subjects and assent was obtained when appropriate.
Statistical analyses
Data were summarized as means (± standard deviation) or medians (range). Hardy-Weinberg equilibrium (HWE) and the χ2 test of HWE were determined for CYP3A genetic polymorphisms. For univariable group comparisons, unordered categorical variables were compared, using a χ2 test or Fisher exact test, when expected cell counts were less than 5. We used the Shapiro-Wilk test for normality and found that the distribution of asthma control scores was highly skewed (P<0.001). Due to the skewed nature of our asthma control scores, a non-parametric Fisher-Pitman permutation test for independent samples was used to compare the association between CYP3A genetic polymorphisms and asthma control scores. The Fisher-Pitman test, which assumes a continuously scaled variable, is as powerful as the independent groups t-test, but without the assumption of a normal distribution.23 Differences were considered significant at the P<0.05 level. The Bonferroni procedure was used to adjust for 3 pairwise group comparisons (3 SNPs) for each CYP3A enzyme.24 Stata 11.2 (StataCorp LP, College Station, TX, USA) was used to perform all statistical analyses.
Results
Among 734 children with asthma who provided saliva samples, patients were predominantly male (61%) and non-Hispanic Whites (53%). The mean age was 8.8 (± 4.3) years (Table II). Four-hundred and thirteen (56%) children with asthma were receiving inhaled glucocorticoids daily, of which FP was prescribed most frequently (65%). Of the 268 children receiving daily FP, the median asthma control score was 4 (range 0-15). There was no difference in the age, sex, or race / ethnicity of children receiving daily FP when compared with the overall asthma cohort. However, despite no difference in mean height (123.2 ± 24.1 cm vs. 126.2 ± 28.4 cm; P=0.5), FP treated patients had a lower mean body weight (31.7 ± 21.2 kg vs. 37.3 ± 21.7 kg; P=0.003).
Table 2.
Population characteristics of asthmatic children and young adults who underwent genotyping for CYP3A polymorphisms
| Variables | Number (%) |
|---|---|
| Total no. of subjects | 734 |
| Subjects receiving daily inhaled glucocorticoids | 413 (56.3) |
| Fluticasone propionate | 268 (36.5) |
| Flunisolide | 10 (1.4) |
| Triamcinolone acetonide | 2 (0.3) |
| Beclomethasone dipropionate | 31 (4.2) |
| Budesonide | 56 (7.6) |
| Ciclesonide | 2 (0.3) |
| Mometasone | 7 (1.0) |
| Fluticasone propionate / salmeterol xinafoate | 82 (11.2) |
| Budesonide / formoterol | 11 (1.5) |
| Age, yrs | 8.8 ± 4.3 |
| Sex | |
| Male | 443 (60.5) |
| Female | 289 (39.5) |
| Race | |
| American Indian / Alaskan Native | 9 (1.2) |
| Asian | 6 (0.8) |
| Black | 24 (3.3) |
| Native Hawaiian / Pacific Islander | 15 (2.0) |
| White | 554 (75.5) |
| Unknown / Not reported | 122 (16.6) |
| Ethnicity | |
| Hispanic | 102 (14.0) |
| Non-Hispanic | 211 (28.9) |
| Unknown / Not reported | 418 (57.2) |
| Weight, kg | 35.1 ± 21.6 |
| Height, cm | 124.9 ±.26.6 |
| Asthma control score | |
| Subjects receiving daily fluticasone | 4.8 ± 3.6 |
| Subjects receiving other inhaled glucocorticoids | 4.9 ± 3.6 |
NOTE: Values are presented as mean + SD where applicable.
DNA quantification
Saliva was collected from all 734 enrolled subjects for DNA extraction. The DNA yield varied substantially among individuals; the average concentration was 32.2 ng/μL (± 53.5 ng/μL) with a median of 17.4 (>1-821.03) ng/μL. Ninety-five percent of the samples had DNA concentrations which met the Applied Biosystems’ recommendations for TaqMan reactions of 1-20 ng of DNA per reaction.25
SNP genotyping
FP was the only inhaled glucocorticoid for which the sample size was sufficient to study the distribution of allelic variants among the 9 SNPs in the CYP3A gene locus. Nine probe hydrolysis (TaqMan) based genotyping assays were performed upon 734 saliva samples with a success rate of 99.96%. Genotyping results were available for all 268 patients who were treated daily with FP, and CYP3A SNPs were compared with their asthma control scores (Table III). Variant allele frequencies ranged from 1% to 33% and differed among individual SNPs. Two SNPs deviated from Hardy-Weinberg Equilibrium; however, we were unable to explain these findings in evaluation of the analytical assays or local referral patterns.
Table 3.
Association of CYP3A genetic polymorphisms and asthma control scores among children receiving daily inhaled fluticasone propionate
| Polymorphism | Reference SNP (rs #) |
Test for Hardy Weinberg Equilibrium (P-value) |
Variant Allele Frequency |
Association with Asthma Control Scores (P-value) ⊗ |
|---|---|---|---|---|
| CYP3A4 | ||||
| CYP3A4*22 | rs35599367 | 0.29 | 0.04 | 0.02 |
| CYP3A4 int 7 | rs2246709 | 0.12 | 0.33 | 0.42 |
| CYP3A4 int 7 | rs4646437 | 0.03 | 0.17 | 0.54 |
| CYP3A5 | ||||
| CYP3A5*3 | rs776746 | 0.33 | 0.15 | 0.51 |
| CYP3A5*6 | rs10264272 | 0.80 | 0.01 | 0.86 |
| CYP3A5*1D | rs15524 | 0.30 | 0.15 | 0.96 |
| CYP3A7 | ||||
| CYP3A7 | rs2687133 | 0.13 | 0.14 | 0.55 |
| CYP3A7*2 | rs2257401 | 0.04 | 0.15 | 0.50 |
| CYP3A7 6 nt 5′ of ex 14 | rs2740565 | 0.18 | 0.16 | 0.69 |
Bonferroni adjustment for multiple comparisons.
Twenty-eight percent of the patients treated with FP daily exhibited CYP3A5 genetic variants. No associations were observed between CYP3A5*3 (P=0.5), CYP3A5*6 (P=0.9), and CYP3A5*1D (P=1.0) alleles and asthma control scores in this patient cohort.
For CYP3A7, allelic variants, CYP3A7*2, and rs2740565 located downstream of the canonical CYP3A7 (6 nucleotides 5′ of exon 14 of CYP3A7.1L) were screened in this population of children with asthma requiring FP therapy. Homozygosity for the variants tested was infrequent, occurring in 2.6-3.4% of subjects. Individuals heterozygous for a CYP3A7 genetic variant accounted for 22.8-23.5% of subjects with asthma. Genotypic status, however, was not significantly associated with asthma control scores among patients screened for any of the CYP3A7 variants.
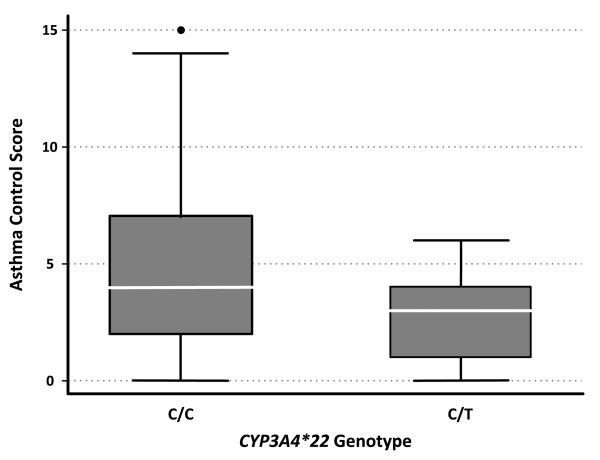
For the SNPs in CYP3A4, no association was found among patients with CYP3A4 rs2246709 or CYP3A4 rs4646437 genetic variants. We also assessed rs4646437 and asthma control by sex, as it has been reported to feature sex-dependent functional changes;19 however, we found no difference by sex. In contrast, there was a significant difference among patients with a CYP3A4*22 variant T-allele, who were more likely to have lower asthma control scores, indicating improved control, when compared with those who featured the C/C genotype (C/T mean: 2.9 (± 2.2) vs. C/C mean: 5.0 (± 3.7); P=0.02) (Figure 2).
Figure 2.
The box plot depicts pediatric asthma control scores among patients receiving daily fluticasone, stratified by CYP3A4*22 genotype. Boxes indicate the median and interquartile ranges. Vertical lines above and below boxes indicate the maximum and minimum values. Asthma control scores are scaled from 0 (well-controlled) to 15 (poorly-controlled). The variant T allele was significantly associated with lower asthma control scores, indicating improved asthma control (P=0.02).
CYP3A4*22
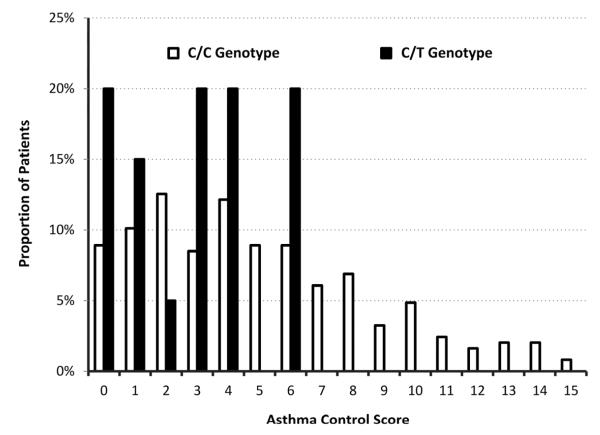
Among the 20 children who featured the CYP3A4*22 variant allele, average asthma control scores were significantly lower compared with the 247 children with asthma who did not feature the SNP (2.9 ± 2.2 vs. 5.0 ± 3.7; P=0.02) (Figure 2). Asthma control scores of 0-6 were recorded for children with a CYP3A4*22 T-allele, and scores among children with the C/C genotype ranged from 0-15 (Figure 3; available at www.jpeds.com). The presence of the variant T-allele was associated with a decreased average asthma control score by 2.10 points (95% CI: 0.46-3.75) out of 15. No subjects were homozygous for the CYP3A4*22 T-allele.
Figure 3.
Asthma control scores stratified by CYP3A4*22 genotype. Children with asthma featured wild-type (C/C) and variant (C/T) alleles. Scores are ranked from 0 (well-controlled) to 15 (poorly-controlled).
Discussion
We report variants within the CYP3A locus which may explain a portion of the variability in therapeutic response to inhaled FP among children with asthma. Despite the predominance of CYP3A5 and CYP3A7 in human lung tissue, we could not identify an association between several CYP3A5 and CYP3A7 polymorphisms and asthma control. Even though CYP3A4 is a minor constituent of the CYP3A enzymes in the lung,7 a polymorphism in CYP3A4 was associated with improved asthma control.
We have shown in this study that improved asthma control was associated with the presence of the CYP3A4*22 allele, which has been shown to decrease hepatic CYP3A4 mRNA expression and enzyme activity.17 Reduced pulmonary enzyme activity would likely prolong the presence of active FP within the airway and increase its effectiveness. Reduced hepatic CYP3A4 expression would be expected to result in higher FP concentrations in the systemic circulation, but such an effect would be dependent on the amount of drug that is systemically available following inhalation. Available data in children indicate that the mean steady-state concentrations of FP in the systemic circulation range from 107 to 183 pg/mL, depending on the particular inhalation chamber used.26 FP is a potent inactivator of CYP3A5, and to a lesser extent CYP3A4, such that drug which was not metabolized in the liver may inhibit its own metabolism.12 Reduced enzyme activity by CYP3A4*22 has been associated with concentration-related toxicity by cyclosporine, including an increased risk of delayed graft function and decreased creatinine clearance in kidney transplant patients.27 Additionally, this SNP has been associated with a decreased dose requirement among kidney transplant patients receiving the immunosuppressive agent tacrolimus, which is primarily a CYP3A5 substrate.28 However, Elens et al found that the combination of CYP3A4*22 and CYP3A5*1 exerted a larger effect on cyclosporine clearance than either SNP individually consistent with the multiple enzymatic pathways involved in metabolic clearance of cyclosporine.27 In the present study, more patients were well-controlled with an asthma score of zero in the group with CYP3A4*22. This finding is in agreement with our hypothesis that reduced enzymatic activity by CYP3A4*22 increases airway exposure to FP. It is also consistent with the previous observation that the clinical effectiveness of statins at conventional doses is increased by CYP3A4*22.
Although the association is evident, the physiologic basis for the association between improved asthma control with FP and CYP3A4*22 is uncertain. Other possible explanations exist. An enteral or swallowed dose of FP is cleared by first-pass metabolism in the liver and gastrointestinal tract.29 In the presence of the CYP3A4*22 allele hepatic CYP3A4 expression would be reduced, which could theoretically increase systemic exposure. This might also increase adverse effects associated with inhaled glucocorticoids such as adrenal suppression and cataracts. Alternatively, this association with CYP3A4*22 may be a consequence of linkage disequilibrium with another SNP, which regulates CYP3A expression in the lung. As a result of CYP3A7′s close proximity to CYP3A4 it is a likely candidate; however, linkage disequilibrium with CYP3A5 is also possible.
CYP3A4*22 is located in intron 6 of CYP3A4 and has not been reported to be in linkage disequilibrium with any known common polymorphisms.17 Mounting evidence suggests that genetic variants in both coding and non-coding regions of genomic DNA can have functional implications.30 Genetic variants can alter mRNA levels and protein levels, thereby resulting in phenotypic changes. Several mechanisms have been proposed as potential ways in which an intronic SNP may be associated with disease in humans. Defects in transcription, alternative splicing of the gene transcript, and alterations of RNA processing are all possible mechanisms by which an intronic polymorphism can lead to dysfunctional proteins in different disease states, including inflammatory processes.31, 32
The molecular mechanism underlying the direct functional significance of CYP3A4*22 is unclear. However, Wang et al reported that in silico folding of single-stranded DNA and RNA was altered by variation at the site of CYP3A4*22, potentially influencing the binding capacity of key regulatory proteins.17 Functional assessment of CYP3A4*22 in pediatric asthma patients is required to more thoroughly understand the basis of its effect upon pediatric asthma control with FP. Further research is needed to quantify the impact of this SNP on CYP3A4 activity and FP biotransformation. Even without these additional studies, the correlation between CYP3A4*22 and improved asthma control with inhaled FP has the potential to improve successful selection of an inhaled glucocorticoid for the treatment of individuals with severe asthma.
The metabolic pathways leading to biotransformation and inactivation of other inhaled glucocorticoids may be influenced in a manner similar to that of FP. For glucocorticoids metabolized by other CYPs, similar mechanisms involving inactivating SNPs in CYP3A5 and CYP3A7 may enhance their effectiveness. For example, beclomethasone dipropionate is primarily metabolized by CYP3A5,33 whose expression level is reduced in patients carrying the CYP3A5*3, leading to a similar increase in effectiveness. However, the number of patients in the current study treated with inhaled glucocorticoids other than FP is too small to permit a meaningful analysis of the correlation between specific inactivating SNPs and asthma control scores.
This study has several limitations. We did not directly measure CYP3A expression or activity; however, previous studies have shown that the CYP3A4*22 SNP decreases total CYP3A4 mRNA levels and enzyme activity in vivo.17 Furthermore, our effect estimates have wide confidence intervals, reflecting the small sample size of our study population. To minimize the effects of population stratification between study groups, we have recruited all participants from the same children’s hospital. Additionally, we did not measure pulmonary function and do not have an objective, physiologic measure of asthma control for the children included in this analysis. Although pulmonary function tests (PFTs) are objective measures that can be tracked with time and during various interventions, they may not accurately reflect the impact of asthma on the patient’s overall health and level of daily functionality.34 In addition, standard spirometry measurements require patient cooperation that is seldom possible in patients at the youngest age range of our study. We therefore utilized self-reported scoring measures of lung function based upon NIH criteria to stratify subjects according to their degree of asthma control. For young children the questionnaire was frequently completed by the parent/caregiver rather than the child. A study which compared asthma symptom agreement between children and parents revealed that parental reporting may result in a more complete dataset, but is also subject to underreporting of asthma symptoms.35 Lastly, we are limited by self-reporting of medication compliance and do not have an objective, pharmacologic measure of drug concentrations such as saliva or blood concentrations. The clinical relevance of this finding requires confirmation with a larger sample and external validation in other populations with asthma. Until other mechanisms are identified, our data suggest that patients with decreased CYP3A4 activity may benefit from improved asthma control with inhaled FP.
Acknowledgments
We thank Bradley W. Thomas and Amber Bagherian for performing the genotyping of our research samples (Leeder Laboratory).
Supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development (R01HD060559) and Primary Children’s Medical Center Research Foundation and the Center for Clinical and Translational Sciences (CTSA 5UL1RR025764-02). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health & Human Development or the National Institutes of Health. The authors declare no conflicts of interest.
Abbreviations
- FP
fluticasone propionate
- SNPs
single nucleotide polymorphisms
- CYP
cytochrome P450
- PFTs
pulmonary function tests
- SD
standard deviation
- CI
confidence interval
- ATS
American Thoracic Society
- NIH
National Institutes of Health
Appendix.
Pediatric asthma control scores were calculated by summing the patient’s responses (assessed on a 4-point scale) for all of the above questions. Overall score: 0 (well-controlled) – 15 (poorly-controlled).
 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Akinbami LJ, Sullivan SD, Campbell JD, Grundmeier RW, Hartert TV, Lee TA, et al. Asthma outcomes: healthcare utilization and costs. J Allergy Clin Immunol. 2012;129:S49–64. doi: 10.1016/j.jaci.2011.12.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Barnes PJ. Efficacy of inhaled corticosteroids in asthma. J Allergy Clin Immunol. 1998;102:531–8. doi: 10.1016/s0091-6749(98)70268-4. [DOI] [PubMed] [Google Scholar]
- [3].Expert Panel Report 3 (EPR-3): Guidelines for the Diagnosis and Management of Asthma-Summary Report 2007. J Allergy Clin Immunol. 2007;120:S94–138. doi: 10.1016/j.jaci.2007.09.043. [DOI] [PubMed] [Google Scholar]
- [4].Chung KF, Caramori G, Adcock IM. Inhaled corticosteroids as combination therapy with beta-adrenergic agonists in airways disease: present and future. Eur J Clin Pharmacol. 2009;65:853–71. doi: 10.1007/s00228-009-0682-z. [DOI] [PubMed] [Google Scholar]
- [5].Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua AA, Himes BE, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365:1173–83. doi: 10.1056/NEJMoa0911353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mager DE, Moledina N, Jusko WJ. Relative immunosuppressive potency of therapeutic corticosteroids measured by whole blood lymphocyte proliferation. J Pharm Sci. 2003;92:1521–5. doi: 10.1002/jps.10402. [DOI] [PubMed] [Google Scholar]
- [7].Pearce RE, Leeder JS, Kearns GL. Biotransformation of fluticasone: in vitro characterization. Drug Metab Dispos. 2006;34:1035–40. doi: 10.1124/dmd.105.009043. [DOI] [PubMed] [Google Scholar]
- [8].Anttila S, Hukkanen J, Hakkola J, Stjernvall T, Beaune P, Edwards RJ, et al. Expression and localization of CYP3A4 and CYP3A5 in human lung. Am J Respir Cell Mol Biol. 1997;16:242–9. doi: 10.1165/ajrcmb.16.3.9070608. [DOI] [PubMed] [Google Scholar]
- [9].Biggs JS, Wan J, Cutler NS, Hakkola J, Uusimaki P, Raunio H, et al. Transcription factor binding to a putative double E-box motif represses CYP3A4 expression in human lung cells. Molecular pharmacology. 2007;72:514–25. doi: 10.1124/mol.106.033795. [DOI] [PubMed] [Google Scholar]
- [10].Leeder JS, Gaedigk R, Marcucci KA, Gaedigk A, Vyhlidal CA, Schindel BP, et al. Variability of CYP3A7 expression in human fetal liver. The Journal of pharmacology and experimental therapeutics. 2005;314:626–35. doi: 10.1124/jpet.105.086504. [DOI] [PubMed] [Google Scholar]
- [11].Leclerc J, Tournel G, Courcot-Ngoubo Ngangue E, Pottier N, Lafitte JJ, Jaillard S, et al. Profiling gene expression of whole cytochrome P450 superfamily in human bronchial and peripheral lung tissues: Differential expression in non-small cell lung cancers. Biochimie. 2010;92:292–306. doi: 10.1016/j.biochi.2009.12.007. [DOI] [PubMed] [Google Scholar]
- [12].Murai T, Reilly CA, Ward RM, Yost GS. The inhaled glucocorticoid fluticasone propionate efficiently inactivates cytochrome P450 3A5, a predominant lung P450 enzyme. Chemical research in toxicology. 2010;23:1356–64. doi: 10.1021/tx100124k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nature genetics. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- [14].Juniper EF, Guyatt GH, Cox FM, Ferrie PJ, King DR. Development and validation of the Mini Asthma Quality of Life Questionnaire. Eur Respir J. 1999;14:32–8. doi: 10.1034/j.1399-3003.1999.14a08.x. [DOI] [PubMed] [Google Scholar]
- [15].Reddel HK, Taylor DR, Bateman ED, Boulet LP, Boushey HA, Busse WW, et al. An official American Thoracic Society/European Respiratory Society statement: asthma control and exacerbations: standardizing endpoints for clinical asthma trials and clinical practice. Am J Respir Crit Care Med. 2009;180:59–99. doi: 10.1164/rccm.200801-060ST. [DOI] [PubMed] [Google Scholar]
- [16].Taylor DR, Bateman ED, Boulet LP, Boushey HA, Busse WW, Casale TB, et al. A new perspective on concepts of asthma severity and control. Eur Respir J. 2008;32:545–54. doi: 10.1183/09031936.00155307. [DOI] [PubMed] [Google Scholar]
- [17].Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bhatnagar V, Garcia EP, O’Connor DT, Brophy VH, Alcaraz J, Richard E, et al. CYP3A4 and CYP3A5 polymorphisms and blood pressure response to amlodipine among African-American men and women with early hypertensive renal disease. American journal of nephrology. 2010;31:95–103. doi: 10.1159/000258688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schirmer M, Rosenberger A, Klein K, Kulle B, Toliat MR, Nurnberg P, et al. Sex-dependent genetic markers of CYP3A4 expression and activity in human liver microsomes. Pharmacogenomics. 2007;8:443–53. doi: 10.2217/14622416.8.5.443. [DOI] [PubMed] [Google Scholar]
- [20].Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–9. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- [21].Quaranta S, Chevalier D, Allorge D, Lo-Guidice JM, Migot-Nabias F, Kenani A, et al. Ethnic differences in the distribution of CYP3A5 gene polymorphisms. Xenobiotica; the fate of foreign compounds in biological systems. 2006;36:1191–200. doi: 10.1080/00498250600944300. [DOI] [PubMed] [Google Scholar]
- [22].Rodriguez-Antona C, Axelson M, Otter C, Rane A, Ingelman-Sundberg M. A novel polymorphic cytochrome P450 formed by splicing of CYP3A7 and the pseudogene CYP3AP1. The Journal of biological chemistry. 2005;280:28324–31. doi: 10.1074/jbc.M502309200. [DOI] [PubMed] [Google Scholar]
- [23].Siegel S, Castellan NJ., Jr. Nonparametric Statistics for the Behavioral Sciences. 2nd ed McGraw-Hill; New York: 1988. [Google Scholar]
- [24].Fisher RA. The design and analysis of experiments. Oliver & Boyd; Edinburgh and London: 1935. [Google Scholar]
- [25].TaqMan SNP. Genotyping Assays -- Protocol. Part Number 4332856 Rev. B. Applied Biosystems. 2004 Jun; [Google Scholar]
- [26].Khan Y, Tang Y, Hochhaus G, Shuster JJ, Spencer T, Chesrown S, et al. Lung bioavailability of hydrofluoroalkane fluticasone in young children when delivered by an antistatic chamber/mask. J Pediatr. 2006;149:793–7. doi: 10.1016/j.jpeds.2006.08.022. [DOI] [PubMed] [Google Scholar]
- [27].Elens L, Bouamar R, Hesselink DA, Haufroid V, van Gelder T, van Schaik RH. The new CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) is associated with an increased risk of delayed graft function and worse renal function in cyclosporine-treated kidney transplant patients. Pharmacogenet Genomics. 2012;22:373–80. doi: 10.1097/FPC.0b013e328351f3c1. [DOI] [PubMed] [Google Scholar]
- [28].Elens L, Bouamar R, Hesselink DA, Haufroid V, van der Heiden IP, van Gelder T, et al. A new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipients. Clin Chem. 2011;57:1574–83. doi: 10.1373/clinchem.2011.165613. [DOI] [PubMed] [Google Scholar]
- [29].Harding SM. The human pharmacology of fluticasone propionate. Respiratory medicine. 1990;84(Suppl A):25–9. doi: 10.1016/s0954-6111(08)80004-2. [DOI] [PubMed] [Google Scholar]
- [30].Cooper DN. Functional intronic polymorphisms: Buried treasure awaiting discovery within our genes. Hum Genomics. 2010;4:284–8. doi: 10.1186/1479-7364-4-5-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Do TN, Ucisik-Akkaya E, Davis CF, Morrison BA, Dorak MT. An intronic polymorphism of IRF4 gene influences gene transcription in vitro and shows a risk association with childhood acute lymphoblastic leukemia in males. Biochim Biophys Acta. 2010;1802:292–300. doi: 10.1016/j.bbadis.2009.10.015. [DOI] [PubMed] [Google Scholar]
- [32].Kwok JB, Hallupp M, Loy CT, Chan DK, Woo J, Mellick GD, et al. GSK3B polymorphisms alter transcription and splicing in Parkinson’s disease. Ann Neurol. 2005;58:829–39. doi: 10.1002/ana.20691. [DOI] [PubMed] [Google Scholar]
- [33].Somers GI, Lindsay N, Lowdon BM, Jones AE, Freathy C, Ho S, et al. A comparison of the expression and metabolizing activities of phase I and II enzymes in freshly isolated human lung parenchymal cells and cryopreserved human hepatocytes. Drug Metab Dispos. 2007;35:1797–805. doi: 10.1124/dmd.107.015966. [DOI] [PubMed] [Google Scholar]
- [34].Spahn JD, Chipps BE. Office-based objective measures in childhood asthma. J Pediatr. 2006;148:11–5. doi: 10.1016/j.jpeds.2005.08.077. [DOI] [PubMed] [Google Scholar]
- [35].Magzamen S, Mortimer KM, Davis A, Tager IB. School-based asthma surveillance: a comparison of student and parental report. Pediatr Allergy Immunol. 2005;16:669–78. doi: 10.1111/j.1399-3038.2005.00304.x. [DOI] [PubMed] [Google Scholar]