Abstract
Isosilybin A (1) is one of the major flavonolignans that constitute silymarin, an extract of the fruits (achenes) of milk thistle [Silybum marianum (L.) Gaertn. (Asteraceae)]. The chemistry of the Silybum flavonolignans has been studied for over four decades, and the absolute configuration of 1 has been determined previously by electronic circular dichroism (ECD) and X-ray crystallography via correlating the relative configuration of the phenylpropanoid moiety to the established absolute configuration of the 3-hydroxyflavanone portion of the molecule. Herein we report the X-ray crystallographic structure of the product of the reaction of 1 with 4-bromobenzoyl chloride, and, thus, the absolute configuration of 1 was established as (2R, 3R, 7″R, 8″R) directly via X-ray crystallography of an analogue that incorporated a heavy atom. The results were consistent with previously reported assignments and verified the absolute configuration of the diastereoisomer of 1, isosilybin B, and the related diastereoisomeric regioisomers, silybin A and silybin B.
We have been studying the chemistry of flavonolignans from milk thistle extract for nearly a decade. This includes isolating gram-scale quantities of individual flavonolignans1 that enabled exploration of milk thistle's prostate cancer chemopreventive activities,2-8 hepatoprotective activities,9-12 and metabolism via cytochrome P450 2C9.13 Continued interest in Silybum flavanolignans and recent discussions with collaborators has prompted us to review the absolute configuration of these constituents. While a wealth of spectroscopic and spectrometric evidence has been amassed, primarily using ECD, MS, NMR and X-ray crystallography data, direct X-ray crystallographic analyses involving a heavy atom to confirm the absolute configuration conclusively has yet to be reported.
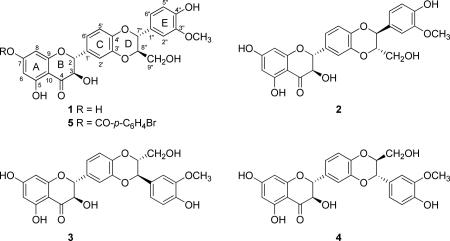
To chronicle milk thistle briefly, Silybum marianum (L.) Gaertn. (Asteraceae) has been utilized for centuries for hepatoprotective properties, and several reviews should be examined to learn more about the history and medicinal properties of this plant14-19 or the nomenclature of compounds isolated therefrom.20 Silymarin is the most commonly examined crude extract of the fruits (technically, achenes), and the bioactive constituents in silymarin have been the subject of more than 100 chemistry manuscripts over the past 40 plus years, largely because their biological activities continue to be studied and the technology to isolate, analyze, and characterize them has evolved progressively. A breakthrough in the chemistry of silymarin was reported in 1974, when Wagner and colleagues21 demonstrated that silymarin was a mixture of flavonolignans, and what was initially referred to as silymarin corresponded to silybin, likely formed from the flavonoid taxifolin and the phenylpropanoid coniferyl alcohol. Indeed, the term ‘flavonolignan’ was first coined by Pelter and Hänsel22 when they characterized the scaffold of these compounds and proposed that the phenylpropanoid could react with taxifolin in two different fashions, thus leading to what would later become the silybin vs isosilybin regioisomers.23 The silybin and isosilybin regioisomers were each shown to be a mixture of the corresponding diastereoisomers at C-7″ and C-8″ of the D-ring, with anti relative configuration across all four compounds (1-4). The purification and structure elucidation of seven of the major flavonolignans in silymarin was first reported in 2003, thereby establishing the absolute configuration of isosilybin A (1), isosilybin B (2), silybin A (3), and silybin B (4) using ECD;24,25 later that same year Lee and Liu26,27 reported the isolation and configuration of 1-4, together with the X-ray crystallographic data for 1. Gram-scale quantities of these compounds, principally 1-4, were made possible through the initial work of Graf et al.1 Kren and colleagues have also made significant progress in the gram-scale isolation and characterization of 3 and 4 via a semi-synthetic approach.28,29
There have been two reports of X-ray crystallographic data for flavonolignans from silymarin, both of which only established relative configurations. Besides the aforementioned report on 1 in 2003,26 a 1983 study by Lotter and Wagner30 examined a crystal of the 1:1 mixture of the diastereoisomers, 3 and 4 (often referred to as silibinin).20 In fact, the tendency for 3 and 4 to co-crystallize can be utilized as a key step toward purification of the individual flavonolignans on a multi-gram scale.1
The absolute configuration of the flavonolignans from milk thistle has been determined largely via comparison to standards using ECD. For example, the absolute configuration at C-2 and C-3 in 1–4 (all with R configurations) were established by comparing ECD spectra with those of known dihydroflavonols31-33 and by chemical synthesis of various isomers.34-36 In a similar manner, the ECD spectra for the phenypropanoid portion of compounds 1–4 were compared with model compounds to determine the configuration at C-7″ and C-8″, thus establishing the absolute configuration of compounds 1–4.24 Complementing that finding, the X-ray crystallographic data for isosilybin A (1) together with the established absolute configurations at C-2 and C-3 through prior ECD studies, and in conjunction with comparison of specific rotations, permitted an independent determination of the absolute configuration of 1–4.26 Given that the configurations at C-2 and C-3 are well established based on comparisons to flavonoids, one could consider deriving the relative configurations at C-7″ and C-8″ via NOESY experiments. However, those pairs of stereogenic centers were too distant to observe appropriate NOESY correlations (data not shown). In summary, the absolute configuration of the four stereogenic centers in 1–4 has been pieced together using different types of data, with particular emphasis on ECD.
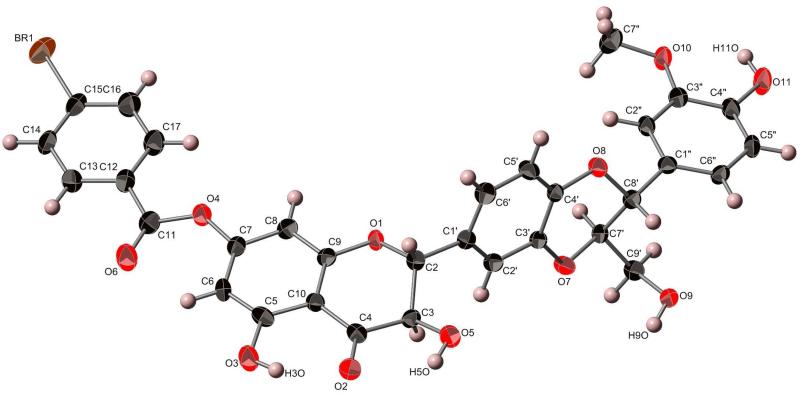
To define the absolute configuration of the stereogenic centers of the four main flavonolignans directly, we first crystallized isosilybin A (1), and the preliminary X-ray crystallography data (not shown) matched that of Lee and Liu.26 Then, by reacting 1 with 4-bromobenzoyl chloride37 and crystallizing the purified product (5; 8 mg) by slow evaporation in MeOH, suitable crystals were isolated for X-ray crystallographic analysis with the presence of a heavy atom, permitting the determination of the absolute configuration of 1 (Figure 1) and confirming the earlier results using ECD24,25 or a combination of ECD, X-ray crystallography, and specific rotation.26,27 The HRMS data for 5 showed m/z = 687.0463, corresponding to C32H2579BrO11Na (calcd 687.0472), and m/z = 689.0444, corresponding to C32H2581BrO11Na (calcd 689.0452). The 1H NMR data of 5 supported incorporation of a C-7 monobromobenzoyl group into 1, based on the chemical shifts and integration (see Supporting Information). The hypothesis for the reaction was that the 4-bromobenzoyl chloride would react with the C-9″ primary hydroxy group; however, there was literature precedence using similar reagents for esterification at one of the phenolic moieties in the flavonolignans, particularly the 7-OH group.38,39
Figure 1.
X-ray crystallographic structure of 7-(4-bromobenzoyl)isosilybin A (5).40
Colorless single crystals of the bromobenzoyl product (5) were, at 193(2) K, orthorhombic, space group P212121, with a = 4.7129(6) Å, b = 16.470(2) Å, c = 38.853(5) Å, V = 3015.8(6) Å3 and Z = 4. Data were collected with a Bruker APEX CCD diffractometer ( radiation, graphite monochromator) at 193(2) K (cold N2 gas stream) using standard CCD techniques, yielding 31 437 integrated reflection intensities of which 5515 were unique, [R(int) = 0.0814]. The structure was solved with the Bruker SHELXTL software using direct methods techniques. All non-solvent non-hydrogen atoms were refined with anisotropic thermal parameters; non-solvent hydrogen atoms were included as idealized isotropic atoms “riding” on their respective carbon or oxygen atoms. The structure contained a region of disordered electron density that was modeled as disordered methanol of crystallization. A total of six partially occupied (1/3 normal occupancy) sites were refined as oxygen or carbon atoms. These atoms were refined isotropically, and no solvent hydrogen atoms were included in the refinement model. The resulting structural parameters were to convergence (R1 = 0.0484 for 4225 I>2σ(I) data, wR2 = 0.1037 for all 5515 reflections, GOF = 1.046, absolute structure (Flack) parameter = 0.02(1)). Crystallographic data were deposited with the Cambridge Crystallographic Data Centre (deposition code: CCDC 893140). Copies of the data can be obtained, free of charge, on application to the Director, CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (deposit @ccdc.cam.ac.uk).
In conclusion, the absolute configuration of isosilybin A (1) has been established as (2R, 3R, 7″R, 8″R) via X-ray crystallographic analysis of the heavy atom analogue, 7-(4-bromobenzoyl)isosilybin A (5), supporting the validity of earlier findings.24-27 Moreover, these data, in conjunction with the well-established ECD data24 (see Supporting Information) confirm the absolute configuration of the diastereoisomer of 1, isosilybin B (2) and the diastereoisomeric regioisomers of 1, silybin A (3) and silybin B (4).
Supplementary Material
ACKNOWLEDGEMENT
This research was supported in part by R01 AT006842 from the National Center for Complementary and Alternative Medicine/National Institutes of Health, Bethesda, MD. At the University of North Carolina at Greensboro, we thank Mr. Tyler Graf for isolating the isosilybin A starting material and Drs. Mitch Croat and Maria Elena Meza Avina for assistance with the synthetic chemistry portions of this manuscript. We also thank Drs. Guido Pauli, James McAlpine, and José Napolitano at the University of Illinois at Chicago and Dr. Stephen J. Polyak at the University of Washington for helpful discussions.
Footnotes
ASSOCIATED CONTENT
Supporting Information. 1H NMR spectra of compounds 1 and 5 and ECD spectra of compounds 1, 2, and 5. These materials are available free of charge on the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Graf TN, Wani MC, Agarwal R, Kroll DJ, Oberlies NH. Planta Med. 2007;73:1495–1501. doi: 10.1055/s-2007-990239. [DOI] [PubMed] [Google Scholar]
- 2.Davis-Searles PR, Nakanishi Y, Kim NC, Graf TN, Oberlies NH, Wani MC, Wall ME, Agarwal R, Kroll DJ. Cancer Res. 2005;65:4448–4457. doi: 10.1158/0008-5472.CAN-04-4662. [DOI] [PubMed] [Google Scholar]
- 3.Deep G, Oberlies NH, Kroll DJ, Agarwal R. Carcinogenesis. 2007;28:1533–1542. doi: 10.1093/carcin/bgm069. [DOI] [PubMed] [Google Scholar]
- 4.Deep G, Oberlies NH, Kroll DJ, Agarwal R. Oncogene. 2008;27:3986–3998. doi: 10.1038/onc.2008.45. [DOI] [PubMed] [Google Scholar]
- 5.Deep G, Oberlies NH, Kroll DJ, Agarwal R. Int. J. Cancer. 2008;123:41–50. doi: 10.1002/ijc.23485. [DOI] [PubMed] [Google Scholar]
- 6.Deep G, Raina K, Singh RP, Oberlies NH, Kroll DJ, Agarwal R. Int. J. Cancer. 2008;123:2750–2758. doi: 10.1002/ijc.23879. [DOI] [PubMed] [Google Scholar]
- 7.Deep G, Gangar SC, Oberlies NH, Kroll DJ, Agarwal R. Mol. Carcinog. 2010;49:902–912. doi: 10.1002/mc.20670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deep G, Gangar SC, Rajamanickam S, Raina K, Gu M, Agarwal C, Oberlies NH, Agarwal R. PLoS One. 2012;7:e34630. doi: 10.1371/journal.pone.0034630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morishima C, Shuhart MC, Wang CC, Paschal DM, Apodaca MC, Liu Y, Sloan DD, Graf TN, Oberlies NH, Lee DY, Jerome KR, Polyak SJ. Gastroenterology. 2010;138:671–681. doi: 10.1053/j.gastro.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polyak SJ, Morishima C, Lohmann V, Pal S, Lee DY, Liu Y, Graf TN, Oberlies NH. Proc. Natl. Acad. Sci. USA. 2010;107:5995–5999. doi: 10.1073/pnas.0914009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagoner J, Negash A, Kane OJ, Martinez LE, Nahmias Y, Bourne N, Owen DM, Grove J, Brimacombe C, McKeating JA, Pecheur EI, Graf TN, Oberlies NH, Lohmann V, Cao F, Tavis JE, Polyak SJ. Hepatology. 2010;51:1912–1921. doi: 10.1002/hep.23587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagoner J, Morishima C, Graf TN, Oberlies NH, Teissier E, Pecheur EI, Tavis JE, Polyak SJ. PLoS One. 2011;6:e16464. doi: 10.1371/journal.pone.0016464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brantley SJ, Oberlies NH, Kroll DJ, Paine MF. J. Pharmacol. Exp. Ther. 2010;332:1081–1087. doi: 10.1124/jpet.109.161927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abenavoli L, Capasso R, Milic N, Capasso F. Phytother. Res. 2010;24:1423–1432. doi: 10.1002/ptr.3207. [DOI] [PubMed] [Google Scholar]
- 15.Deep G, Agarwal R. Cancer Metast. Rev. 2010;29:447–463. doi: 10.1007/s10555-010-9237-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramasamy K, Agarwal R. Cancer Lett. 2008;269:352–362. doi: 10.1016/j.canlet.2008.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal R, Agarwal C, Ichikawa H, Singh RP, Aggarwal BB. Anticancer Res. 2006;26:4457–4498. [PubMed] [Google Scholar]
- 18.Comelli MC, Mengs U, Schneider C, Prosdocimi M. Integr. Cancer Ther. 2007;6:120–129. doi: 10.1177/1534735407302349. [DOI] [PubMed] [Google Scholar]
- 19.Gazak R, Walterova D, Kren V. Curr. Med. Chem. 2007;14:315–338. doi: 10.2174/092986707779941159. [DOI] [PubMed] [Google Scholar]
- 20.Kroll DJ, Shaw HS, Oberlies NH. Integr. Cancer Ther. 2007;6:110–119. doi: 10.1177/1534735407301825. [DOI] [PubMed] [Google Scholar]
- 21.Wagner H, Diesel P, Seitz M. Arzneimittelforschung. 1974;24:466–471. [PubMed] [Google Scholar]
- 22.Pelter A, Hänsel R. Tetrahedron Lett. 1968:2911–2916. [Google Scholar]
- 23.The term ‘flavonolignan’ may be somewhat inappropriate, as lignans result from the dimerization of two phenylpropanoids via a carbon-carbon bond. Given that these compounds represent a chimera of a flavonoid and a phenylpropanoid, and that the product results in a 1,4-dioxane moiety (i.e. carbon-oxygen bonds), an alternate term, such as ‘flavonolignoid’, may be more appropriate.
- 24.Kim NC, Graf TN, Sparacino CM, Wani MC, Wall ME. Org. Biomol. Chem. 2003;1:1684–1689. doi: 10.1039/b300099k. [DOI] [PubMed] [Google Scholar]
- 25.Kim NC, Graf TN, Sparacino CM, Wani MC, Wall ME. Org. Biomol. Chem. 2003;1:3470. doi: 10.1039/b300099k. [DOI] [PubMed] [Google Scholar]
- 26.Lee DYW, Liu YZ. J. Nat. Prod. 2003;66:1171–1174. doi: 10.1021/np030163b. [DOI] [PubMed] [Google Scholar]
- 27.Lee DYW, Liu YZ. J. Nat. Prod. 2003;66:1632–1632. [Google Scholar]
- 28.Monti D, Gazak R, Marhol P, Biedermann D, Purchartova K, Fedrigo M, Riva S, Kren V. J. Nat. Prod. 2010;73:613–619. doi: 10.1021/np900758d. [DOI] [PubMed] [Google Scholar]
- 29.Kren V, Gazak R, Purchartova K, Marhol P, Biedermann D, Sedmera P. J. Mol. Catal. B Enzym. 2009;61:247–251. [Google Scholar]
- 30.Lotter H, Wagner H. Z. Naturforsch. C. 1983;38:339–341. [Google Scholar]
- 31.Gaffield W. Tetrahedron. 1970;26:4093–&. [Google Scholar]
- 32.Pelter A, Hänsel R. Chem. Ber. 1975;108:790–802. [Google Scholar]
- 33.Arnone A, Merlini L, Zanarotti A. J. Chem. Soc., Chem. Commun. 1979:696–697. [Google Scholar]
- 34.Merlini L, Zanarotti A, Pelter A, Rochefort MP, Hänsel R. J. Chem. Soc., Chem. Commun. 1979:695–695. [Google Scholar]
- 35.Merlini L, Zanarotti A, Pelter A, Rochefort MP, Hänsel R. J. Chem. Soc., Perkin Trans. 1. 1980:775–778. [Google Scholar]
- 36.Tanaka H, Shibata M, Ohira K, Ito K. Chem. Pharm. Bull. 1985;33:1419–1423. doi: 10.1248/cpb.33.1419. [DOI] [PubMed] [Google Scholar]
- 37.Deyrup ST, Swenson DC, Gloer JB, Donald TW. J. Nat. Prod. 2006;69:608–611. doi: 10.1021/np050460b. [DOI] [PubMed] [Google Scholar]
- 38.Wang F, Huang KX, Yang LX, Gong JX, Tao QF, Li HB, Zhao Y, Zeng S, Wu XM, Stöckigt J, Li XK, Qu J. Bioorg. Med. Chem. 2009;17:6380–6389. doi: 10.1016/j.bmc.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 39.Gazak R, Valentova K, Fuksova K, Marhol P, Kuzma M, Medina MA, Oborna I, Ulrichova J, Kren V. J. Med. Chem. 2011;54:7397–7407. doi: 10.1021/jm201034h. [DOI] [PubMed] [Google Scholar]
- 40.The numbering of the compound in the crystallographic structure corresponds to the numbering scheme in the CIF file as deposited with the Cambridge Crystallographic Data Centre, deposition code: CCDC 893140. However, the numbering shown in the structural diagram preserves the biosynthetic consideration of the compound, where the eastern side of the dioxane originates from a phenypropanoid moiety.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.