Abstract
The Golgi receives the entire output of newly synthesized cargo from the endoplasmic reticulum (ER), processes it in the stack largely through modification of bound oligosaccharides, and sorts it in the trans-Golgi network (TGN). GRASP65 and GRASP55, two proteins localized to the Golgi stack and early secretory pathway, mediate processes including Golgi stacking, Golgi ribbon linking, and unconventional secretion. Previously we have shown that GRASP depletion in cells disrupts Golgi stack formation. Here we report that knockdown of the GRASP proteins, alone or combined, accelerates protein trafficking through the Golgi membranes but also has striking negative effects on protein glycosylation and sorting. These effects are not caused by Golgi ribbon unlinking, unconventional secretion, or ER stress. We propose that GRASP55/65 are negative regulators of exocytic transport and that this slowdown helps to ensure more complete protein glycosylation in the Golgi stack and proper sorting at the TGN.
Introduction
The Golgi apparatus plays a central role in the trafficking and processing of membrane and secretory proteins in the exocytic and endocytic pathways as well as in multiple recycling routes. It is present in almost all eukaryotic cells and is composed of stacks of flattened cisternae that are laterally linked to form a perinuclear ribbon-like structure in mammalian cells1. The Golgi matrix proteins, a group of proteins localized at the cytoplasmic face of the Golgi membranes, play important roles in Golgi stack and ribbon formation, membrane tethering, unconventional secretion, Golgi biogenesis during cell division, as well as cell cycle progression, apoptosis, and directed cell migration2–9. Among these Golgi matrix proteins, two homologues, GRASP65 (Golgi reassembly stacking protein of 65 kDa) and GRASP55 (55 kDa) were first discovered as Golgi stacking factors10, 11. Antibodies against GRASP65 or GRASP55 inhibited post-mitotic stacking of newly formed cisternae both in vitro10, 11 and in vivo12. Depletion of GRASP65 or GRASP55 in mammalian cells by RNA interference (RNAi) reduced the number of cisternae per stack13–15, and simultaneous depletion of both GRASPs resulted in complete disassembly of the Golgi stacks14. In Drosophila S2 cells, depletion of the sole Drosophila GRASP protein dGRASP and its interacting protein, dGM130, resulted in the disassembly of the Golgi stacks into single cisternae and vesicles16.
In addition to Golgi stacking, GRASP65 and GRASP55 have been shown to have other functions, including ribbon linking, cargo transport, unconventional secretion, cell cycle progression, apoptosis, and Golgi remodeling in migrating cells12, 13, 17–21. GRASP65 and GRASP55 form homodimers and dimers from adjacent membranes further form trans-oligomers that tether membranes into close proximity12, 22. GRASPs are localized between the cisternae, where their oligomers hold cisternal membranes into stacks11, 14, 15. It was also proposed that GRASPs localize to the rims of the Golgi stacks and link Golgi stacks into a ribbon17, 20. GRASP65 may also exert its tethering function through its interacting protein GM130. This interaction is thought to participate in the docking of p115 tethered COPII vesicles23. It has also been shown that the GM130/GRASP65 complex recycles between the cis-Golgi and the late ER-Golgi intermediate compartment (ERGIC), the membrane station that dynamically protrudes from the Golgi and undergoes homotypic fusion with intermediate compartments24. Although the GRASP proteins play essential roles in Golgi structure formation, how they regulate Golgi functions has not been well studied.
In our previous study, we inhibited the function of GRASP65 by microinjecting anti-GRASP65 antibodies into cells and we observed an increased rate of protein transport, probably due to the acceleration of vesicle formation from unstacked cisternae25. However, the microinjection technique used in this previous study did not allow us to perform the necessary biochemical and cell biology experiments to examine protein glycosylation, sorting and other cellular activities. In the present study, we knocked down GRASP55, GRASP65, or both proteins by RNAi and determined the changes in protein trafficking, glycosylation, and sorting. Our results reveal essential roles for GRASP55/65 in protein glycosylation and sorting.
Results
GRASP depletion does not cause apoptosis or ER stress
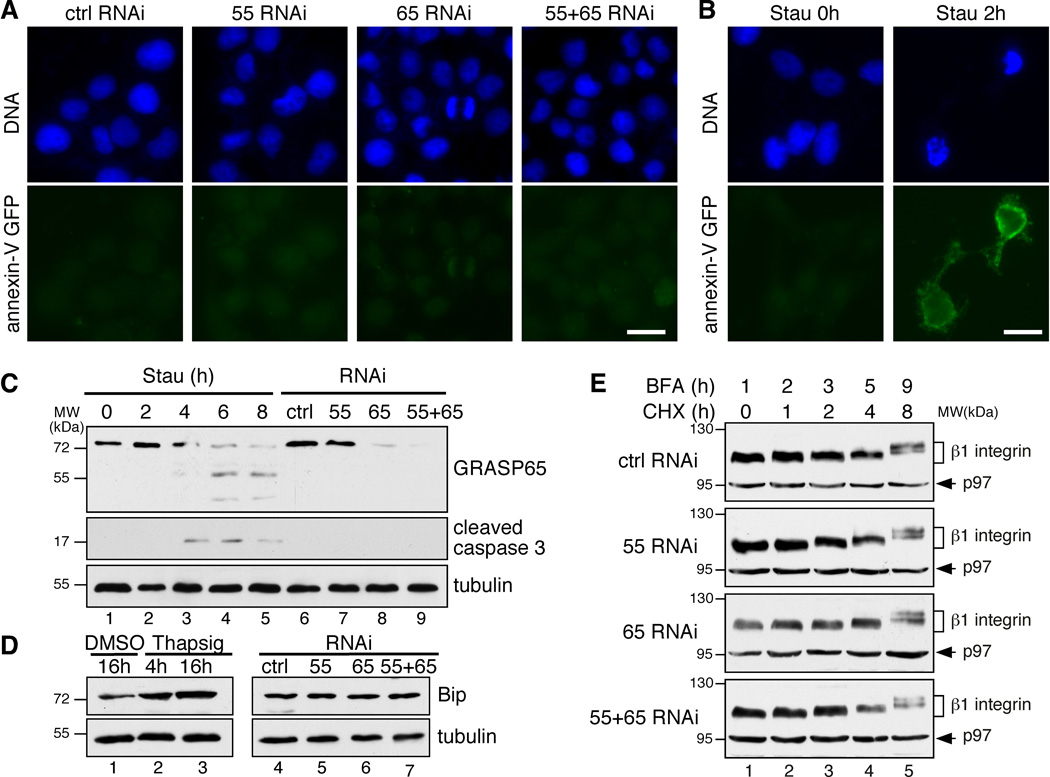
To determine the biological roles of GRASP55/65, we knocked down GRASP55, GRASP65, or both proteins in rtTA HeLa m2 cells with RNAi as previously described14, 15. The depletion was efficient, determined by immunofluorescence microscopy and Western blot (Supplementary Figure S1). GRASP depletion had no significant effect on the cellular levels of coat proteins, Rabs and tethers, SNAREs, membrane fusion proteins, Golgi enzymes and other Golgi structural proteins in the cell except GM130, which is reduced when both GRASP55 and GRASP65 were depleted, consistent with a previous observation14. To determine if GRASP depletion causes apoptosis in our system as previously reported13, we stained non-permeabilized cells with annexin V–GFP, a widely used marker for early apoptosis26, 27. Cells treated with GRASP RNAi were not stained with annexin V, while cells treated with staurosporine, a known apoptosis inducer, were readily stained (Figure 1A–B). Furthermore, neither nuclear fragmentation nor cleaved caspase 3 was detected in the GRASP-depleted cells (Figure 1A, 1C). These results demonstrate that GRASP depletion does not cause apoptosis in the cells in the present study.
Figure 1. Depletion of GRASP55/65 does not cause apoptosis or ER stress.
(A) HeLa cells transfected with the indicated RNAi were stained with annexin V-GFP before fixation. Note that no obvious annexin V staining was observed on GRASP-depleted cells. (B) HeLa cells treated with 2 µM staurosporine (Stau) for 0 h or 2 h were stained with annexin V-GFP before fixation. Scale bars in A-B, 10 µm. (C) HeLa cells transfected with the indicated RNAi or treated with staurosporine (ST) were analyzed by Western blot. Note that the cleaved caspase 3 was detected after 2 h of staurosporine treatment, but not following the depletion of the GRASPs. (D) Depletion of GRASPs does not cause ER stress. HeLa cells transfected with the indicated RNAi or treated with DMSO or 2 µM thapsigargin (Thapsig) for 4 h or 16 h were analyzed by SDS-PAGE and Western blot. Note that the protein level of Bip increased after thapsigargin treatment, but not following GRASP depletion. (E) GRASP depletion does not result in ERAD. HeLa cells transfected with indicated RNAi were first treated with 5 µg/ml BFA for 1 h followed by the addition of 100 µM cycloheximide (CHX) into the medium and incubated for the indicated time period. Shown are Western blots of cell lysate for β1 integrin and p97 on the same gels. Molecular weight standards (kDa) are indicated on the left in C–E.
GRASP depletion also does not cause ER stress. Unlike thapsigargin-treated cells, Bip expression was not increased by GRASP depletion (Figure 1D). We then determined whether GRASP depletion increases protein degradation in the ER. Cells were first treated with Brefeldin A (BFA) to trap membrane and secretory proteins in the ER, then treated with cycloheximide (CHX) to block protein synthesis, and protein levels were measured by Western blot. The reduction of β1 integrin, Lamp2, and cathepsin D over time in GRASP-depleted cells was comparable to control RNAi-treated cells (Figure 1E, Supplementary Figure S2A–B). In contrast, ER stress induced by thapsigargin treatment reduced the level of β1 integrin in the cell by ER-associated degradation (ERAD), an effect that was rescued by the proteasome inhibitor MG132 (Supplementary Figure S2C). These results indicate that GRASP depletion does not cause ER stress or ERAD.
GRASP depletion accelerates integrin trafficking
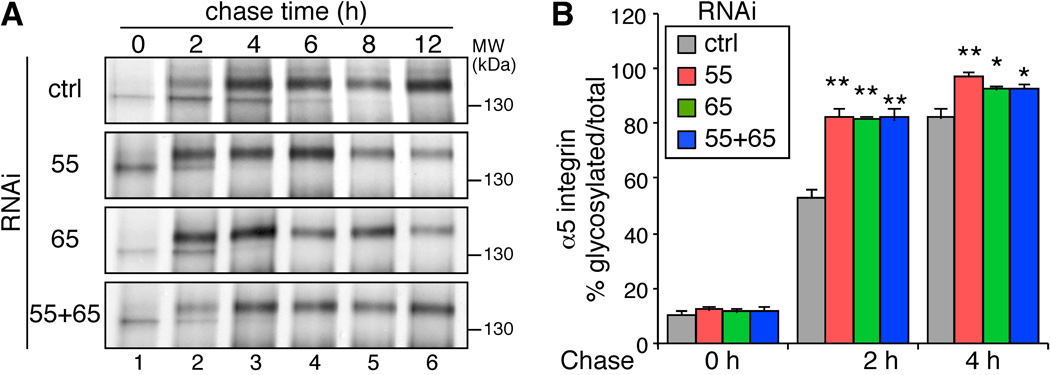
To determine the roles of GRASP55/65 in protein trafficking, we performed pulse-chase experiments to examine the effects of GRASP depletion on the trafficking of α5 integrin, a type I transmembrane glycoprotein on the cell surface. Cells were labeled with 35S-labeled methionine and cysteine followed by chases for different time periods. α5 integrin was then immunoprecipitated and analyzed by autoradiography. After 0 h of chase, α5 integrin in all cells existed predominantly in their immature forms (Figure 2A). After 2 h of chase, the mature form of α5 integrin, indicated by its higher molecular weight due to glycan processing in the Golgi, began to appear. Approximately 53% of the α5 integrin was detected as the mature form in the cells treated with control RNAi, while the proportion of the mature form increased to over 80% in cells depleted of GRASP55, GRASP65, or both. After 4 h and 6 h of chase, detectable amounts of immature α5 integrin existed in the control cells, whereas almost all of the α5 integrin had matured in the GRASP-depleted cells (Figure 2). These results demonstrate that GRASP depletion increases trafficking of plasma membrane proteins through the conventional trafficking route.
Figure 2. Depletion of GRASPs accelerates α5 integrin trafficking.
(A) GRASP depletion increases α5 integrin trafficking. HeLa cells transfected with the indicated RNAi were labeled with 250 µCi/ml TRANS 35S-LABEL for 1 h, chased for the indicated time periods, and lysed. α5 integrin were immunoprecipitated and analyzed by SDS-PAGE and autoradiography. Note that the mature form of α5 integrin appeared more rapidly in GRASP-depleted cells. (B) The ratio of the mature form vs. total α5 integrin at the indicated chase time for one representative result out of three independent experiments. Shown are the quantitation results from three independent experiments. P-value was determined by Student’s t-test; *, p < 0.05; **, p<0.01; ***, p<0.001 in this and all following figures. Note that the maturation rate of α5 integrin increased in GRASP-depleted cells.
GRASP depletion accelerates VSV-G traffic through the Golgi
To further dissect the effect of GRASP depletion on protein trafficking, we monitored the transport of the vesicular stomatitis virus glycoprotein (VSV-G) temperature-sensitive mutant (ts045)28. Cells were infected with VSV-G ts045-GFP adenovirus at 40.5°C for 16 h and then shifted to 32°C to release VSV-G from the ER to the Golgi (Supplementary Figure S3). After 40.5°C block, the VSV-G molecules in all cells were retained in the ER. After 15 min incubation at 32°C, VSV-G started to accumulate in the Golgi and this process was accelerated in GRASP-depleted cells. After 30 min, the accumulation in GRASP-depleted cells was complete; whereas in control RNAi treated cells, a significant amount of VSV-G was found in the ER. After 45 min, VSV-G containing vesicles emerged in GRASP-depleted cells but not in control RNAi treated cells. After 60 min, VSV-G began to appear at the plasma membrane in GRASP-depleted cells, but not in control cells. These results demonstrate that GRASP depletion increases VSV-G trafficking through the Golgi.
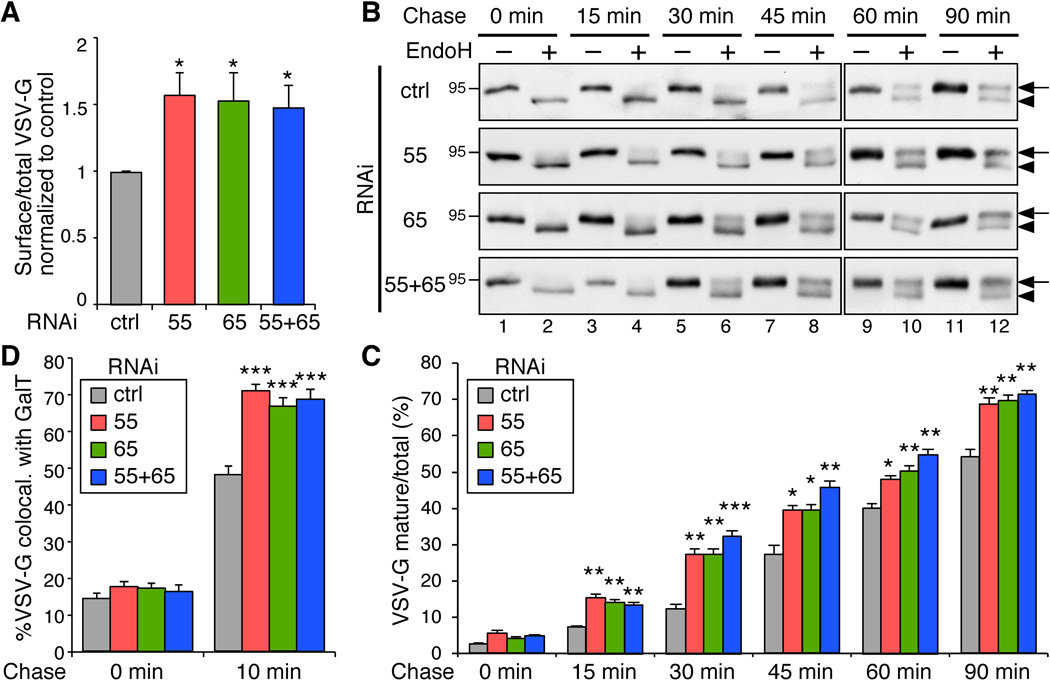
To quantitatively measure the traffic rate of VSV-G, we labeled VSV-G on the cell surface with an antibody against its extracellular domain and assessed the ratio of surface VSV-G to total VSV-G GFP signal by flow cytometry 2 h after 32°C release. As shown in Figure 3A, the ratio of surface-to-total VSV-G was increased in the cells depleted of GRASP55, GRASP65, or both. These results are consistent with the previous observation that CD8 trafficking is accelerated in NRK cells in which GRASP65 function is blocked by microinjected antibodies25.
Figure 3. GRASP depletion accelerates VSV-G trafficking through the Golgi membranes.
(A) VSV-G traffic to the cell surface is accelerated by GRASP depletion. HeLa cells stably expressing VSV-G-ts045-GFP under an inducible promoter were transfected with control (ctrl), GRASP55 (55), GRASP65 (65), or both GRASP55 and GRASP65 (55+65) RNAi. The cells were incubated with doxycycline at 40.5°C for 16 h to induce VSV-G expression and shifted to 32°C for 120 min to release the VSV-G from the ER. The cells were collected, and the VSV-G on the surface was stained with an antibody against its extracellular domain followed by incubation with fluorescently labeled secondary antibodies. Total VSV-G and surface-bound VSV-G were assessed by flow cytometry. The ratio of surface-to-total VSV-G normalized against control cells (mean±SEM) are shown based on three independent experiments. (B) GRASP depletion accelerates VSV-G trafficking. HeLa cells transfected with indicated RNAi were infected with VSV-G ts045-GFP adenovirus and incubated at 40.5°C for 16 h. Cells were shifted to 32°C, incubated for indicated time period (chase), and lysed. Proteins were treated with EndoH and analyzed by Western blot for VSV-G-GFP. Arrows indicate the EndoH resistant form and arrowheads indicate the EndoH sensitive form of VSV-G. Molecular weight markers are indicated on the left. (C) Quantitation of (B) to indicate the percentage of VSV-G in EndoH resistant form. Note that the difference between the GRASP-depleted cells and control RNAi treated cells is most dramatic after 15–45 min chase when VSV-G travels through the Golgi membranes. (D) GRASP depletion accelerates VSV-G trafficking through the Golgi membranes. HeLa cells transfected with indicated RNAi were infected with VSV-G ts045-GFP adenovirus and incubated at 40.5°C for 16 h. Cells were shifted to 15°C for 2 h and then to 20°C for 0 and 10 min followed by immunofluorescence microscopy. The colocalization of VSV-G with GalT was quantified with ImageJ. Shown are representative results (mean±SEM from 20 different cells) from three independent experiments.
To specifically analyze the effect of GRASP depletion on VSV-G trafficking through the Golgi membranes, we collected cell lysates at different points of 32°C release and treated the samples with endoglycosidase H (endoH), an enzyme that removes mannose rich (ER/cis-Golgi form) but not complex (Golgi and post Golgi form) N-linked oligosaccharides from glycoproteins. The change of a glycoprotein from endoH sensitive to endoH resistant has been widely used to monitor protein trafficking through the Golgi, while proteins transported by unconventional secretion bypass the Golgi and thus are not modified by Golgi enzymes18, 19. Western blot showed that VSV-G trafficking is increased in GRASP-depleted cells compared to control cells (Figure 3B–C). In control RNAi-treated cells, the endoH resistant form of VSV-G started to appear after 30 min chase and became stronger over time. In GRASP-depleted cells, the endoH resistant form of VSV-G appeared earlier (e.g. after 15 min chase) and increased faster than that in control cells. The difference between GRASP-depleted cells and control cells was most obvious after 30 min and 45 min chase, the time period when VSV-G traveled through the Golgi stack (Supplementary Figure S3).
To pinpoint the effect of GRASP depletion on VSV-G trafficking across the Golgi membranes, we first accumulated VSV-G in the ERGIC by incubating cells at 15°C29 and then shifted the cells to 20°C to allow VSV-G to travel from the ERGIC to the trans Golgi30, 31. Cells were analyzed at different time points of 20°C incubation by fluorescence microscopy for colocalization of VSV-G with galactosyltransferase-1 (GalT), a trans-Golgi marker. As shown in Figure 3D, after 10 min incubation, 48.2 ± 2.2% of VSV-G colocalized with GalT in control cells, while it was increased to 70.8 ± 1.8%, 66.8 ± 1.9% and 68.5 ± 2.6% in cells depleted of GRASP55, GRASP65 or both GRASPs, respectively (Figure 3D). Taken together, these results demonstrate that GRASP depletion increases protein trafficking through the Golgi membranes.
GRASP depletion enhances coat - membrane interaction
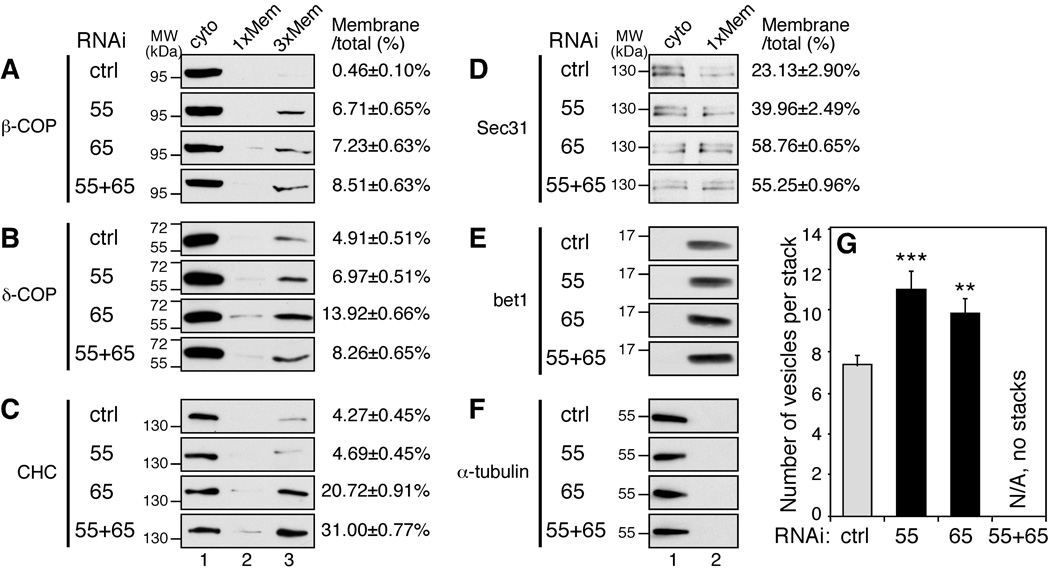
To determine the mechanism of the enhanced protein trafficking by GRASP depletion, we assessed the effect of GRASP depletion on vesicle formation. By subcellular fraction (Figure 4A–F), we found that membrane association of both β -COP and δ-COP, two subunits of the coatomer complex, significantly increased upon GRASP depletion. In addition to COPI, membrane association of clathrin heavy chain (CHC) and the COPII component Sec31 also increased. These results are consistent with our previous data that unstacked Golgi membranes generate COPI vesicles more rapidly in an in vitro budding assay25. In addition, quantitation of EM images of GRASP-depleted cells showed that GRASP depletion reduced the number of cisternae per stack14, 15, but the average number of vesicles adjacent to each distinguishable Golgi stack in single GRASP-depleted cells was significantly higher than in control RNAi treated cells (Figure 4G). Cells depleted of both GRASP55 and GRASP65 did not usually contain distinguishable Golgi stacks, with a large number of vesicles and single cisternae in the perinuclear region14. Collectively, these results demonstrate that GRASP depletion causes Golgi cisternal unstacking and enhances protein trafficking by stimulating vesicle formation.
Figure 4. GRASP depletion enhances membrane association of coat proteins.
HeLa cells transfected with indicated RNAi were subjected to subcellular fractionation. Proteins in the cytosol and membrane fractions were analyzed by Western blot for β-COP (A), δ-COP (B), clathrin heavy chain (CHC, C), sec31 (D), bet1 (E) and α-tubulin (F). In some panels 3 times of the membrane fractions were loaded onto the SDS-PAGE (lane 3). Shown are representative images from 3 independent experiments. Quantification results (mean±SEM) are shown on the right to indicate the percentage of the protein found in the membrane fractions. (G) GRASP depletion causes vesiculation of the Golgi. Quantitation of EM images of GRASP-depleted cells for the number of vesicles adjacent to a Golgi stack. Shown are representative results (mean±SEM from 20 different cells) from three independent experiments. Cells with both GRASP55 and GRASP65 depleted did not usually contain distinguishable Golgi stacks and thus were not quantified.
GRASP depletion impairs proper protein glycosylation
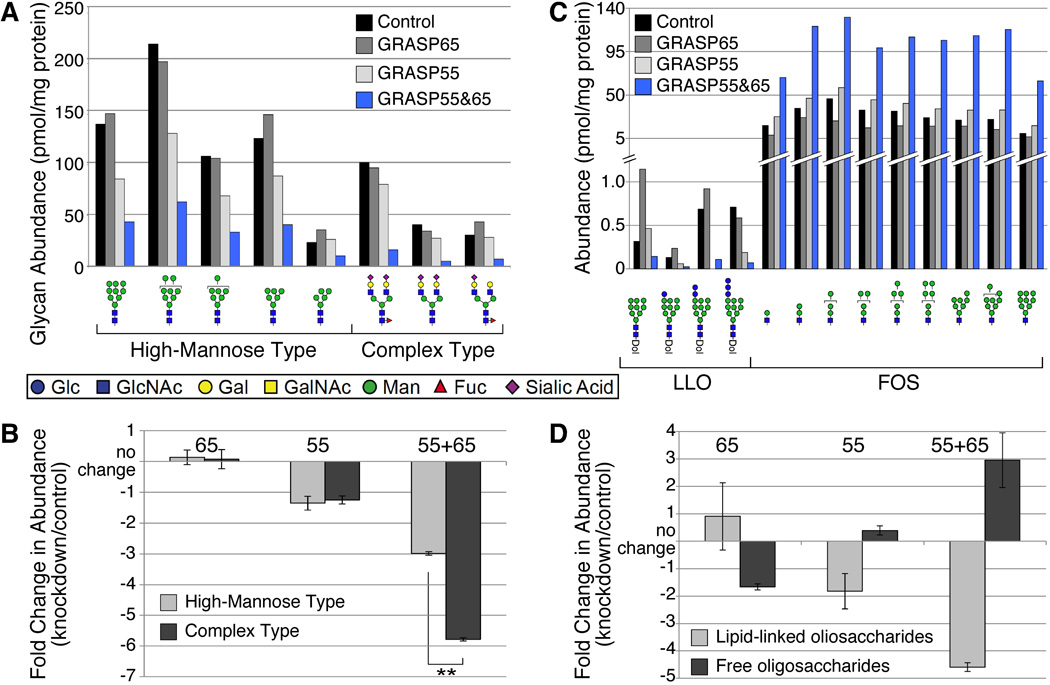
Since GRASP proteins slow the flow of cargo proteins, it was of interest to examine protein glycosylation in GRASP-depleted cells. To directly assess glycomic changes associated with GRASP depletion, we harvested N-linked glycans from control and GRASP-depleted cells, verified the structure of the most highly abundant high-mannose and complex glycans by multiple-stage mass spectrometry (MSn), and quantified them relative to an external glycan standard (Figure 5A). Depletion of both GRASP55 and GRASP65 resulted in a substantial decrease in N-linked glycoprotein glycans compared to that associated with equivalent amounts of proteins from control cells. Single GRASP knockdowns showed somewhat variable increases or decreases in the abundance of individual glycans, although knockdown of GRASP55 was more consistently decreased in the major glycans.
Figure 5. Glycomic analysis of total protein glycosylation in GRASP-depleted cells.
HeLa cells expressing RNAi targeting the indicated GRASP proteins were collected and processed for glycomic analysis by mass spectrometry. (A) The major high-mannose and complex glycans were quantified relative to an external standard and normalized to protein content. Double knockdown cells exhibit significantly decreased glycan abundance. Glycan structural representations are consistent with the nomenclature proposed by the Consortium for Functional Glycomics: GlcNAc, blue square; Man, green circle; Gal, yellow circle; SA (sialic acid), purple diamond; Fuc, red triangle. (B) The fold-change for each glycan indicated in panel A was calculated (knockdown/control). Average fold-changes ± SEM are plotted; negative values indicate decreased abundance relative to control. Single knockdowns exhibit relatively minor alteration in the pool of major high-mannose and complex glycans, but the double knockdown cells are significantly reduced in both high-mannose and complex glycans. In the double knockdown cells, the fold-reduction in complex glycans is greater than the reduction in high-mannose glycans (Student’s t-test, **, p<0.001). (C) Lipid-linked oligosaccharides (LLO) and free oligosaccharides (FOS) were quantified by mass spectrometry relative to an external standard and normalized to protein content (Dol, dolichol pyrophosphate). Total LLO levels are significantly lower than FOS levels, note change in y-axis scale. All quantifiable LLO species were reduced in the GRASP double knockdown cells, while some LLOs increased and some decreased relative to control in the single knockdown cells. The abundance of all quantifiable FOS species was dramatically increased in double knockdown cells, while single knockdown had relatively little effect on FOS levels. (D) Fold-changes in LLO and FOS, calculated as in panel B, reveals the inverse relationship expected for precursor and product. Comparison to the fold-change in glycoprotein glycan abundance (panel B) emphasizes that LLO availability limits glycosylation. When LLOs are decreased by knockdown, glycoprotein glycosylation is also decreased (55 alone and 55+65) and vice versa (65 alone), although the impact of either single knockdown is much smaller than the double knockdown.
To assess the efficacy of Golgi processing pathways in knockdown cell lines, the fold change in knockdown cells relative to control was calculated for each quantified high-mannose or complex glycan. By this parameter, the double knockdown exhibited a decrease in both high-mannose and complex glycans but the decrease in complex glycans was significantly greater than the decrease in high-mannose glycans (Student’s t-test, p<0.01, Figure 5B), consistent with the notion that GRASP depletion affects glycan processing in the Golgi. Fold changes for these glycan classes in single knockdowns did not reach statistical significance. Thus, GRASP depletion exerted two significant effects on glycoprotein processing: 1) decreased global N-linked glycoprotein glycosylation, and 2) decreased N-linked glycan complexity.
Decreased N-glycosylation suggests altered co-translational initiation of glycoprotein glycosylation in the ER of GRASP-depleted cells. To determine whether decreased glycosylation resulted from reduced availability of N-linked oligosaccharide precursor or increased precursor hydrolysis, we measured the levels of dolichol-linked oligosaccharide (lipid-linked oligosaccharide, LLO) and free oligosaccharide (FOS) (Figure 5C). Double knockdown cells exhibited over 4-fold decrease in LLO and 3-fold increase in FOS (Figure 5D). Single knockdown cells exhibited intermediate, less pronounced changes. These results indicate that GRASP proteins influence early secretory pathway function, a previously undetected activity of these proteins.
GRASP depletion alters cell surface glycan presentation
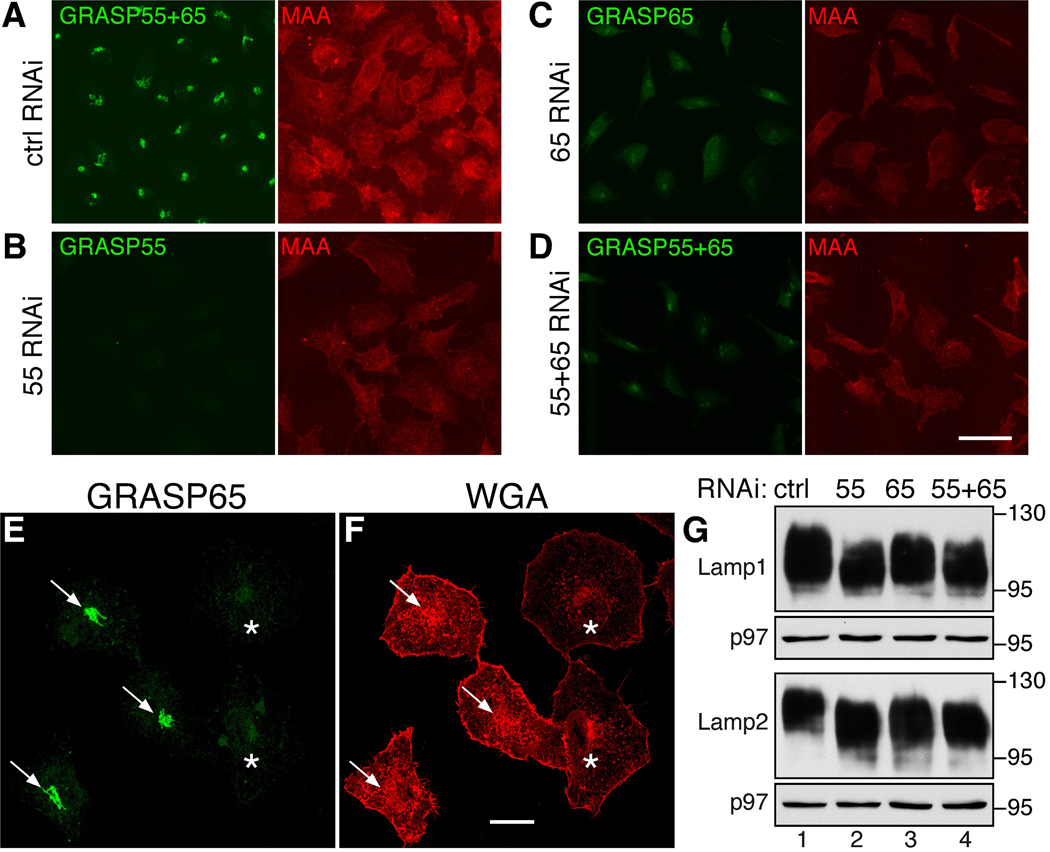
MS analysis of the total cellular glycome does not distinguish between intracellular and cell surface glycan presentation. To determine whether GRASP depletion affects the profile of glycans on the cell surface, we stained non-permeabilized cells with fluorescently labeled lectins. The Maackia amurensis (MAA) lectin binds to α(2,3) sialic acid (SA), a terminal capping monosaccharide residue on glycoproteins and glycosphingolipids32. The intensity of MAA staining decreased in cells depleted of GRASP55, GRASP65, or both, compared to control cells (Figure 6A vs. B–D). To verify this result, we mixed and co-cultured control and GRASP55/65-depleted cells on the same coverslip and stained these cells for Wheat germ agglutinin (WGA), a lectin that selectively binds to SA and N-acetylglucosamine (GlcNAc). The cell surface signal for WGA was significantly reduced in GRASP-depleted cells compared to control cells (Figure 6E–F). The effect of GRASP depletion on protein glycosylation was also confirmed by analysis of individual glycoproteins by SDS-PAGE. We observed higher mobility of Lamp1 and Lamp2 on the gel upon GRASP depletion (Figure 6G). Similar effects were observed by flow cytometry (Supplementary Figure S4A–B) for cells surface-stained by MAA and the Sambucus nigra (SNA) lectin that specifically binds to α(2,6) SA32. The staining pattern of Maclura pomifera (MPA), which binds to the O-linked core 1 disaccharide (Gal β 1-3GalNAc or T-antigen), did not change significantly (Supplementary Figure S4C), indicating that core 1 O-glycosylation is not significantly affected by GRASP depletion. These results demonstrate that GRASP depletion resulted in reduced protein glycosylation, but not reduced protein levels.
Figure 6. Depletion of GRASP results in altered cell-surface glycosylation.
(A–D) Surface staining of fluorophore-conjugated lectin MAA specific for α(2,3) sialic acid was reduced upon the depletion of GRASPs. Non-permeabilized HeLa cells treated with the indicated RNAi were exposed to TRITC-conjugated MAA. The cells were then fixed, stained with GRASP antibodies, and visualized using a confocal microscope. Scale bar, 50 µm. (E–F) HeLa cells transfected with control RNAi (arrows) and with RNAi for GRASP65 and 55 (asterisks) were mixed, cultured together and stained for GRASP65 (green) and WGA (red). Scale bar, 20 µm. (G) GRASP depletion impacts Lamp1 and Lamp2 glycosylation. Indicated GRASP-depleted cells were analyzed for Lamp1, Lamp2 and p97 by Western blots. Molecular weight markers are shown on the right. Note the shift of the bands after GRASP depletion.
GRASP depletion does not impact Golgi enzymes
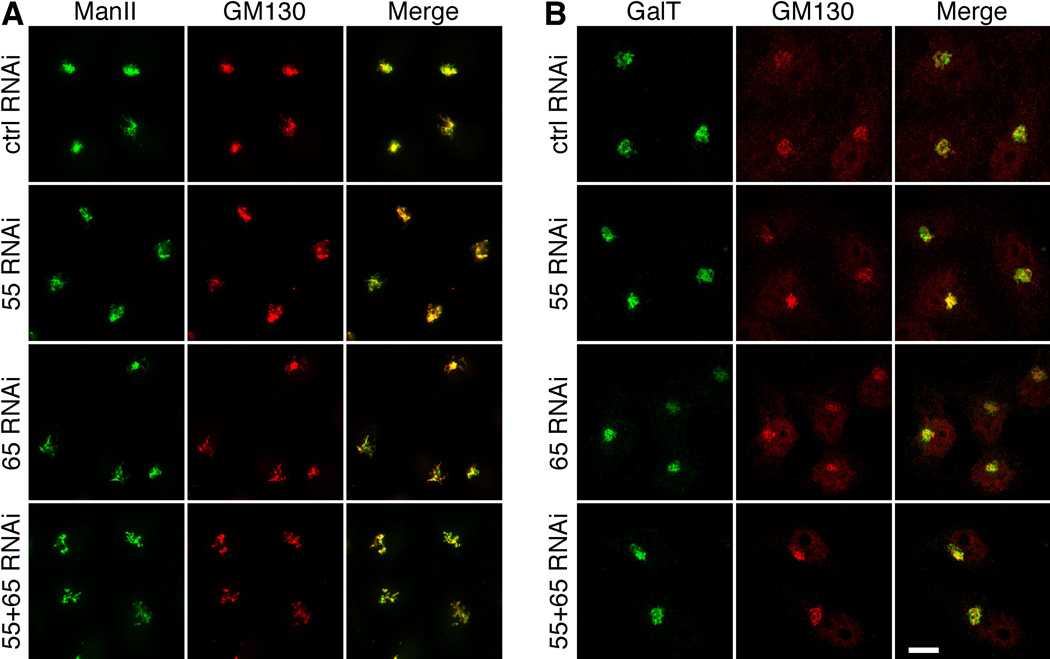
To determine whether the defects in glycoprotein glycosylation were caused by changes in glycan processing enzymes in the Golgi, we determined the expression level and localization of α-mannosidase II (ManII) and GalT by Western blot and microscopy. Our results showed that GRASP depletion did not affect the protein expression level and localizations of these Golgi enzymes (Figure 7, Supplementary Figures S1, S5).
Figure 7. GRASP depletion does not affect the localization of Golgi enzymes.
HeLa cells were treated with the indicated RNAi and analyzed by immunofluorescence microscopy for Mannosidase II (ManII, A), Galactosyltransferase (GalT, B) and GM130. Note that Golgi enzymes colocalized with GM130 in all cells. Scale bar, 10 µm.
GRASP depletion results in cathepsin D secretion
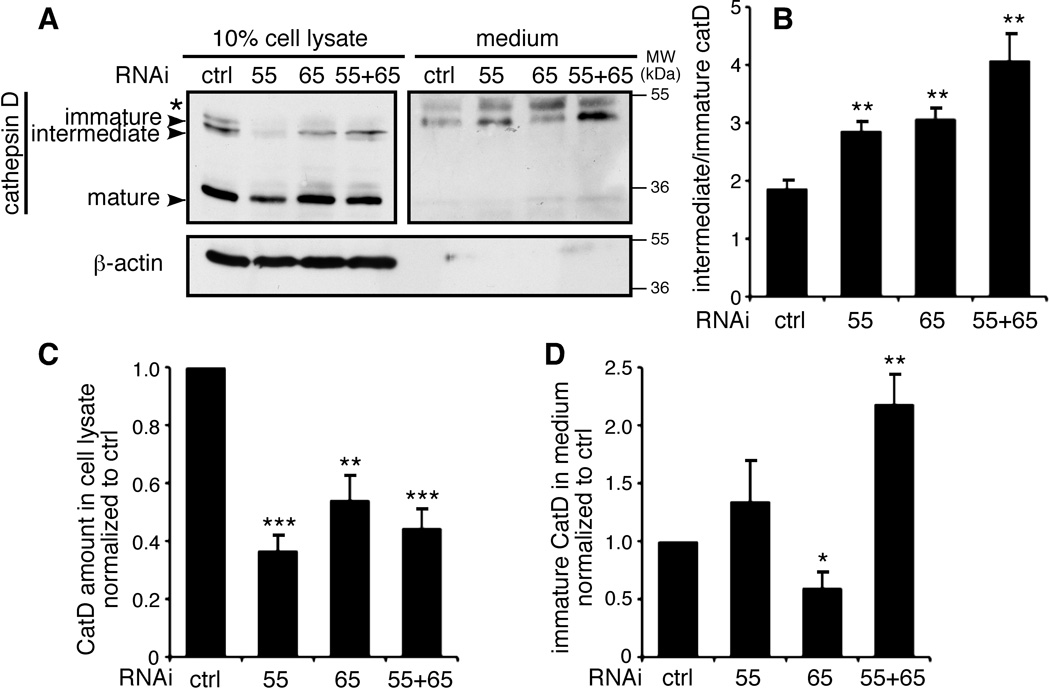
We then determined the effect of GRASP depletion on protein sorting. Cathepsin D is synthesized in the ER as a 53-kDa polypeptide precursor; it is then transported through the Golgi to the lysosome, where the precursor is converted to a 47-kDa intermediate and, subsequently, the 31-kDa mature form33, 34. Cathepsin D has previously been used as a marker for protein sorting35. Cells were incubated in serum-free media for 2 h, and cathepsin D in the cell lysate and media was detected by Western blot. Three bands of cathepsin D were detected in the cell lysate, with the major band being the 31-kDa mature form; in contrast, only the 53-kDa immature form was detected in the media. Depletion of both GRASP55 and GRASP65 increased the amount of immature cathepsin D in the media by two fold (Figure 8, A and D). This was not due to cell lysis, as β-actin was not detected in the medium of these cells. These results suggest that disruption of the Golgi structure results in the missorting of cathepsin D to the extracellular space instead of the endosomes/lysosomes. In addition, the ratio between the intermediate and the immature form increased from 2 in control cells to 3–4 after GRASP depletion, indicating that the transport to the endosomes/lysosomes is enhanced when GRASP proteins are depleted (Figure 8A–B).
Figure 8. GRASP depletion impairs cathepsin D sorting.
(A) Missorting of cathepsin D in cells by GRASP depletion. HeLa cells transfected with the indicated RNAi were washed with PBS and incubated in serum-free DMEM for 2 h. The secreted proteins in the medium were TCA precipitated and normalized based on the total protein in the cell lysate. Equal portions of the cell lysate and medium were analyzed by Western blot for cathepsin D. Immature form: 53 kDa; intermediate form: 47 kDa; mature form: 31 kDa. The asterisk indicates a nonspecific band that appears in the medium. (B) Ratio of intermediate vs. immature cathepsin D in cells transfected with the indicated RNAi by quantifying the related bands on the Western blots in (A). (C) The amount of cathepsin D in the cell lysate normalized to the cells transfected with control RNAi. (D) The amount of immature cathepsin D in the medium normalized to the cells transfected with control RNAi. The data are presented as the mean±SEM of four independent experiments.
The enhanced transport is not through Golgi ribbon unlinking
GRASP65 and GRASP55 have been reported to play a role in Golgi ribbon formation17, 20. Therefore we determined whether Golgi ribbon unlinking was the cause for enhanced trafficking and reduced glycosylation by GRASP-depletion. We induced Golgi ribbon unlinking by the depletion of golgin-8436 and by nocodazole treatment37, 38, none of which induces Golgi unstacking. Both Golgin-84 depletion and nocodazole treatment caused significant Golgi ribbon unlinking and reduced VSV-G trafficking (Supplementary Figure S6), an opposite effect of GRASP depletion. Golgin-84 depletion also reduced glycosylation of Lamp1 and Lamp2, while Nocodazole treatment did not affect cell surface glycosylation. Since depletion of GRASPs did not cause significant fragmentation of the Golgi or mislocalization of Golgi enzymes in our experimental system (Figure 7), these results rule out the possibility that the acceleration of protein trafficking by GRASP depletion is through Golgi ribbon unlinking.
GRASP depletion does not induce unconventional secretion
Previous studies indicate that GRASP65 and GRASP55 are involved in unconventional protein trafficking18, 39, 40. Under stress conditions, some proteins bypass the Golgi to the cell surface in a GRASP-dependent manner. Given that GRASP depletion does not cause ER stress (Figure 1, Supplementary Figure S2), GRASP depletion accelerates rather than slows down trafficking of the marker proteins (Figures 2–3), marker proteins used in this study (e.g. integrins and VSV-G) travel through the Golgi and are glycosylated by Golgi enzymes (Figures 2–3, Supplementary Figure S3), membrane association of coat proteins involved in conventional traffic is enhanced in GRASP-depleted cells (Figure 4), and GRASP depletion reduces the abundance of high-mannose glycans (Figure 5), unconventional secretion is unlikely to be the explanation of the observed effects. Finally, the role of GRASP65 and GRASP55 in unconventional secretion depends on their direct interaction with cargo proteins18. Therefore we determined whether the marker proteins used in this study bind GRASP65 or GRASP55. As shown in Supplementary Figure S7, β1 integrin coimmunoprecipitated with α 5 integrin, but not with GRASP55 or GRASP65. Similarly, Lamp1, Lamp2 and VSV-G did not interact with GRASP proteins, although GRASP65 coprecipitated with GM130. Taken together, these results demonstrate that the enhanced trafficking by GRASP deletion is not caused by unconventional secretion.
Discussion
Previous work has revealed multiple functions for GRASP55 and GRASP65 in Golgi structure formation and function, including Golgi cisternal stacking, ribbon linking, membrane tethering, protein trafficking, unconventional secretion and apoptosis12, 13, 17–21. However, a systematic determination of these functions in the same cellular context has not been reported. In this study, we depleted GRASP55, GRASP65 or both and explored the functional consequences. Depletion of GRASP proteins does not cause ER stress and apoptosis, but accelerates protein trafficking through the Golgi, decreases glycoprotein glycosylation and glycan complexity, and impairs protein sorting.
GRASP depletion increases protein trafficking. We showed that the trafficking of VSV-G and α5 integrin was accelerated in GRASP-depleted cells using microscopy, EndoH digestion, flow cytometry and radioactive pulse-chase labeling. Notably, in the VSV-G experiment, the difference in VSV-G ts045-GFP transport between the control and GRASP RNAi-treated cells was most obvious at 30–45 min post temperature shift, the time when VSV-G travels through the Golgi. In contrast, surface VSV-G protein was more prominent 90–120 min after release in GRASP depleted cells. The effect was significant though less obvious over the course of the longer chases, this may explain why previous studies did not detect the effect of GRASP65 or GRASP55 depletion on VSV-G transport13, 41. Specifically, the EndoH treatment result pinpoints that VSV-G travels through the Golgi membranes faster in GRASP-depleted cells. Similarly, GRASP depletion also increased cathepsin D transport to the endosomal/lysosomal system.
The enhanced protein trafficking by GRASP depletion is not caused by Golgi ribbon unlinking or unconventional secretion. Instead, it might be attributable, at least in part, to Golgi cisternal unstacking. Previously, we inhibited GRASP65 function by microinjection of GRASP65 antibodies into mitotic cells and monitored the transport of CD8 in post-mitotic daughter cells. We showed that the Golgi become largely unstacked and CD8 arrived at the plasma membrane faster in cells with disassembled Golgi25. These observations were confirmed in GRASP-depleted cells in the present study. We hypothesize that when Golgi cisternae are fully stacked, vesicles can only form and fuse at the rims; once the cisternae begin to unstack, more membrane area becomes accessible, thereby increasing the rate of vesicle budding and cargo transport through the Golgi25. In support of this hypothesis, membrane association of coat proteins was increased in cells with unstacked Golgi membranes (Figure 4); unstacking also increased the rate of COPI vesicle formation from Golgi membranes in an in vitro budding assay25. On the other hand, vesicles that travel from one cisterna to the next in intact Golgi stacks may be held by tethering factors that reside on the surface of Golgi membranes, facilitating the fusion of vesicles with Golgi membranes. When the Golgi membranes become fully unstacked by the depletion of both GRASPs, the resulting membrane disorder may impair the function of these tethers. Thus, complete unstacking induced by double GRASP knockdown may induce a significantly different organellar state than partial unstacking in the single GRASP knockdown, perhaps explaining why we did not detect additive effects on protein trafficking when comparing the results of double and single GRASP depletion experiments. In general, our results do not contradict a role for GRASPs as Golgi stacking factors, but also indicate additional functions for these proteins10–12, 14, 15.
GRASP depletion impairs proper protein glycosylation. By lectin binding and MS-based glycomic analysis, we found that GRASP depletion, especially the depletion of both GRASP65 and GRASP55, decreases glycoprotein glycosylation and glycan complexity (Figure 5B). A reasonable explanation for the decrease in glycoprotein glycan complexity is that as cargo moves more rapidly through Golgi compartments, resident enzymes have less time to recognize and modify their substrates. As a result, glycoproteins contain more trimmed high-mannose glycans and less complex structures in the GRASP knockdown cells. Double knockdown has a more significant effect on glycan complexity than either single knockdown, despite the fact that trafficking rates are comparable in single and double knockdowns. Thus, glycan complexity must be affected by additional factors. For instance, Golgi organization is much more severely disturbed in double knockdown cells, which may reflect impaired function of Golgi localized membrane tethers. Since the expression and localization of a subset of Golgi enzymes are unaffected, it is more likely that altered glycosylation reflects changes in cargo trafficking, but not mislocalization or underexpression of Golgi enzymes.
It is not immediately obvious why total N-linked glycan amount should be reduced in double knockdown cells. Therefore, we examined the abundance and fate of LLO, the precursor donor substrates that present the Glc3Man9GlcNAc2 glycan for en bloc transfer onto N-linked sequons of nascent glycoproteins. We detected decreased LLO abundance but increased FOS in GRASP double knockdown cells (Figure 5). These results suggest that GRASP-depletion increases LLO hydrolysis in the absence of ER stress or elevated ERAD. The role of GRASPs appears to be specific for N-glycosylation, as core 1 O-glycosylation in the ER is not significantly affected by GRASP depletion (Supplementary Figure S4C). A role for GRASP in the early secretory pathway is consistent with our observation that upon GRASP depletion, the distribution of Sec31, a COPII coat, was also shifted towards the membrane-bound state (Figure 4), although its expression level was not affected (Supplementary Figure S1B). In addition, the trafficking of VSV-G from the ER to the Golgi is enhanced upon GRASP depletion (Supplementary Figure S3). In Drosophila, dGRASP is localized to ER exit sites16; in mammalian cells, GRASP65 is localized to the Golgi and ERGIC24, but has not been detected in the ER, which argues against a direct role for GRASPs in ER organization and function. Recent studies indicate novel influences of several protein kinases and small metabolic intermediates that, taken together, suggest the existence of novel regulatory mechanisms which might underlie the pathophysiology of some congenital disorders of glycosylation in humans42–46. Therefore, we speculate that a feedback pathway monitors Golgi processing and adjusts the flux of protein trafficking and processing through early secretory compartments.
Neither GRASP proteins nor Golgi stacking may be essential for protein trafficking and cell survival. The Golgi membranes in the budding yeast S. cerevisiae do not form stacks under normal growth conditions47. In yeast and other fungi, N-glycosylation in the Golgi mainly involves the addition of mannoses48, while in multi-cellular organisms N-glycosylation is more complex. Therefore, the multi-function of GRASP proteins and Golgi stacking may be features that were gained during evolution and have essential functions in multi-cellular organisms. Depletion of both GRASP55 and GRASP65 caused missorting of cathepsin D precursors to the extracellular space. This is possibly because of defective glycosylation, or due to the complete disruption of the Golgi stack and the sorting machinery in it. A similar observation has been reported in human mammary cancer cells upon estrogen induction49. It is possible that stack formation ensures that sorting of cargo molecules occurs only when cargo molecules reach the trans-Golgi network, but not in other cisternae.
Our results revealed that GRASP55/65 negatively regulate exocytic transport to ensure more complete glycosylation in the stack and better sorting at the TGN. In the Golgi stacks, GRASPs function as stacking factors. Depletion of the GRASP proteins results in Golgi cisternal unstacking14, 15, increases membrane surface for vesicle formation, and accelerates protein trafficking25, but impairs protein glycosylation and sorting. Our results further suggest that Golgi cisternal stacking functions as a quality control mechanism for protein sorting and modifications by reducing the trafficking speed of cargo through the Golgi stack.
Methods
Antibodies and reagents
All reagents used were from Sigma, Roche, or Calbiochem, unless otherwise stated. The following antibodies were used: monoclonal antibodies against Bet1, Gos28 and GM130 from BD Transduction Laboratories; monoclonal antibodies against β-actin (Sigma), clathrin heavy chain (CHC, Hong Chen), cleaved caspase 3 (Cell Signaling), ARF1 and β-COP50, human β1,4 Galactosyltransferase I (CB002, CellMab AB), GFP antibody (Sigma), α 5 integrin (BIIG2, DSHB), β1 integrin (P5D2, DSHB), Lamp1 (H4A3, DSHB), Lamp2 (H4B4, DSHB), α /β-SNAP and syntaxin 651, α-tubulin (DSHB), and VSV-G extracellular domain (D. Sheff); polyclonal antibodies against Bip (P. Arvan), cathepsin D (sc-6486, Santa Cruz), human GRASP55 (Proteintech Group, Inc.), β1 integrin C-terminus (Abcam, EP1041Y), ManII (Kelley Moremen), Sec16 (X. Chen); polyclonal antibodies against δ- and ε-COP, GM130, and GRASP65, golgin-84, NSF, p47, p97, p115, Rab1, Sec31, and syntaxin 5 were previously described51, 52.
Lectin staining
For immunofluorescence analysis, HeLa cells growing on cover slides were fixed in 3.7% paraformaldehyde. After blocking with 1% BSA (in PBS), surface glycosylated proteins were labeled with TRITC-conjugated lectins, such as MAA or WGA (EY laboratories), in the dark for 30 min at 4°C. After washing with 1% BSA, the cells were permeabilized with 0.3% Triton X-100 and processed for immunofluorescence with the indicated antibodies.
For flow cytometry analysis, HeLa cells were harvested in PBS containing 20 mM EDTA, blocked with cold BSA buffer, and labeled with FITC-conjugated lectins (EY laboratories) in the dark at 4°C, for 30 min. After washing, the cells were fixed in 3.7% paraformaldehyde and analyzed on a FACScalibur flow cytometer (BD Biosciences).
Glycomic analysis
HeLa cells transfected with control or GRASP RNAi were washed with cold PBS, scraped and collected by centrifugation. Proteins were harvested from the cell pellets as previously described53. Briefly, the cell pellets were resuspended in ice-cold water and disrupted by Dounce homogenization on ice. The homogenate was adjusted to 4:8:3::choloroform:methanol:water before removal from ice. After 18 hours of end-over-end agitation (Nutator), the insoluble proteinacious material was collected by centrifugation. Following re-extraction of the pellet three times with 4:8:3 and once with ice-cold 80% acetone (v/v, aqueous) the final pellet was dried under a gentle stream of nitrogen and stored as a fine white powder. All supernatants were combined and dried under nitrogen for analysis of LLO and FOS. Protein content of the powder was determined by BCA assay (Pierce). N-linked glycans were harvested from the protein powder as previously described53. Generally, 2 mg of protein powder (dry weight) was resuspended and digested with trypsin/chymotrypsin. Glycopeptides were enriched by Sep-Pak C18 cartridge chromatography and subjected to digestion with PNGase F for 18 h at 37°C. Released oligosaccharides were separated from residual peptide by Sep-Pak C18 clean-up and permethylated prior to mass spectrometric analysis54.
For analysis of LLO and FOS, the dried supernatants from the cell extraction were resuspended in 50% aqueous methanol and loaded onto a Sep-Pak C18 column previously washed with methanol and equilibrated in water. FOS were collected in the run-through and a subsequent water wash, which were combined and dried. The collected FOS fraction was permethylated for analysis by MS54. The LLOs remained bound to the Sep-Pak C18 column and were recovered by elution with methanol and 10:10:3::chloroform:methanol:water as previously described55. Briefly, after drying, the LLO fraction was resuspended in 10:10:3 and applied to a DEAE-cellulose column. Following washes with 10:10:3 to remove neutral lipids, the LLOs were eluted in 10:10:3 containing 300 mM ammonium acetate. The eluted LLOs were recovered by phase partition into the organic layer, which was dried and resuspended in 0.1 M HCl in isopropanol. The LLO oligosaccharides were released from dolichol pyrophosphate by heating at 50°C for 1 h. After cooling and drying under nitrogen, the released LLO oligosaccharides were resuspended in water and recovered in the aqueous phase after addition of n-butanol. The LLO oligosaccharides in the aqueous phase were dried by lyophilization and permethylated for analysis by MS54.
Permethylated N-linked, FOS, and LLO glycans were analyzed by nanospray ionization mass spectrometry using an ion trap instrument (NSI-LTQ Orbi Discoverer, Thermo-Fisher) by dissolving in 1 mM NaOH in 50% methanol and directly infusing into the instrument at a syringe flow rate of 0.40–0.60 µl/min. For fragmentation by CID in MS/MS and MSn modes 40% collision energy was applied. The total ion mapping (TIM) functionality of the XCalibur software package (version 2.0) was utilized to detect and quantify the prevalence of individual glycans in the total glycan profile. Most permethylated oligosaccharides were identified as singly, doubly, and triply charged species by NSI-MS. TIM peaks for all charge states of a given ion with m/z ≤ 2000 were summed together for quantification. Glycan quantification was performed relative to a known quantity of external standards (maltotriose and maltotetraose, Supelco) that was permethylated with heavy methyliodide (13CH3I) and spiked into the sample matrix.
Pulse-chase labeling and immunoprecipitation
HeLa cells growing in 60 mm dishes were incubated in methionine/cysteine-free DMEM (Invitrogen) for 1 h and labeled with 1 ml of medium containing 250 µCi/ml TRANS 35S-LABEL [35S] (MP BIOMEDICALS) for 1 h. After washing with PBS, the cells were collected either immediately on ice or after incubation in 4 ml complete growth medium containing 2 mM L-cysteine and L-methionine for the indicated time periods. To lyse the cells, 0.8 ml lysis buffer (PBS containing 1 mM CaCl2, 0.5 mM MgCl2, 1% TritonX-100, protease inhibitor cocktail, and 1 µM pepstatin A) was added. After centrifugation at 14,000 rpm for 15 min, the protein concentration of the supernatant was measured using a Bio-Rad protein assay (Bio-Rad Laboratories, CA). The cell lysates (2 mg) were immunoprecipitated with 50 ng of the α5 integrin antibody BIIG2 and 60 ng of the β1 integrin antibody P5D2 overnight at 4°C. The antibodies were precipitated with 30 µl protein G beads (Roche Diagnostics GmbH, Germany) at 4°C for 2 h. The beads were washed five times with lysis buffer, and the immunoisolated materials were eluted by boiling for 5 min in nonreducing SDS sample buffer. Integrins were resolved using 6.5% nonreducing SDS-PAGE.
To determine whether the marker proteins interact with GRASP55 or GRASP65, HeLa cells, or HeLa cells infected with VSV-G ts045-GFP adenovirus for 20 h, were lysed in 20 mM Tris-HCl, pH7.4, 150 mM NaCl, 1% Triton X-100, 25 mM β-glycerolphosphate and protease inhibitors. Lysate was cleared by centrifugation and mixed with antibodies to β1 integrin, Lamp1, Lamp2, GM130 or GFP and subsequently isolated by protein A or protein G beads followed by Western blot analysis.
Annexin V assay
HeLa cells grown on cover slides were incubated with annexin V-EGFP diluted 1:100 in the binding buffer provided by the manufacturer (BioVision, K104-25) at room temperature in the dark. After washing with PBS, the cells were fixed, permeabilized, and stained with Hoechst 33342. Images were taken using a Zeiss Observer Z1 inverted microscope with a 40× lens.
Quantification and statistics
In all figures, the quantification results are expressed as the mean±SEM (standard error of the mean) from three independent experiments, unless otherwise stated. The statistical significance of the results was assessed using a Student’s t-test. *, p<0.05, **, p<0.01, ***, p<0.001. For glycomic analysis, individual biological samples were analyzed twice, once without external standard and once following addition of standard. The 5 most abundant high-mannose glycans and the 3 most abundant complex glycans were quantified relative to the external standard. Fold changes relative to normal cells were calculated for each glycan and statistical differences between the fold-changes for the group of high-mannose and complex glycans were determined using a Student’s t-test, **, p<0.01.
Supplementary Material
Acknowledgements
We thank Drs. Peter Arvan, Xiao-wei Chen, Cunming Duan, Fed Gorelick, Victor Hsu, Hiroyasu Kamei, A. Roberto Lara-Lemus, Dennis Larkin, Jennifer Lippincott-Schwartz, Olga Makarova, Kelley Moremen, Hemmo Meyer, Joachim Seemann, David Sheff, and Graham Warren for antibodies and cDNA constructs, and David Sheff and Heike Fölsch for VSV-G-GFP adenovirus. The monoclonal antibodies BIIG2, P5D2, H4A3 and H4B4 were obtained from the Developmental Studies Hybridoma Bank (DSHB). We also thank Dr. Ming Liu for technical help with the pulse-chase assay, Drs. Kimberly Wearne and Irwin Goldstein for suggestions for the lectin experiments, Drs. Reid Glimore, Mark Lehrman and Kelley Moremen for stimulating discussions, Dr. Graham Warren for critical reading of the manuscript, and members of the Wang Lab for suggestions and reagents. This work was supported by the National Institutes of Health (GM087364), the American Cancer Society (RGS-09-278-01-CSM), the NIH-funded Michigan Alzheimer’s Disease Research Center (P50 AG08761), an anonymous donation to Y. Wang, and a University of Michigan Rackham Predoctoral Fellowship to Y. Xiang. The authors acknowledge the support and access to instrumentation provided through grants from the National Center for Research Resources (P41RR018502) and the National Institute of General Medical Sciences (P41GM103490) from the National Institutes of Health.
Footnotes
Author Contributions
Y.W., Y.X., X.Z., and M.T. designed the research; Y.X., X.Z., K.A., T.K., and D.N. performed the experiments; Y.W., Y.X., X.Z., D.N., K.A., T.K., and M.T. analyzed the data; and Y.W. and Y.X. wrote the paper.
Statement of Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J Cell Biol. 1999;144:1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr FA, Short B. Golgins in the structure and dynamics of the Golgi apparatus. Curr Opin Cell Biol. 2003;15:405–413. doi: 10.1016/s0955-0674(03)00054-1. [DOI] [PubMed] [Google Scholar]
- 3.Jackson CL. Membrane traffic: Arl GTPases get a GRIP on the Golgi. Curr Biol. 2003;13:R174–R176. doi: 10.1016/s0960-9822(03)00116-7. [DOI] [PubMed] [Google Scholar]
- 4.Matteis M, Mironov A, Beznoussenko G. The Golgi ribbon and the function of the Golgins. In: Mironov A, Pavelka M, editors. The Golgi Apparatus. Springer Vienna; 2008. pp. 223–246. [Google Scholar]
- 5.Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramirez IB, Lowe M. Golgins and GRASPs: holding the Golgi together. Semin Cell Dev Biol. 2009;20:770–779. doi: 10.1016/j.semcdb.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Vinke FP, Grieve AG, Rabouille C. The multiple facets of the Golgi reassembly stacking proteins. Biochem J. 2011;433:423–433. doi: 10.1042/BJ20101540. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Seemann J. Golgi biogenesis. Cold Spring Harb Perspect Biol. 2011;3:a005330. doi: 10.1101/cshperspect.a005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang Y, Wang Y. New components of the Golgi matrix. Cell Tissue Res. 2011;344:365–379. doi: 10.1007/s00441-011-1166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 11.Shorter J, et al. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. Embo J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. Embo J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang D, Yuan H, Wang Y. The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression. Traffic. 2010;11:827–842. doi: 10.1111/j.1600-0854.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondylis V, Spoorendonk KM, Rabouille C. dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol Biol Cell. 2005;16:4061–4072. doi: 10.1091/mbc.E04-10-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feinstein TN, Linstedt AD. GRASP55 Regulates Golgi Ribbon Formation. Mol Biol Cell. 2008;19:2696–2707. doi: 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gee HY, Noh SH, Tang BL, Kim KH, Lee MG. Rescue of DeltaF508-CFTR Trafficking via a GRASP-Dependent Unconventional Secretion Pathway. Cell. 2011;146:746–760. doi: 10.1016/j.cell.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Giuliani F, Grieve A, Rabouille C. Unconventional secretion: a stress on GRASP. Curr Opin Cell Biol. 2011;23:498–504. doi: 10.1016/j.ceb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- 21.D'Angelo G, et al. GRASP65 and GRASP55 sequentially promote the transport of C-terminal valine bearing cargoes to and through the golgi complex. J Biol Chem. 2009;284:34849–34860. doi: 10.1074/jbc.M109.068403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Satoh A, Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. Embo J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marra P, et al. The GM130 and GRASP65 Golgi proteins cycle through and define a subdomain of the intermediate compartment. Nat Cell Biol. 2001;3:1101–1113. doi: 10.1038/ncb1201-1101. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Wei JH, Bisel B, Tang D, Seemann J. Golgi Cisternal Unstacking Stimulates COPI Vesicle Budding and Protein Transport. PLoS ONE. 2008;3:e1647. doi: 10.1371/journal.pone.0001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin SJ, et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhoven B, Schlegel RA, Williamson P. Mechanisms of phosphatidylserine exposure, a phagocyte recognition signal, on apoptotic T lymphocytes. J Exp Med. 1995;182:1597–1601. doi: 10.1084/jem.182.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Presley JF, et al. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 29.Saraste J, Kuismanen E. Pre- and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell. 1984;38:535–549. doi: 10.1016/0092-8674(84)90508-7. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths G, Simons K. The trans Golgi network: sorting at the exit site of the Golgi complex. Science. 1986;234:438–443. doi: 10.1126/science.2945253. [DOI] [PubMed] [Google Scholar]
- 31.Yang JS, et al. COPI acts in both vesicular and tubular transport. Nat Cell Biol. 2011;13:996–1003. doi: 10.1038/ncb2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wearne KA, Winter HC, Goldstein IJ. Temporal changes in the carbohydrates expressed on BG01 human embryonic stem cells during differentiation as embryoid bodies. Glycoconj J. 2008;25:121–136. doi: 10.1007/s10719-007-9064-x. [DOI] [PubMed] [Google Scholar]
- 33.Gieselmann V, Hasilik A, von Figura K. Processing of human cathepsin D in lysosomes in vitro. J Biol Chem. 1985;260:3215–3220. [PubMed] [Google Scholar]
- 34.Zaidi N, Maurer A, Nieke S, Kalbacher H. Cathepsin D: a cellular roadmap. Biochem Biophys Res Commun. 2008;376:5–9. doi: 10.1016/j.bbrc.2008.08.099. [DOI] [PubMed] [Google Scholar]
- 35.von Blume J, et al. Actin remodeling by ADF/cofilin is required for cargo sorting at the trans-Golgi network. J Cell Biol. 2009;187:1055–1069. doi: 10.1083/jcb.200908040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diao A, Rahman D, Pappin DJ, Lucocq J, Lowe M. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J Cell Biol. 2003;160:201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Storrie B, et al. Recycling of golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J Cell Biol. 1998;143:1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Zeijl MJ, Matlin KS. Microtubule perturbation inhibits intracellular transport of an apical membrane glycoprotein in a substrate-dependent manner in polarized Madin-Darby canine kidney epithelial cells. Cell Regul. 1990;1:921–936. doi: 10.1091/mbc.1.12.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruns C, McCaffery JM, Curwin AJ, Duran JM, Malhotra V. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J Cell Biol. 2011;195:979–992. doi: 10.1083/jcb.201106098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinseth MA, et al. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 2007;130:524–534. doi: 10.1016/j.cell.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 41.Duran JM, et al. The Role of GRASP55 in Golgi Fragmentation and Entry of Cells into Mitosis. Mol Biol Cell. 2008;19:2579–2587. doi: 10.1091/mbc.E07-10-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao N, et al. Mannose-6-phosphate regulates destruction of lipid-linked oligosaccharides. Mol Biol Cell. 2011;22:2994–3009. doi: 10.1091/mbc.E11-04-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baas S, et al. Sugar-free frosting, a homolog of SAD kinase, drives neural-specific glycan expression in the Drosophila embryo. Development. 2011;138:553–563. doi: 10.1242/dev.055376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farhan H, et al. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J Cell Biol. 2010;189:997–1011. doi: 10.1083/jcb.200912082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao N, Shang J, Lehrman MA. Analysis of glycosylation in CDG-Ia fibroblasts by fluorophore-assisted carbohydrate electrophoresis: implications for extracellular glucose and intracellular mannose 6-phosphate. J Biol Chem. 2005;280:17901–17909. doi: 10.1074/jbc.M500510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones MA, et al. DDOST mutations identified by whole-exome sequencing are implicated in congenital disorders of glycosylation. American journal of human genetics. 2012;90:363–368. doi: 10.1016/j.ajhg.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol. Biol. Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wildt S, Gerngross TU. The humanization of N-glycosylation pathways in yeast. Nat Rev Microbiol. 2005;3:119–128. doi: 10.1038/nrmicro1087. [DOI] [PubMed] [Google Scholar]
- 49.Capony F, et al. Increased secretion, altered processing, and glycosylation of pro-cathepsin D in human mammary cancer cells. Cancer Res. 1989;49:3904–3909. [PubMed] [Google Scholar]
- 50.Xiang Y, Seemann J, Bisel B, Punthambaker S, Wang Y. Active ADP-ribosylation factor-1 (ARF1) is required for mitotic Golgi fragmentation. J Biol Chem. 2007;282:21829–21837. doi: 10.1074/jbc.M611716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X, et al. Quantitative proteomics analysis of cell cycle regulated Golgi disassembly and reassembly. J Biol Chem. 2010;285:7197–7207. doi: 10.1074/jbc.M109.047084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang D, et al. The ubiquitin ligase HACE1 regulates Golgi membrane dynamics during the cell cycle. Nature Communications. 2011;2:501. doi: 10.1038/ncomms1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aoki K, et al. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 54.Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydrate Research. 1984;131:209–217. [Google Scholar]
- 55.Gao N, Lehrman MA. Non-radioactive analysis of lipid-linked oligosaccharide compositions by fluorophore-assisted carbohydrate electrophoresis. Methods in enzymology. 2006;415:3–20. doi: 10.1016/S0076-6879(06)15001-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.