SUMMARY
Toxin endoribonucleases of toxin/antitoxin (TA) systems regulate protein production by selectively degrading mRNAs but have never been shown to control other TA systems. Here we demonstrate that toxin MqsR of the MqsR/MqsA system enriches toxin ghoT mRNA in vivo and in vitro, since this transcript lacks the primary MqsR cleavage site 5′-GCU. GhoT is a membrane toxin that causes the ghost cell phenotype, and is part of a type V TA system with antitoxin GhoS that cleaves specifically ghoT mRNA. Introduction of MqsR primary 5′-GCU cleavage sites into ghoT mRNA reduces ghost cell production and cell death likely due to increased degradation of the altered ghoT mRNA by MqsR. GhoT also prevents cell elongation upon the addition of low levels of ampicillin. Therefore, during stress, antitoxin GhoS mRNA is degraded by toxin MqsR allowing ghoT mRNA translation to yield another free toxin that forms ghost cells and increases persistence. Hence, we show that GhoT/GhoS is the first TA system regulated by another TA system.
Keywords: Toxin/antitoxin, Persistence, MqsR, GhoT/GhoS, Differential mRNA decay
INTRODUCTION
Bacterial toxin/antitoxin (TA) systems play prominent roles in cell physiology including altruistic behavior such as phage inhibition (Pecota and Wood, 1996), global gene regulation (Wang and Wood, 2011), and tolerance to antibiotics (Lewis, 2010). For antitoxins, both MqsA (Wang et al., 2011) and DinJ (Hu et al., 2012) have been shown to repress the stationary-phase sigma factor RpoS and thereby influence the response to stress and biofilm formation as global regulators. Toxins in TA systems also play a role in gene regulation in that mRNA endoribonuclease toxins cleave specific mRNAs and thereby cause differential mRNA decay (Wang and Wood, 2011). For example, upon antibiotic stress, toxin MazF degrades most mRNAs with ACA sequences, but its activity also results in the preferential synthesis of a subset of small proteins whose mRNAs are not degraded (Amitai et al., 2009). These enriched proteins are necessary both for toxicity and for survival (Amitai et al., 2009). Therefore, both toxins and antitoxins may be considered global regulators.
The endoribonuclease toxin MqsR (motility quorum sensing regulator) (Ren et al., 2004; González Barrios et al., 2006; Brown et al., 2011) of the MqsR/MqsA TA system is also a global regulator (González Barrios et al., 2006; Kim et al., 2010). It cleaves specific mRNA primarily at 5′-GCU sites (Yamaguchi et al., 2009; Christensen-Dalsgaard et al., 2010) and leads to enrichment of mRNAs that code for stress-associated proteins CstA, CspD, RpoS, Dps, and HokD (Kim et al., 2010). The binding of MqsR to its antitoxin MqsA potently inhibits MqsR toxicity (Brown et al., 2011).
MqsR/MqsA is a type II TA system (protein toxin inhibited by the binding of a protein antitoxin), and type II TA systems are one of the most wide-spread forms of TA systems (Hayes and Van Melderen, 2011). In type I TA systems (RNA-RNA), the antitoxin RNA prevents the translation of toxin RNA through complementary base pairing (Hayes and Van Melderen, 2011), in type III TA systems (RNA-protein), the antitoxin RNA inhibits the protein toxin (Hayes and Van Melderen, 2011), in type IV TA systems (protein-protein), the protein antitoxin interferes with binding of the toxin to its target (Masuda et al., 2012), and in type V systems (protein-RNA), the antitoxin is a specific RNase that cleaves the toxin mRNA (Wang et al., 2012).
There are 14 Escherichia coli mRNA transcripts that do not contain the MqsR-preferred 5′-GCU cleavage site, and six of these (pheL, tnaC, trpL, yciG, ygaQ and ralR) are differentially regulated in biofilms (Domka et al., 2007). One of these 14 transcripts that lacks 5′-GCU sites is ghoT (yjdO, ghost cells (Wang et al., 2012)). GhoT is a membrane toxin that produces the ghost-cell phenotype (lysed cells with damaged membranes) (Wang et al., 2012). The antitoxin for GhoT, GhoS, cleaves specifically ghoT mRNA thereby halting ghoT expression; hence, this TA system is the first type V system in that an enzyme antitoxin inactivates the mRNA of the toxin (Wang et al., 2012). The structure of antitoxin GhoS revealed it is related to the CRISPR-associated-2 (CAS2) sequence-specific endoribonuclease, and a whole-transcriptome study showed the specificity of GhoS as a ribonuclease (Wang et al., 2012). Although mqsR is more prevalent than ghoT, ghoT-related genes are found in Escherichia, Shigella, Salmonella, Citrobacter, and Proteus spp.
Persister cells are a small fraction of bacteria that exhibit tolerance to antibiotics without genetic change (Lewis, 2007); it is believed they survive antibiotic treatment by becoming metabolically dormant through TA systems (Lewis, 2010). The evidence relating TA systems to persistence includes the finding that when MqsR is inactivated, persister cell formation is reduced (Kim and Wood, 2010); production of MqsR also increases persistence. Similarly, deletion of the TisB toxin of the TisAB/IstR-1 type I TA system decreases persistence (Dörr et al., 2010). Subsequent studies have also demonstrated the importance of TA systems for persistence by showing that cells lacking 10 mRNA endoribonuclease toxins including MqsR form less persister cells (Maisonneuve et al., 2012). In persister cells, mqsR is also the most induced gene (Shah et al., 2006).
Our previous studies showed that the antitoxin MqsA is rapidly degraded by Lon protease which frees toxin MqsR upon stress (Wang et al., 2011). The importance of Lon for persister cell formation has been demonstrated by showing both that cells lacking lon have reduced persister cell formation and that removal of the 10 toxins causes the lon mutant to no longer affect persistence (Maisonneuve et al., 2012).
We hypothesized that since ghoT mRNA lacks the primary MqsR 5′-GCU sites, it would be enriched under conditions when MqsR is produced. Here we demonstrate that MqsR does enrich ghoT transcripts and thereby activates expression of this membrane toxin which is involved in persister cell formation.
RESULTS
MqsR requires GhoT to prevent cell elongation
reviously, we examined the 14 E. coli transcripts that lack 5′-GCU sites to determine if these transcripts were related to the ability of MqsR to increase persistence (Wang et al., 2012). Among them, deleting gene ghoT had one of the largest negative effects on MqsR-mediated persistence; corroborating this result, production of GhoT increased persistence 48-fold (Wang et al., 2012). We repeated these persister experiments by varying the ampicillin concentration and found similar results at both 35 μg/mL and 100 μg/mL ampicillin for MqsR production in the wild-type and in the ghoT mutant so the results are not influenced by the antibiotic concentration. In addition, we determined that the minimum inhibitory concentration for ampicillin was the same for both strains (approximately 5 μg/mL). Hence, we investigated further the relationship between MqsR and GhoT in E. coli to determine how GhoT influences persister cell formation when MqsR is produced.
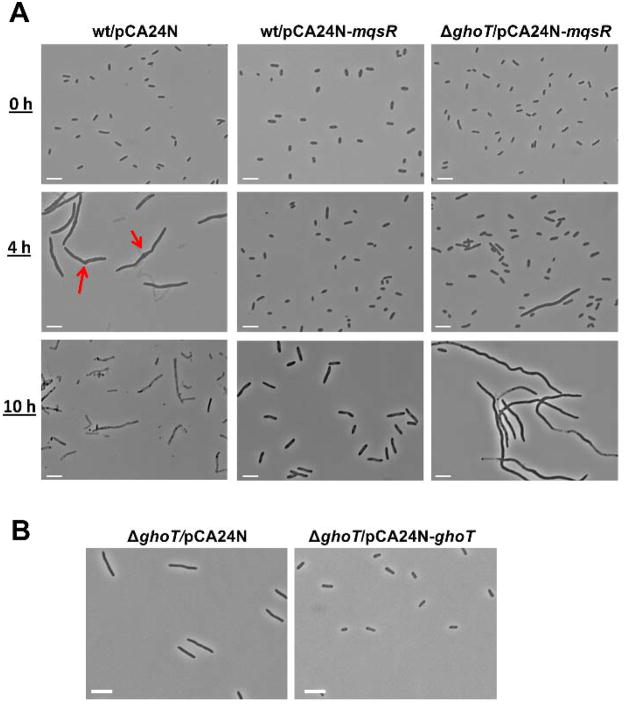
To initially probe the relationship between MqsR and GhoT, we used microscopy. High concentrations of ampicillin (100 μg/mL, 10 × MIC) are commonly used to assess persister cell formation in E. coli (Balaban et al., 2004; Keren et al., 2004; Shah et al., 2006; Kim et al., 2010). When added at 100 μg/mL, ampicillin rapidly lyses non-persister cells, which makes it difficult to probe for changes in cell morphology; hence, here we used a lower ampicillin concentration (20 μg/mL, 2 × MIC), which causes cell elongation rather than lysis allowing for the observation of the response to β-lactam antibiotics (Rolinson, 1980). We reasoned that since MqsR increases persister cells (Kim and Wood, 2010), MqsR should prevent cells from elongating upon addition of ampicillin since these persisters would be tolerant of the antibiotic. As expected, we found that addition of 20 μg/mL ampicillin to wild-type cells (BW25113/pCA24N) caused cell elongation of most cells (23 ± 3 μm) after 4 h by inhibiting cell division (Donachie and Begg, 1970; Staugaard et al., 1976), and bulges or collars were observed at the division site (red arrows in Fig. 1A) as reported by others (Donachie and Begg, 1970; Rolinson, 1980). After 10 h, the majority of the cells lysed (Fig. 1A, left panel). In contrast, the majority of cells producing MqsR (BW25113/pCA24N-mqsR) were not elongated (1.5 ± 0.6 μm) after 4 h with ampicillin, and the cells remained intact with sizes increased moderately after 10 h (3.5 ± 0.9 μm) (Fig. 1A, middle panel). This dramatic increase in persister cells is due to production of the toxin MqsR. For cells producing MqsR in the absence of ghoT (ΔghoT/pCA24N-mqsR), a few cells were significantly elongated after 4 h with ampicillin, and the majority of cells were significantly elongated after 10 h (38 ± 7 μm) (Fig. 1A, right panel). Hence, MqsR prevents cell elongation upon the addition of ampicillin primarily via its control of GhoT.
Fig. 1. GhoT decreases cell size in the presence of ampicillin.
(A) Cell morphology after 0 h, 4 h, and 10 h with ampicillin (20 μg/mL) for cells producing MqsR (wt/pCA24N-mqsR) and cells producing MqsR but without ghoT (ΔghoT/pCA24N-mqsR). Red arrows point to the bulges or collars with ampicillin. (B) Cell morphology after 4 h with ampicillin (20 μg/mL) for cells without GhoT (ΔghoT/pCA24N) and cells with GhoT (ΔghoT/pCA24N-ghoT). Two independent cultures of each strain were evaluated and representative images were shown. Scale bars indicate 5 μm.
We also investigated how cells, with or without GhoT, respond to ampicillin challenge. After 4 h with 20 μg/mL ampicillin, cells producing GhoT (ΔghoT/pCA24N-ghoT) were not elongated (1.7 ± 0.5 μm) whereas those that lacked GhoT (ΔghoT/pCA24N) were elongated (5 ± 1 μm) (Fig. 1B). Note that normal cells prior to antibiotic treatment were about 1.8 μm. This result corroborates that GhoT prevents cell elongation with ampicillin stress. Since persister cells are not dividing and are metabolically dormant with smaller sizes than non-persister cells when exposed to bactericidal antibiotics including ampicillin (Lewis, 2010), these micrographs provide additional evidence that MqsR helps to maintain a dormant state in the presence of ampicillin through GhoT action and that deletion of ghoT reduces the ability of MqsR to maintain persistence.
MqsR regulates production of GhoT in vivo through differential ghoS mRNA decay
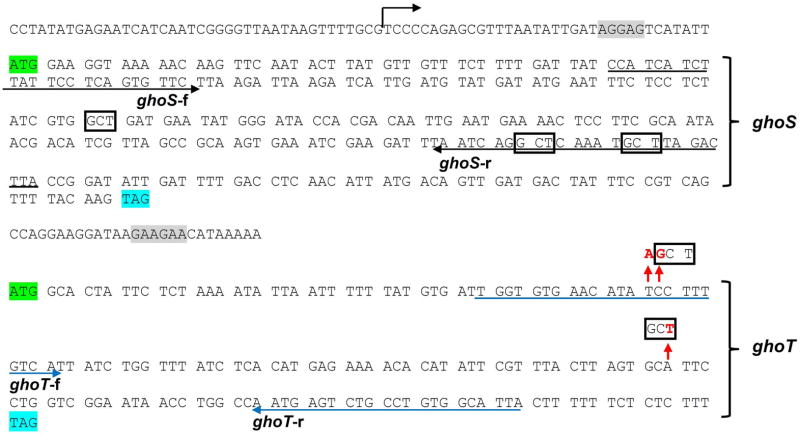
Given the link between MqsR and GhoT, we investigated in vivo whether MqsR can regulate ghoST mRNA stability at a post-transcriptional level by using quantitative real time reverse transcription PCR (qRT-PCR) using a primer pair within the coding portion of each gene (Table 2). We found that when MqsR was produced (BW25113/pBS(Kan)-mqsR vs. BW25113/pBS(Kan)), there was a 5 ± 1-fold increase in the 3′ ghoT portion of the ghoST mRNA compared to the 5′ ghoS portion, which suggests that the ghoST transcript is preferentially degraded at the 5′ end when MqsR is active (Table 3 lists the qRT-PCR results). This result is not unexpected since the region of the ghoST transcript that codes for ghoS (5′ end) contains three of the primary MqsR 5′-GCU cleavage sites, whereas the region that codes for ghoT (3′ end) does not contain any 5′-GCU cleavage sites (Fig. 2). Therefore, one mechanism by which MqsR regulates GhoT activity is by selectively degrading the ghoST transcript in a manner that leaves the portion of the transcript for toxin GhoT intact, while that of GhoS is degraded.
Table 2.
Oligonucleotides used for RT-PCR, qRT-PCR, mutant verification, site directed mutagenesis (target mutated nucleotides are underlined) and in vitro transcription (for the forward primers, the T7 promoter sequence is underlined and the bases incorporated into RNA during transcription are in bold). f indicates forward primer and r indicates reverse primer.
| Purpose/Name | Sequence (5′ to 3′) |
|---|---|
| RT-PCR and qRT-PCR | |
| ghoS-f | CCATCATCTTATTCCTCAGTGTTCT |
| ghoS-r | TAAGTCTAAGCATTTGAGCCTGATT |
| ghoT-f | TGGTGTGAACATATCCTTTGTCA |
| ghoT-r | TAATGCCACAGGCAGACTCA |
| ralR-f | AAACCATGTCCATTTTGTGGT |
| ralR-r | TCCAGTGGTTCGTTTATTCCA |
| ompA-f | CACTGGCTGGTTTCGCTACCG |
| ompA-r | ACCCATTTCAAAGCCAACATC |
| ompF-f | AAGCGCAATATTCTGGCAGT |
| ompF-r | TGCCACCGTAACTGTTTTCA |
| yciG-f | ACATCGTGGTGGTTCAGGAA |
| yciG-r | TACCACCGCTTTGTTGACCG |
| Deletion mutant verification | |
| GhoS-veri-f | CTCCTATATGAGAATCATCAATCGGGG |
| GhoS-veri-r | GACAAAGGATATGTTCACACCAATCAC |
| GhoT-veri-f | GACCTCAACATTATGACAGTTGATGAC |
| GhoT-veri-r | GCTTCGTTCATCGTTCCGCAAATCCAG |
| Site-directed mutagenesis | |
| GhoT-GCU-1-f | GTGATTGGTGTGAACATAAGCTTTGTCATTATCTGGTTTATCTC |
| GhoT-GCU-1-r | GAGATAAACCAGATAATGACAAAGCTTATGTTCACACCAATCAC |
| GhoT-GCU-2-f | CACATATTCGTTTACTTAGTGCTTTCCTGGTCGGAATAACCTGG |
| GhoT-GCU-2-r | CCAGGTTATTCCGACCAGGAAAGCACTAAGTAAACGAATATGTG |
| T7 in vitro transcription | |
| PT7-GhoT-f | TAATACGACTCACTATAGGGAGAATGGCACTATTCTCTAAAATATTAATTTTT |
| PT7-GhoT-r | CTAAAAGAGAGAAAAAAGTAATGC |
| PT7-GhoS-f | TAATACGACTCACTATAGGGAGAATGGAAGGTAAAAACAAGTTCAATACTTAT |
| PT7-GhoS-r | TCATCAACTGTCATAATGTTGAG |
Table 3. Summary of the qRT-PCR results.
Experimental conditions and the target mRNA are indicated along with the number of 5′-GCU sites in the target mRNA. The fold changes indicate the changes in enrichment (not necessarily changes in transcription) after differential mRNA decay by RNase MqsR and were calculated as described earlier (Pfaffl, 2001) relative to the house-keeping gene (rrsG), which was used to normalize the data. Values less than one are indicated as negative fold changes (i.e., mRNA levels were reduced relative to other mRNA). The specificity of the qRT-PCR products was verified by a melting curve analysis.
| Conditions | Target mRNA | Fold change | 5′-GCU site |
|---|---|---|---|
| BW25113/pBS(Kan)-mqsR vs. BW25113/pBS(Kan) | ghoT | 40 ± 1 | 0 |
| ghoS | 8 ± 1 | 3 | |
| 1 mM IPTG for 60 min | ompA | −4.5 ± 1 | 27 |
| ompT | 3 ± 2 | 14 | |
| ompF | −2 ± 1 | 20 | |
| kilR | 10.8 ± 0.4 | 0 | |
| ralR | 12 ± 1 | 0 | |
| yciG | 11 ± 1 | 0 | |
|
| |||
| BW25113 wild-type | mqsR | 18 ± 2 | 1 |
| ghoT | 40 ± 1 | 0 | |
| 2 μg/mL nalidixic acid for 90 min vs. no nalidixic acid | ompA | 1 ± 1 | 27 |
|
| |||
| BW25113 wild-type | mqsR | 20 ± 2 | 1 |
| ghoT | 48 ± 2 | 0 | |
| 10 μg/mL azlocillin for 90 min vs. no azlocillin | ompA | −1 ± 1 | 27 |
|
| |||
| BW25113 wild-type | mqsR | 4.6 ± 0.6 | 1 |
| 20 mM H2O2 for 5 min vs. no H2O2 | ghoT | 6 ± 2 | 0 |
Fig. 2. DNA sequence of ghoST.
The positions with introduced mutations in ghoT are indicated by red arrows with the changed nucleotide in red font. Native 5′-GCT sites in ghoS are boxed in black. Start codons are highlighted in green and stop codons are highlighted in blue. The putative transcription start site of the ghoST operon is indicated by a black arrow, and the putative RBS sites are highlighted in grey. Primers from ghoS (ghoS-f and ghoS-r) the primers from ghoT (ghoT-f and ghoT-r) are underlined with their direction indicated.
If MqsR degrades other transcripts in addition to that of ghoS, but not ghoT, ghoT mRNA should be enriched by comparison with these other transcripts. We investigated this using qRT-PCR (Table 3) and found that production of MqsR enriches ghoT mRNA 40 ± 1-fold compared to 5′-GCU-containing transcripts ompA (27 5′-GCU sites) and ompF (20 5′-GCU sites), which have been shown to be degraded by MqsR by in vivo and in vitro (Yamaguchi et al., 2009), as well as ompT (14 5′-GCU sites). As expected, we found that three other mRNAs that lack 5′-GCU sites were enriched during production of MqsR: kilR by 10.8 ± 0.4 -fold, ralR by 12 ± 1-fold, and yciG by 11 ± 1-fold. Also, ribosomal RNA rrsG was not affected by production of MqsR (confirmed by an intact band of 16S rRNA with agarose gel electrophoresis) since this RNA should be protected by its incorporation into ribosomes.
Similarly, we investigated ghoT mRNA levels under external stress conditions, where MqsR was induced at its normal level from the chromosomal locus. Upon addition of sub-lethal concentrations of nalidixic acid or azlocillin to wild-type cells, mqsR mRNA was induced 18 ± 2-fold and 20 ± 2-fold, respectively (Table 3). Under these conditions, ghoT mRNA was enriched by 40 ± 1-fold and 48 ± 2-fold, respectively, whereas 5′-GCU-containing ompA mRNA was not enriched. Upon oxidative stress (20 mM hydrogen peroxide for 5 min), due to the rapid degradation of MqsA by Lon (Wang et al., 2012), mqsR transcription was induced 4.5 ± 0.6-fold, which, due to the increase in MqsR protein, enabled ghoT mRNA to increase 6 ± 2-fold. Therefore, these three stress conditions which induce mqsR transcription result in concomitant enrichment of ghoT mRNA relative to cellular mRNA.
MqsR preferentially cleaves ghoS mRNA in vitro
Using purified toxin MqsR, we also found that MqsR cleaves ghoS mRNA far more efficiently than ghoT mRNA (Fig. 3). The relatively small amount of degradation of native ghoT mRNA (Fig. 3, lane 5) appears to be related to purified MqsR cleavage at the three less favorable GCA sites based on the sizes of the fragments generated.
Fig. 3. MqsR cleaves ghoS mRNA more efficiently than ghoT mRNA in vitro.
Two-micrograms of in vitro synthesized ghoS mRNA (278 nt, lanes 2, 3), ghoT mRNA (180 nt, lanes 4, 5) and mutated ghoT-GCU mRNA with two GCU sites (180 nt, lanes 6, 7) were incubated either without (−) or with (+) 30 ng MqsR protein for 15 min at 37°C. The arrows indicate the uncleaved ghoS and ghoT transcripts. Lane 1, low range ssRNA ladder.
To further demonstrate that the lack of ghoT mRNA degradation was due to the absence of MqsR cleavage sites, we added two of the primary MqsR 5′-GCU cleavage sites to ghoT mRNA and tested whether the mutated ghoT-GCU mRNA was degraded by MqsR. As it is currently unknown whether MqsR cleaves mRNA at specific bases of a codon similar to RelE, which preferentially cleaves after the second or third base of a codon (Hurley et al., 2011), we introduced both a non-codon 5′-GCU and a codon 5′-GCU site into ghoT mRNA. Both changes are silent: at aa position 18, TCC (Ser) was changed to AGC (Ser) to create the non-codon 5′-GCU site and at aa position 37, GCA (Ala) was changed to GCT (Ala) to create a codon 5′-GCU site (Fig. 2; pCA24N-ghoT-GCU). Using purified toxin MqsR, we found that MqsR cleaves the mutated ghoT mRNA with two 5′-GCU sites (Fig. 3). Compared to the wild-type ghoT mRNA that lacks GCU sites, multiple cleavage products were clearly observed when the ghoT-GCU mRNA was used as a template. Furthermore, we also found that the sizes of the dominant cleavage products agree well with those that are predicted when the ghoT-GCU mRNA is cleaved at 5′-GCU sites (56, 59, 65, 115 and 121 nt) (Fig. 3, lane 7). These in vitro results demonstrate that MqsR preferentially cleaves 5′-GCU sequences, and that it functions without ribosomes (Yamaguchi et al., 2009).
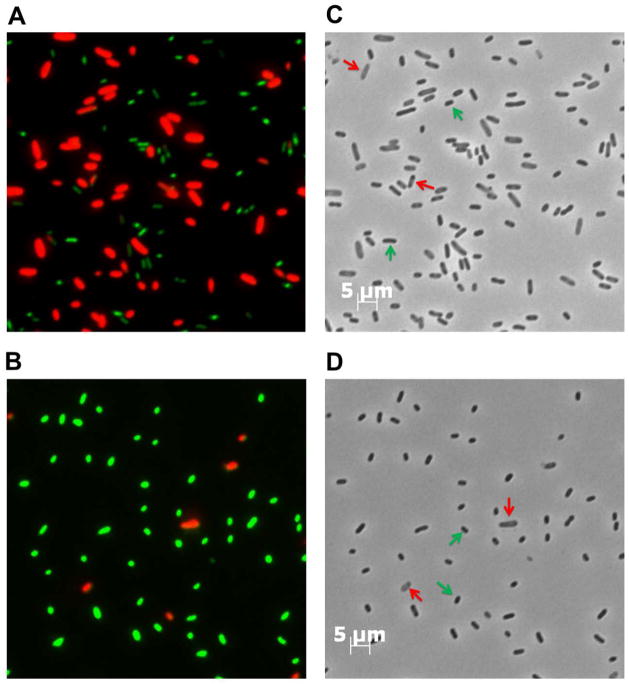
Furthermore, using a ghoT deletion strain (BW25113 ΔghoT) to ensure that no endogenous ghoT transcript was produced, we found that producing MqsR resulted in three-fold fewer ghost cells for those cells transformed with pCA24N-ghoT-GCU vs. those with pCA24N-ghoT (Fig. 4). Live/Dead staining was also used to differentiate between cells with intact membranes (live) and cells with damaged membranes (dead). A four-fold increase in the number of cells with intact membranes was observed in cells transformed with mutant ghoT DNA with two GCT (5′-GCU in the transcribed mRNA) sites (Fig. 4) as compared to wild-type ghoT which confirms that adding the two 5-GCU sites decreased production of ghost cells. Therefore, both in vitro and in vivo studies demonstrate that MqsR regulates GhoT activity by GCU-dependent cleavage of ghoS in the ghoST mRNA transcript upon stress.
Fig. 4. Addition of 5′-GCU to ghoT mRNA reduces MqsR toxicity.
Live/dead staining via fluorescence microscopy showing that the production of wild-type GhoT (A) causes ghost cell morphology and leads to cell death (red), while production of mutant ghoT mRNA (containing 2 introduced 5′-GCU sites) (B) has less ghost cells and less cell death (live cells are green). Panels C and D show the corresponding phase contrast microscopy images. Red arrows indicate representative ghost cells, and green arrows indicate representative live cells. Images were taken 3 h after the addition of 0.5 mM IPTG to cultures of ΔghoT ΔKm/pBS(Kan)-mqsR/pCA24N-ghoT (A and C) and ΔghoTΔKm/pBS(Kan)-mqsR/pCA24N-ghoT-GCU (B and D) when the turbidity reached 0.5, which allows production of MqsR through its leaky Plac promoter followed by the production of GhoT through PT5-lac promoter. Two independent cultures of each strain were evaluated and representative images were shown. Scale bars on the right column indicate 5 μm which is the same for all 4 panels.
DISCUSSION
Toxin MqsR regulates toxin GhoT via post-transcriptional differential mRNA cleavage resulting in a regulatory hierarchy where one TA system controls another. The nine lines of evidence that demonstrates this are that (i) when MqsR is produced (BW25113/pBS(Kan)-mqsR vs. BW25113/pBS(Kan)), there is a 5 ± 1-fold increase in the 3′ ghoT portion of the ghoST mRNA compared to the 5′ ghoS portion, (ii) when MqsR is produced, ghoT mRNA is enriched 40 ± 1-fold compared to 5′-GCU-containing transcripts ompA and ompF, which have been shown to be degraded by MqsR by in vivo and in vitro (Yamaguchi et al., 2009), (iii) the addition of sub-lethal concentrations of nalidixic acid induces mqsR mRNA 18 ± 2-fold, which results in concomitant enrichment of ghoT mRNA 40 ± 1-fold while there is no enrichment of ompA mRNA, (iv) the addition of sub-lethal concentrations of azlocillin induces mqsR mRNA 20 ± 2-fold, which results in the concomitant enrichment of ghoT mRNA 48 ± 2-fold while there is no enrichment of the ompA mRNA, (v) upon oxidative stress, mqsR transcription was induced 4.5 ± 0.6-fold, which, in turn, enabled ghoT mRNA to increase 6 ± 2-fold, (vi) upon addition of two 5′-GCU sites to ghoT mRNA, the number of ghost cells and dead cells were reduced due to the presence of the primary cleavage sites for MqsR, (vii) MqsR cleaves more readily mutated ghoT mRNA with two introduced 5′-GCU sites in vitro, (viii) MqsR preferentially cleaves ghoS mRNA more efficiently than ghoT mRNA in vitro, and (ix) MqsR-derived persistence depends on GhoT activity. Although it is possible that other endoribonuclease toxins might also impact the stability of ghoST mRNA under stressed conditions in vivo, the in vitro studies demonstrate the importance of toxin MqsR on the stability of ghoST mRNA. Therefore, MqsR is the first identified toxin that regulates a second TA system, GhoT/GhoS, illustrating how MqsR, along with MqsA (Wang et al., 2011), function as global regulators of environmental stress in E. coli. These results also illustrate how cells have interwoven TA systems into their genetic fabric to control cell physiology.
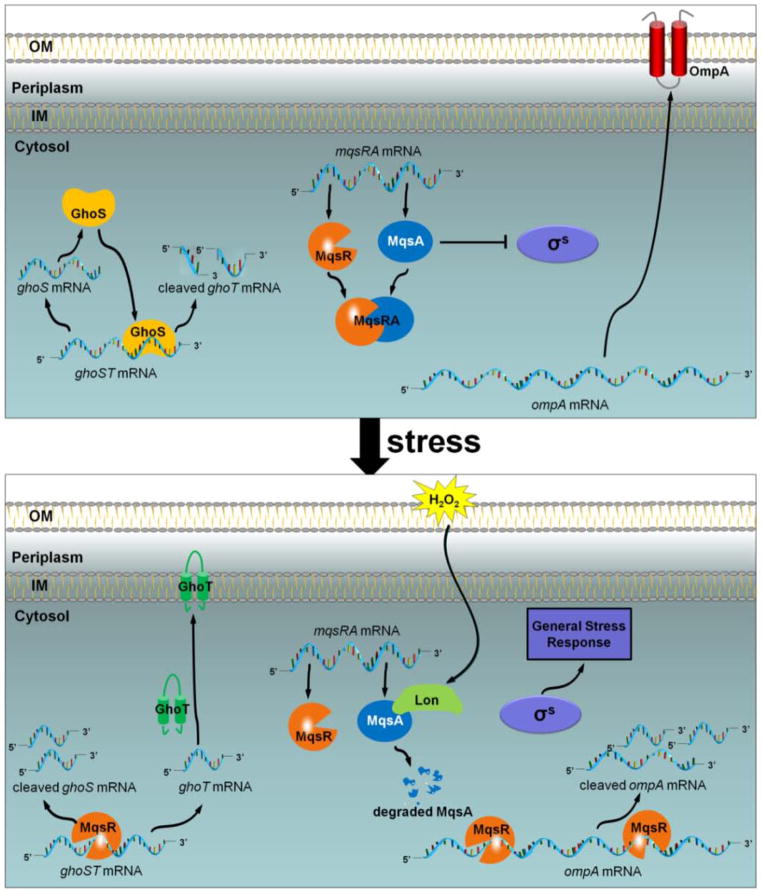
Our model of the response of E. coli to oxidative stress, as mediated by the MqsR/MqsA and GhoT/GhoS TA systems, is shown in Fig. 5. In the absence of stress, the antitoxin MqsA binds to the toxin MqsR to block its RNase activity. Furthermore, MqsA also binds to the rpoS promoter region via a palindrome to inhibit its transcription (Wang et al., 2011). In the absence of stress, the antitoxin GhoS is produced which prevents the expression of ghoT by cleaving its transcript, and the translation of both proteins is coupled. Under stress, Lon protease degrades MqsA which frees the toxin MqsR (Wang et al., 2011) and further induces mqsR expression. MqsR then degrades unprotected mRNAs primarily at 5′-GCU sites, such as ompA and the 5′ end of ghoST mRNA (i.e., the coding region of ghoS, which contains three 5′-GCU sites). As ghoST is more efficiently degraded in the ghoS coding region, while the ghoT coding region stays intact, GhoT protein levels increase. This allows GhoT to exert its effects on the cell membrane, ultimately increasing persistence.
Fig. 5. Schematic of MqsR-mediated control GhoT/GhoS.
Without stress (upper panel), MqsR and MqsA bind and form a complex that prevents MqsR toxicity while GhoS cleaves ghoT mRNA to prevent GhoT toxicity. Upon oxidative stress (lower panel), MqsA is degraded by protease Lon, which derepresses rpoS and triggers the general stress response. The liberated MqsR functions as sequence-specific endoribonuclease to cleave ghoS mRNA, ompA mRNA, and other unprotected mRNAs primarily at 5′-GCU sites. As a result, MqsR rapidly enriches ghoT mRNAs which does not contain 5′-GCU sites as well as other stress-related transcripts and leads to the formation of persister cells.
Therefore, our data further demonstrate the complexity and interconnectivity of TA systems by showing one TA system (MqsR/MqsA) may control another (GhoT/GhoS). This interconnectivity is to be expected given both the prevalence of mRNA cleavage sites of toxin RNases and their differential mRNA decay; hence, production of some proteins is facilitated during stress and this now includes preferential production of toxins via enrichment of their mRNA. For GhoT, its production allows the cell to increase its persistence, probably by disrupting ATP generation. The physiological consequence of enrichments of other transcripts including kilR, ralR, and yciG remains to be elucidated (these genes encode a cell division inhibitor, a protein that alleviates restriction of unmodified DNA and promotes methylation, and an unknown protein, respectively). Nevertheless, it is also clear now that a hierarchy of TA systems exists in the cells (e.g., MqsR/MqsA controlling GhoS/GhoT). This hierarchy was predicted (Bukowski et al., 2011) and now has been demonstrated.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids are listed in Table 1. To remove the kanamycin marker for ΔghoS and ΔghoT, pCP20 was used to express the FLP recombinase (Datsenko and Wanner, 2000). To construct BW25113 ΔghoS ΔmqsRA, P1 transduction (Maeda et al., 2008) was used to transfer the ΔmqsRA deletion to BW25113 ΔghoS. Luria-Bertani (LB) at 37°C was used except where indicated. Kanamycin (50 μg ml−1) was used for pre-culturing isogenic knockout mutants and for maintaining pBS(Kan)-based plasmids, chloramphenicol (30 μg ml−1) was used for maintaining pCA24N-based plasmids, and ampicillin (100 μg ml−1) was used to maintain plasmid pCP20. The persistence assay was conducted as described (Wang et al., 2012) with 35 and 100 μg/mL ampicillin.
Table 1.
Bacterial strains and plasmids used in this study. Chloramphenicol (30 μg/mL) and kanamycin (50 μg/mL) were used to maintain pCA24N-based plasmids and for pBS(Kan)-based plasmids, and ampicillin (100 μg ml−1) was used to maintain plasmid pCP20.
| Bacterial strains/Plasmids | Source | |
|---|---|---|
| E. coli K12 strains | ||
| BW25113 | lacIqrrnBT14ΔlacZWJ16 hsdR514ΔaraBADAH33 ΔrhaBADLD78 | (Baba et al., 2006) |
| BW25113 ΔghoT | BW25113 ΔghoT Ω KmR | (Baba et al., 2006) |
| BW25113 ΔghoT ΔKm | BW25113 ΔghoT | this study |
| BW25113 ΔghoS | BW25113 ΔghoS Ω KmR | (Baba et al., 2006) |
| BW25113 ΔghoS ΔKm | BW25113 ΔghoS | this study |
| BW25113 ΔmqsRA | BW25113 ΔmqsRA Ω KmR | (Kim et al., 2010) |
| BW25113 ΔghoS ΔmqsRA | BW25113 ΔghoS ΔmqsRA Ω KmR | this study |
| Plasmids | ||
| pCP20 | AmpR & CmR; temperature-sensitive replication, thermal induction of FLP recombinase synthesis | (Datsenko and Wanner, 2000) |
| pCA24N | CmR; lacIq, pCA24N | (Kitagawa et al., 2005) |
| pCA24N-mqsR | CmR; lacIq, pCA24N PT5-lac::mqsR+ | (Kitagawa et al., 2005) |
| pCA24N-ghoT | CmR; lacIq, pCA24N PT5-lac::ghoT+ | (Kitagawa et al., 2005) |
| pCA24N-ghoT-GCU | CmR; lacIq, pCA24N PT5-lac::ghoT + with two 5′-GCU sites in ghoT mRNA | this study |
| pBS(Kan) | KmR; pBS(Kan) | (Canada et al., 2002) |
| pBS(Kan)-mqsR | KmR; pBS(Kan) Plac::mqsR+ | (Kim et al., 2010) |
| pET30a-mqsR | KmR; pET30a(Kan) PT7::mqsR+ | (Brown et al., 2012) |
AmpR, CmR, and KmR are ampicillin, chloramphenicol and kanamycin resistance, respectively.
Microscopy
For the measurement of the cell size after exposure to ampicillin, overnight cultures of BW25113/pCA24N, BW25113/pCA24N-mqsR and ΔghoT/pCA24N-mqsR were inoculated into LB with chloramphenicol (30 μg ml−1) at a turbidity of 0.1, and 1 mM IPTG and ampicillin (20 μg ml−1) were added. Cell samples were taken at 0 h, 4 h and 10 h, and washed with 0.85% NaCl. For the measurement of the cell size after exposure to ampicillin for GhoT overproduction, overnight cultures of ΔghoT/pCA24N and ΔghoT/pCA24N-ghoT were inoculated into LB with chloramphenicol at a turbidity of 0.1, and 1 mM IPTG was then added to induce ghoT expression for 2 h. Cells were washed and resuspended in LB, and ampicillin (20 μg ml−1) was added to the cell suspension. Cell size was measured with a Zeiss Axiovert microscope with a 40× dry objective.
To evaluate cell membrane integrity with GhoT, the Live/Dead BacLight™ Bacterial Viability Kit (Molecular Probes) was used. The kit contains SYTO 9 and propidium iodide to differentiate between cells with intact membranes (live) and cells with damaged membranes (dead). After inducing ghoT and mqsR for 3 h with 0.5 mM IPTG, BW25113 ΔghoT ΔKm/pBS(Kan)-mqsR/pCA24N-ghoT and BW25113 ΔghoT ΔKm/pBS(Kan)-mqsR/pCA24N-ghoT-GCU were harvested by centrifugation (15,000×g, 10 min), washed, and re-suspended in 0.85% NaCl to reach a turbidity of 0.3 at 600 nm. Cells were then stained with 0.15 mM propidium iodine and 0.025 mM SYTO 9 dye for 1 h at ambient temperature. An aliquot of each cell suspension was also treated with 70% isopropyl alcohol to use as a dead cell control. Bacterial cells were imaged using a fluorescence microscope (Zeiss Axiovert) with 40 × dry objective.
qRT-PCR
For qRT-PCR, 50 ng of total RNA was used for qRT-PCR using the Power SYBR® Green RNA-to-CT™ 1-Step Kit and the StepOne™ Real-Time PCR System (Applied Biosystems). Primers were annealed at 60°C, and rrsG (Wang et al., 2009) was used to normalize the data.
Site-directed mutagenesis
Site-directed mutagenesis (Wang et al., 2011) was used to introduce two 5′-GCU sites into the coding region of ghoT in pCA24N-ghoT to create pCA24N-ghoT-GCU (Fig. 2) using two primer pairs, GhoT-GCU-1 and GhoT-GCU-2, respectively (Table 2). DNA sequencing using the BigDye Terminator Cycle Sequencing kit was performed to confirm the targeted mutations at these sites.
MqsR endoribonuclease assay
Refolding and purification of MqsR was performed as described (Brown et al., 2012) after producing MqsR from pET30a-mqsR. For the synthesis of ghoS, ghoT and ghoT-GCU mRNAs, PCR products were obtained using the primers shown in Table 2 and were used as templates for in vitro transcription with T7 RNA polymerase. The T7 RNA polymerase promoter sequence was included in the forward primers. PCR products were purified using the PureLink PCR purification kit (Invitrogen), and 1 μg of the PCR product was used as the template for the in vitro RNA reaction with the AmpliScribe T7-Flash transcription kit (Epicentre). The reaction mixture for the MqsR endoribonuclease cleavage assay (10 μL) contained 2 μg mRNA, 100 mM KCl, 2.5 mM MgCl2, and either 30 ng of purified MqsR protein in MqsR buffer (10 mM Tris pH 7.5, 100 mM NaCl, 0.5 mM TCEP) or an equivalent volume of MqsR buffer (for those reaction without MqsR protein). The reaction mixture was incubated at 37°C for 15 min and quenched by the addition of an equal volume of 2× sample loading buffer (Invitrogen). The reaction products were resolved by electrophoresis with RNA denaturing gels (15% polyacrylamide with 7 M urea, Invitrogen).
Acknowledgments
This work was supported by the NIH (R01 GM089999 to T.W.), the NSF (CAREER award MCB 0952550 to R.P.), the NSFC (31270214 to X.W.), and the 1000-Youth Elite Program from China (to X. W.). We are grateful for the Keio and ASKA strains provided by the Genome Analysis Project in Japan and for the help of Brian Kwan with a persistence assay. T.W. is the Biotechnology Endowed Professor at the Pennsylvania State University.
References
- Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet. 2009;5:e1000390. doi: 10.1371/journal.pgen.1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Brown BL, Wood TK, Peti W, Page R. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J Biol Chem. 2011;286:2285–2296. doi: 10.1074/jbc.M110.172643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BL, Lord DM, Grigoriu S, Peti W, Page R. The E. coli toxin MqsR destabilizes the transcriptional repression complex formed between the antitoxin MqsA and the mqsRA operon promoter. J Biol Chem. 2012 doi: 10.1074/jbc.M112.421008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowski M, Rojowska A, Wladyka B. Prokaryotic toxin-antitoxin systems-the role in bacterial physiology and application in molecular biology. Acta Biochim Pol. 2011;58:1–9. [PubMed] [Google Scholar]
- Canada KA, Iwashita S, Shim H, Wood TK. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J Bacteriol. 2002;184:344–349. doi: 10.1128/JB.184.2.344-349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen-Dalsgaard M, Jørgensen MG, Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol Microbiol. 2010;75:333–348. doi: 10.1111/j.1365-2958.2009.06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka J, Lee J, Bansal T, Wood TK. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol. 2007;9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Donachie WD, Begg KJ. Growth of the bacterial cell. Nature. 1970;227:1220–1224. doi: 10.1038/2271220a0. [DOI] [PubMed] [Google Scholar]
- Dörr T, Vulić M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F, Van Melderen L. Toxins-antitoxins: diversity, evolution and function. Crit Rev Biochem Mol Biol. 2011;46:386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- Hu Y, Benedik MJ, Wood TK. Antitoxin DinJ influences the general stress response through transcript stabilizer CspE. Environmental Microbiology. 2012;14:669–679. doi: 10.1111/j.1462-2920.2011.02618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Cruz JW, Ouyang M, Woychik NA. Bacterial toxin RelE mediates frequent codon-independent mRNA cleavage from the 5′ end of coding regions in vivo. J Biol Chem. 2011;286:14770–14778. doi: 10.1074/jbc.M110.108969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Wood TK. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun. 2010;391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Wang X, Zhang XS, Grigoriu S, Page R, Peti W, Wood TK. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ Microbiol. 2010;12:1105–1121. doi: 10.1111/j.1462-2920.2009.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Micro. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- Maeda T, Sanchez-Torres V, Wood TK. Metabolic engineering to enhance bacterial hydrogen production. Microb Biotechnol. 2008;1:30–39. doi: 10.1111/j.1751-7915.2007.00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci USA. 2012;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Masuda H, Tan Q, Awano N, Wu KP, Inouye M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol Microbiol. 2012;84:979–989. doi: 10.1111/j.1365-2958.2012.08068.x. [DOI] [PubMed] [Google Scholar]
- Pecota DC, Wood TK. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol. 1996;178:2044–2050. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol. 2004;64:515–524. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- Rolinson GN. Effect of beta-lactam antibiotics on bacterial cell growth rate. J Gen Microbiol. 1980;120:317–323. doi: 10.1099/00221287-120-2-317. [DOI] [PubMed] [Google Scholar]
- Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staugaard P, van den Berg FM, Woldringh CL, Nanninga N. Localization of ampicillin-sensitive sites in Escherichia coli by electron microscopy. J Bacteriol. 1976;127:1376–1381. doi: 10.1128/jb.127.3.1376-1381.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wood TK. Toxin/antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Wood TK. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J. 2009;3:1164–1179. doi: 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Hong SH, Ma Q, Brown BL, Pu M, et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol. 2011;7:359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol. 2012;8:885–861. doi: 10.1038/nchembio.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Park JH, Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem. 2009;284:28746–28753. doi: 10.1074/jbc.M109.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]