Abstract
Purpose
To evaluate the reproducibility of spectroscopic measurements from the anterior cingulate (AC) and the dorsolateral prefrontal cortex (DLPFC) regions at 7 T using a 32-channel head coil.
Materials and Methods
Spectra were acquired in four healthy subjects each scanned twice using a STEAM sequence, and a MEGA-PRESS-IVS sequence for GABA editing. STEAM spectra were quantified using LCModel, whereas MEGA-PRESS-IVS data were analyzed using peak integrals determined using in-house software. Mean coefficient of variation (CV) and mean absolute difference between visits were calculated.
Results
For the AC STEAM dataset, the mean CV between visits was 6.2% for prominent metabolites such as NAA, tCr, and tCho and 6.3% for low SNR metabolites such as Glu, Gln, and GABA. The mean CV between visits for the DLPFC STEAM dataset was 8.5% for high prominent metabolites and 21% for lower SNR metabolites. In the AC, the reproducibility measures for GABA were superior for STEAM compared to MEGA-PRESS-IVS (mean CV of 3.5% versus 13.6%), but the opposite pattern was observed in the DLPFC region (mean CV of 16.2% versus 13.4%).
Conclusion
7 T MRS of the AC and DLPFC using both short TE STEAM and MEGA-PRESS-IVS sequences provide excellent reproducibility of twelve metabolites, including GABA.
Keywords: magnetic resonance spectroscopy, reproducibility, 7 T, STEAM, GABA
Introduction
High field human MR systems operating at 7 T are becoming available in academic medical centers worldwide. For studies of the brain using proton MR spectroscopy (1H MRS), they offer the advantages of increased frequency separation and SNR (1,2) compared to lower field strengths (3–6). However, there are also technical challenges associated with high field strengths, such as increased susceptibility effects, inhomogeneous transmit (B1) fields, and increased chemical shift dispersion (CSD). Therefore, it is important to establish reproducibility of 7 T MRS in normal subjects before undertaking studies in neurological or psychiatric diseases. To date, there has only been one study that examined the reproducibility of 7 T 1H MRS STEAM in the anterior cingulate and the insula regions (3). Here, we expand upon this previous research by investigating the reproducibility of short TE STEAM (7) and MEGA-PRESS-IVS (8) for GABA acquired from the anterior cingulate (AC) and the dorsolateral prefrontal cortex (DLPFC) regions. These regions were chosen because of their significant involvement in psychiatric disorders such as schizophrenia (9,10). Additionally, we report reproducibility measurements for N-acetylaspartylglutamate (NAAG), an important modulator of glutamate neurotransmission (11,12).
Materials and Methods
All experiments were performed on a 7 T scanner (Philips ‘Achieva’, Best, The Netherlands) equipped with a 32-channel receive head coil and a quadrature transmit coil (Nova Medical, Orlando, FL). The nominal peak B1 of the transmit coil was 15 µT. Written informed consent was obtained after local institutional review board approval. Spectroscopic data were acquired from the AC (Figure 1a) and DLPFC (Figure 1b) regions in four healthy subjects (2 male/2 female, mean age: 24.1 ± 2.3) each scanned twice (average duration between scans: 8 ± 2 days). Voxel placement was based on high-resolution sagittal 3D MPRAGE scans which were also reformatted in axial and coronal views. To ensure reliable placement, the AC voxel was prescribed on the midline, angulated parallel to anterior segment of the corpus callosum, and using the anterior tip of the genu as the landmark depicting the voxel’s most anterior point. For the DLPFC, the voxel was oriented parallel to cortical surface in the left frontal lobe, and placed as anterior as possible without crossing the midline of the two hemispheres. Screen shots of the voxel location in all 3 planes were recorded for visit 1, and then visually matched during visit 2. STEAM spectroscopic parameters were: TR/TM/TE = 3000/25/14-ms, 32 averages, scan time = 1 min 36 sec, 2048 complex points, 3-kHz spectral bandwidth, VOI 27-cm3and VAPOR water suppression (13). Prior to acquisition, localized power optimization and projection-based 2nd order shimming were performed (14). A non-suppressed water reference scan (4 averages) was acquired for phase and eddy current correction as well as quantification. In addition, spectra were also acquired using a MEGA-PRESS-IVS sequence for quantification of GABA from both regions at each visit from the same four subjects for the AC and from three out of the four healthy subjects (2 male/1 female, mean age: 23.3 ± 2) for the DLPFC. The MEGA-PRESS-IVS parameters were: TR/TE = 3000/70-ms, 20 dynamics (10 with ‘on’ and 10 with ‘off’ editing pulses, respectively, 8 averages per dynamic), scan time = 8 minutes, 2048 complex points, and 3-kHz spectral width. For editing, 14-ms Gaussian editing pulses were used, the ‘on’ pulse being applied at 1.9 ppm and the ‘off’ pulse at 1.5 ppm, in order to suppress the co-editing macromolecule which has a chemical shift of 1.7 ppm (15). To suppress regions of the PRESS voxel where the modulation pattern is unfavorable for GABA-editing, the IVS technique was used. The PRESS voxel size was increased in the anterior-posterior direction from 30-mm to 38-mm, and an 8-mm thick saturation pulse was placed in the anterior region. 8 mm corresponds to the chemical shift displacement between the 3.0 ppm and 1.9 ppm resonances of GABA with the slice selective 180° pulses used in the PRESS sequence. Note that an IVS pulse is only applied in one direction, corresponding to the 2nd slice selective 180° pulse (TE2), during which the editing pulses are applied (TE2 >> TE1).
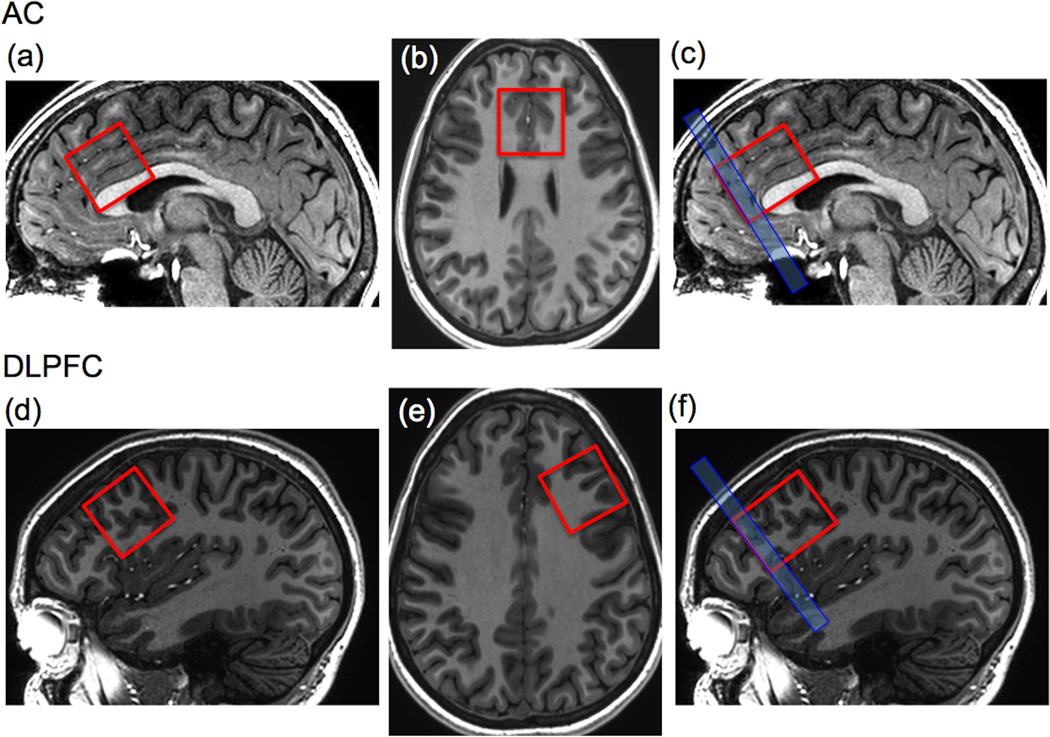
Figure 1.
Depiction of 30x30x30 mm AC voxel on (A) sagittal and (B) axial T1W images for the STEAM pulse sequence, and (C) for the MEGA-PRESS-IVS experiment showing the 8 mm anterior elongation of the PRESS voxel and location of 8 mm thick IVS pulse. (D) and (E) corresponding STEAM voxel location for the DLPFC voxel, and (F) voxel and IVS placement for the DLPFC MEGA-PRESS-IVS sequence.
For the STEAM spectroscopy data, basis set spectra were simulated using the GAVA software package (16). The GAVA software was modified to yield a Lorentzian line shape instead of its default Gaussian shape. The basis set consisted of the following twenty metabolites: Alanine (Ala), Aspartate (Asp), Creatine (Cr), γ-aminobutyric acid (GABA), glucose (Glc), glutamate (Glu), glutamine (Gln), glycine (Gly), glycerophosphocholine (GPC), glutathione (GSH), lactate (Lac), myo-Inositol (mI), N-acetylaspartate (NAA), N-acetylaspartylglutamate (NAAG), phosphocholine (PCh), phosphocreatine (PCr), phosphorylethanolamine (PE), serine (Ser), scyllo-Inositol (sI), and taurine (Tau). The basis set was imported into LCModel (17) and used for spectroscopic quantification. The LCModel parameters, ‘WCONC’ and ‘ATTH2O’, representing the voxel water content and T2 relaxation time correction factor respectively, were set to 55556 and 0.74, respectively. ATTH2O was altered to reflect the T2 of water at 7 T and the TE used in this study, while WCONC was set to the value of pure water, recognizing that both the metabolites and the bulk of the water signal are intracellular. In LCmodel, water scaling was performed using the water reference. All metabolite concentrations were reported in institutional units, and metabolites with CRLBs > 20% were rejected from further analysis. Spectra were apodized with a Chapman-Richards function (18) using an in-house IDL program (Exelis Visual Information Solutions, Boulder, CO, USA). For the GABA MEGA-PRESS-IVS spectroscopy data, all peak areas were measured using manual integration (3.1 to 2.9 ppm range) after baseline correction (cubic spline with manual definition of baseline points) using in-house software (‘XXX’). In addition to descriptive statistics, reproducibility was measured via mean coefficient of variation (CV) and mean absolute differences between visit 1 and visit 2. Differences in CRLB’s between visits were investigated using t-tests. Reproducibility was considered excellent if both measures were less than 10% for the major metabolites in the spectrum, or less than 20% for the smaller signals.
Results
Figure 2 shows representative STEAM and MEGA-PRESS-IVS spectra from the AC and DLPFC in one subject, while figure 3 shows examples of the LCModel fitting results for each region. In the AC, mean LCModel spectral linewidths for visits 1 and 2 were 0.026 ppm and 0.029 ppm, respectively, while mean SNR values for visits 1 and 2 were 49 and 46. The following metabolites were consistently detected in all participants at each visit with CRLBs ≤ 20%: Asp, Cr, GABA, Glu, Gln, Gly, mI, NAA, NAAG, PCh, PCr, and PE as well as the combined metabolites reported by LCModel: tNAA (NAA+NAAG), tCho (GPC+PCh), tCr (Cr+PCr), Glx (Glu+Gln), and mI+Gly (Table 1). Ala, Glc, GPC, GSH, Tau, and Ser could not be consistently detected in all subjects for both visits, and are thus not included in Table 1. The mean CV between visits was below 17% for all metabolites, and the dominant metabolites of NAA, Glu, tCho, tCr, and mI all had between visit CVs of 7.5% or less. As expected, NAA, Glu, tCr, tCho, mI, and their respective combinations had excellent mean absolute differences between visits of 10.2% or less. The reproducibility for Gln was good with a CV of 11.5% and a mean absolute difference of 14.8%, and the ratio of Gln to Glu of approximately 22–24% was within the range previously reported in the literature (12). The reproducibility of GABA (measured from the STEAM spectra) and NAAG was excellent, with absolute differences of 5.0% and 14.7% respectively, and mean CVs of 3.5% and 11.2%.
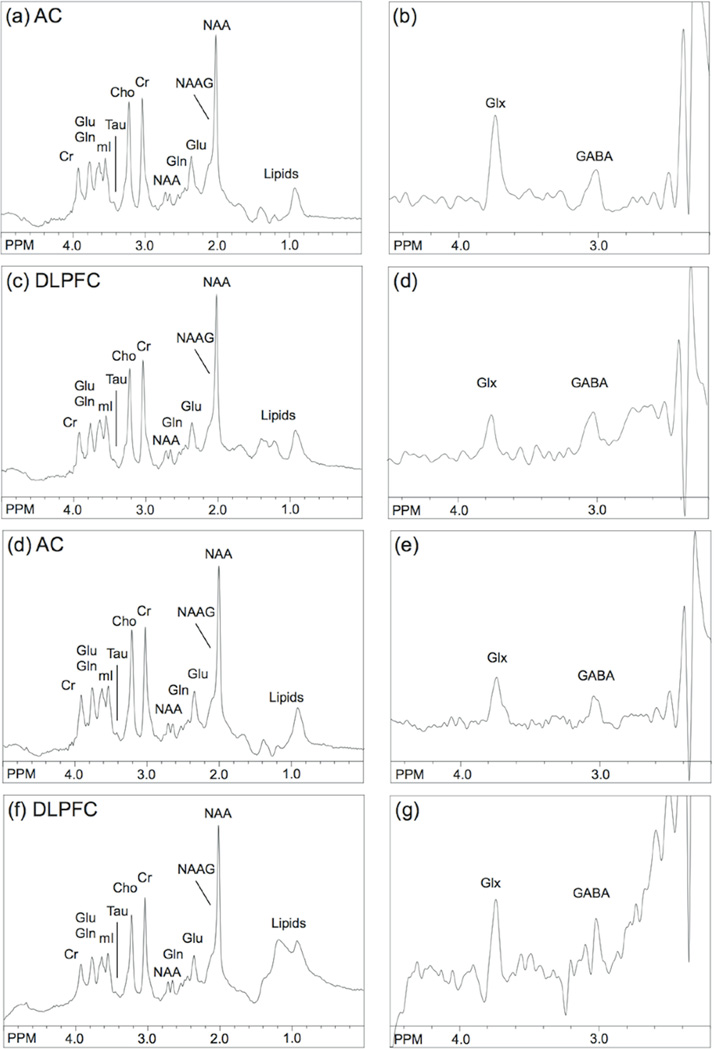
Figure 2.
Representative STEAM (A) and MEGA-PRESS-IVS (B) spectra from the AC region, and corresponding STEAM (C) and MEGA-PRESS-IVS (D) spectra from the DLPFC region in one subject. (E–G) show the corresponding spectra from the one-week follow-up in the same subject.
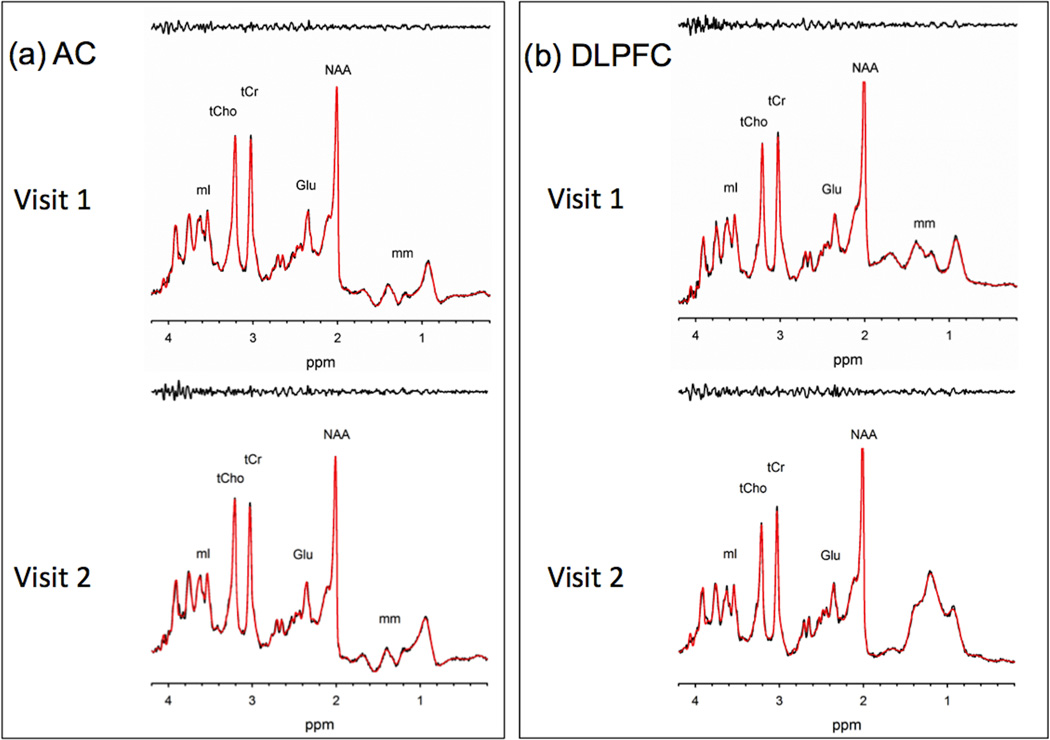
Figure 3.
Results of LCModel fitting results for 14-ms STEAM spectra from (A) AC and (B) DLPFC in one subject. (C) and (D) are the fitting results from the one-week follow-up in the same subject.
Table 1.
LCModel Metabolite concentrations from the Anterior Cingulate with CRLB ≤ 20%
| Metabolites | Visit | Mean Conc. | (IU) STD. | Mean CRLB (%) | Mean CV (%) | Mean Abs Diff |
|---|---|---|---|---|---|---|
| Asp | 1 | 5.08 | 0.9 | 10.3 | 16.9 | 21.0 |
| 2 | 4.00 | 0.7 | 12.0 | |||
| Cr | 1 | 5.28 | 0.3 | 3.8 | 9.8 | 12.4 |
| 2 | 4.64 | 0.4 | 3.3 | |||
| GABA | 1 | 3.20 | 0.1 | 9.3 | 3.5 | 5.0 |
| 2 | 3.22 | 0.3 | 8.3 | |||
| Glu | 1 | 15.88 | 0.3 | 3.0 | 4.0 | 5.4 |
| 2 | 15.02 | 0.4 | 2.5 | |||
| Gln | 1 | 3.94 | 0.6 | 7.5 | 11.5 | 14.8 |
| 2 | 3.36 | 0.6 | 7.5 | |||
| Gly | 1 | 1.40 | 0.2 | 16.8 | 7.3 | 10.0 |
| 2 | 1.36 | 0.2 | 12.0 | |||
| mI | 1 | 6.23 | 0.5 | 5.3 | 6.4 | 9.7 |
| 2 | 6.63 | 1.1 | 4.5 | |||
| NAA | 1 | 12.62 | 1.5 | 2.3 | 4.3 | 6.3 |
| 2 | 13.04 | 0.8 | 1.8 | |||
| NAAG | 1 | 3.49 | 0.3 | 6.3 | 11.2 | 14.7 |
| 2 | 3.13 | 0.4 | 4.8 | |||
| PCh | 1 | 1.94 | 0.5 | 6.3 | 13.5 | 22.2 |
| 2 | 2.17 | 0.2 | 3.0 | |||
| PCr | 1 | 4.11 | 0.5 | 4.8 | 6.3 | 8.9 |
| 2 | 4.13 | 0.5 | 3.8 | |||
| PE | 1 | 5.42 | 0.5 | 6.3 | 2.9 | 4.0 |
| 2 | 5.20 | 0.5 | 5.5 | |||
| tNAA | 1 | 16.11 | 1.6 | 1.8 | 2.6 | 3.6 |
| 2 | 16.17 | 0.9 | 1.5 | |||
| Glx | 1 | 19.82 | 0.8 | 2.8 | 5.3 | 7.2 |
| 2 | 18.38 | 0.4 | 2.5 | |||
| tCr | 1 | 9.39 | 0.7 | 2.0 | 7.5 | 10.2 |
| 2 | 8.77 | 0.6 | 1.8 | |||
| tCho | 1 | 2.33 | 0.1 | 3.3 | 6.9 | 9.3 |
| 2 | 2.17 | 0.2 | 2.8 | |||
| mI+Gly | 1 | 7.63 | 0.6 | 4.0 | 5.3 | 7.9 |
| 2 | 7.99 | 1.1 | 3.0 |
IU - institutional units; STD. - standard deviation
In the DLPFC region (Figure 2), the following metabolites were consistently detected in all participants at each visit with CRLBs ≤ 20%: Asp, Cr, GABA, Glu, mI, NAA, NAAG, PCr, and PE as well as the combined metabolites reported by LCModel: tNAA, tCho, tCr, Glx, and mI+Gly (Table 2). Similar to the AC results, Ala, Glc, GPC, GSH, Tau, and Ser could not be consistently detected in all subjects for both visits, and thus are not included in Table 2. Mean LCModel spectral line widths for visits 1 and 2 were 0.029 ppm and 0.031 ppm, respectively, while mean SNR values for visits 1 and 2 were 50 and 42. For Gln, Gly, and PCh, there was one visit where a participant (different each time) had a CRLB > 20%, but less < 50%. This negatively affected the reproducibility measures for these metabolites, with: mean absolute differences of 33.5%, 27.9%, and 147% and mean CVs of 33.0%, 16.9%, and 44.1% for Gln, Gly, and PCh, respectively. The mean CV between visits was below 45% for all metabolites, and the dominant metabolites of NAA, Glu, tCho, tCr, and mI all had between visit CVs of 13.6% or less and mean absolute differences of 19.7% or less. The reproducibility of GABA (measured from the STEAM spectrum) and NAAG was good with absolute differences of 25.1% and 17.7%, respectively, and the mean CVs were very similar at 16.2% and 14.2%. GABA had one of the larger mean absolute differences among all of the metabolites with 25%. In this region, the superior reproducibility of PE for both measures over NAA was unexpected.
Table 2.
LCModel Metabolite concentrations from the DLPFC with CRLB ≤ 20%
| Metabolites | Visit | Mean Conc. | (IU)STD. | Mean CRLB (%) | Mean CV (%) | Mean Abs Diff (%) |
|---|---|---|---|---|---|---|
| Asp | 1 | 2.92 | 1.0 | 14.3 | 22.1 | 127.0 |
| 2 | 3.65 | 0.7 | 12.8 | |||
| Cr | 1 | 3.63 | 0.5 | 5.0 | 10.0 | 13.9 |
| 2 | 3.57 | 0.6 | 4.3 | |||
| GABA | 1 | 2.08 | 0.5 | 11.0 | 16.2 | 25.1 |
| 2 | 2.24 | 0.6 | 10.5 | |||
| Glu | 1 | 10.56 | 1.1 | 4.0 | 13.6 | 17.4 |
| 2 | 9.57 | 1.3 | 3.5 | |||
| Gln | 1 | 2.65 | 0.5 | 9.3 | 33.0 | 33.5 |
| 2 | 1.91 | 1.1 | 20.8 | |||
| Gly | 1 | 1.00 | 0.2 | 14.0 | 16.9 | 27.9 |
| 2 | 0.95 | 0.2 | 17.0 | |||
| mI | 1 | 4.86 | 1.1 | 5.5 | 13.2 | 18.6 |
| 2 | 4.52 | 0.6 | 5.3 | |||
| NAA | 1 | 10.03 | 0.9 | 2.3 | 6.4 | 9.6 |
| 2 | 10.98 | 1.1 | 1.8 | |||
| NAAG | 1 | 2.85 | 0.3 | 6.3 | 14.2 | 17.7 |
| 2 | 2.37 | 0.6 | 6.3 | |||
| PCh | 1 | 0.96 | 0.5 | 22.0 | 44.1 | 147.0 |
| 2 | 1.47 | 0.2 | 3.5 | |||
| PCr | 1 | 2.97 | 0.2 | 5.8 | 5.4 | 7.5 |
| 2 | 2.86 | 0.1 | 5.0 | |||
| PE | 1 | 3.75 | 0.2 | 6.8 | 4.4 | 6.0 |
| 2 | 3.73 | 0.4 | 7.0 | |||
| tNAA | 1 | 12.88 | 0.6 | 1.8 | 3.4 | 5.0 |
| 2 | 13.35 | 1.0 | 1.5 | |||
| Glx | 1 | 13.21 | 0.9 | 3.0 | 14.5 | 17.5 |
| 2 | 11.48 | 2.4 | 3.5 | |||
| tCr | 1 | 6.60 | 0.4 | 2.3 | 5.7 | 7.8 |
| 2 | 6.42 | 0.7 | 2.3 | |||
| tCho | 1 | 1.38 | 0.2 | 3.8 | 13.5 | 19.7 |
| 2 | 1.47 | 0.2 | 3.5 | |||
| mI+Gly | 1 | 5.86 | 1.3 | 4.5 | 9.3 | 12.8 |
| 2 | 5.47 | 0.7 | 4.0 |
IU - institutional units; STD. - standard deviation; BOLD – not found in all subjects
For the MEGA-PRESS-IVS experiments, very similar reproducibility for GABA was shown for both regions (Table 3). For the AC region, GABA measured using the short TE STEAM sequence showed better mean CV (3.5% versus 13.6%) and mean absolute differences (5.0% versus 16.5%) than the MEGA-PRESS-IVS sequence. However, for the DLPFC, the MEGA-PRESS-IVS sequence gave a slightly better mean CV (13.4% versus 16.2%) and mean absolute difference (18.1% versus 25.1%) than the short TE STEAM sequence.
Table 3.
GABA concentrations using MEGA-PRESS-IVS in the AC and DLPFC
| Region | Visit | GABA/H20 (x10−6 IU) |
STD. | Mean CV (%) |
Mean Abs Diff (%) |
|---|---|---|---|---|---|
| AC | 1 | 4.85 | 0.8 | 13.6% | 16.5% |
| 2 | 4.39 | 0.6 | |||
| DLPFC | 1 | 7.62 | 0.2 | 13.4% | 18.1% |
| 2 | 7.33 | 0.2 |
IU - institutional units, STD. - Standard Deviation
For both STEAM and MEGA-PRESS-IVS spectra, there were no significant differences in CRLB’s between the 1st and 2nd time points (t-test, P > 0.05).
Discussion
This study demonstrates that localized single voxel spectroscopy of the human brain performed at 7 T with signal reception using a 32-channel head coil, with scan times as short as 2 minutes, gives highly reproducible results in two different cortical gray matter regions. CV’s of the major metabolites in the spectrum are well under 10%, and even those of smaller, more difficult to estimate signals (e.g. GABA, NAAG, Gln) were also under 20% (except for Gln in DLPFC). Similar reproducibility was also observed for GABA at 7 T using the MEGA-PRESS-IVS sequence, albeit with a much longer scan time (8 min). The MEGA-PRESS-IVS experiment has lower SNR, since the editing technique inherently does not detect the full GABA signal, with most of the central component of the 3.0 ppm ‘pseudo-triplet’ being suppressed in MEGA-PRESS, as well as T2 losses due to the longer TE of the editing sequence. However, its potential advantage over non-edited STEAM spectra is that it should be a relatively pure measure of GABA, uncontaminated by overlap with signals from other molecules (such as creatine, macromolecules, etc.). The CV’s and mean absolute differences of metabolites reported here may be useful for input into sample size calculators for future studies comparing disease groups; note the the expected power to detect differences will be much higher for the major metabolite signals which are highly reproducible (e.g. tNAA) than for small signals such as GABA or Gln.
The high reproducibility of the current results are not unexpected, since previous studies have demonstrated (e.g. comparing single voxel MRS at 4 and 7 T) increased SNR, increased detectability of metabolites with low concentrations, and improved CRLBs for all metabolites that were quantified (2). To achieve excellent MRS reproducibility at 7 T, however, a number of factors concerning data acquisition must be addressed, including shimming, B1 homogeneity, and RF pulse calibration. At lower field strengths such as 1.5T, adjustment of only first-order linear shim currents is generally sufficient for obtaining acceptable line widths; however, the increased magnetic susceptibility effects at higher field strengths necessitate adjustment of higher order shim currents to obtain sufficient line widths (1,19,20). In this study, the ‘FASTMAP’ technique in combination with shim corrections up to 3rd order was employed to obtain line widths in the range 0.026 to 0.031 ppm (7.8 Hz to 9.2 Hz) (14).
Also at high fields, the transmit B1 field is more inhomogeneous compared to lower field strengths, and higher RF power is typically required than at lower field strengths to obtain the same B1 level. B1 inhomogeneity may directly affect the RF flip angle and efficiency of the localization sequence. In the current study localized power-optimization directly on the localized volume was used to ensure accurate flip angle calibration in each voxel; however, it was noted that the maximal achievable B1 field was both lower and more variable in the DLPFC than the AC with the currently used transmit coil; this may well explain the more variable SNR and CV values reported for the DLPFC compared to the AC.
In a previous study at 7 T (3), the metabolite CV values from the AC were very similar to the values presented here for Glu, Gln, mI, tCho, and NAA; for GABA, the current CV of 3.5% from the STEAM sequence was lower than the 10% reported in the prior study, but for tCr the prior study had a better CV (3% vs. 7.5%). These differences may be in part due to differences in data acquisition (e.g. voxel size, scan time, receive coil used) as well as the basis sets used for LCModel analysis; the current study included 20 metabolites in the basis set, whereas a basis set of 10 metabolites (including an absence of NAAG) was used in reference (3).
For comparing GABA reproducibility between localization sequences, it is important to note that spectral fitting techniques were also different between the two approaches: for STEAM, the LCModel was used, while for MEGA-PRESS-IVS, manual peak integration was used. For quantification of the STEAM spectra, the GABA resonances are fit using a complete GABA spectrum incorporated into LCModel’s basis set; however, for quantification of the MEGA-PRESS-IVS spectra, only the GABA signal at 3.01 ppm is fit using peak integration since it is a single, non-overlapped resonance. Manual integration of the short TE STEAM spectra is expected to be unreliable because of the many overlapping resonances. It is important to recognize that the overall reproducibility depends on both the data acquisition and analysis techniques. We also note that we expect chemical shift displacement (CSD) effects to be insignificant with either localization sequence; in the case of MEGA-PRESS, the IVS pulses should ensure that the GABA signal is coming from the prescibed location, and in the case of the STEAM sequence high bandwidth frequency modulated slice selective excitation pulses were used (3.5 kHz bandwidth) which minimize CSD effects.
Compared to a previous study at 3T (21) using the PRESS sequence, 8-channel head coil and a longer scan time but small voxel size, CV values for the AC from the current study at 7 T were either similar, or in some cases better, than those found at 3T. While it was possible to quantify Glu, Gln, and GABA at 3T, only the CV for Glu was comparable to the current study, with the CV for Gln and GABA being much superior at 7 T. Differences in these values may be attributable to the improved spectral resolution and SNR of these compounds at the higher field strength.
To our knowledge, there are no studies that have examined reproducibility of GABA at 7 T. There are four studies that assessed reproducibility of GABA+ (i.e. the combined GABA and macromolecule, often 50% larger than the ‘pure’ GABA signal recorded in this 7 T study) acquired using MEGA-PRESS at 3T in vivo. Using similarly sized voxels, one study found occipital and sensorimotor cortex CVs averaged across time and subjects of 9.1% and 12%, respectively (22), while another determined inter-individual CV’s that ranged from 19.2%–21.7%, depending on referencing and fitting methodology from the occipital lobe (23). While acquired from different regions, these CVs are comparable to those acquired in the current study. Another study at 3T acquired GABA spectra using MEGA-PRESS from the left dorsolateral prefrontal cortex (24). Again, the voxel sizes were comparable; however, the 3T study used twice the number of acquisitions (320 versus 160 in the current study). The within-session CVs reported at 3T ranged from 7–12% depending on the fitting software utilized, which are similar to the current study’s GABA CV of 13.4% in the DLPFC. A fourth study (28) examining the reproducibility of GABA in the anterior cingulate cortex and right frontal white matter using a modified MEGA-PRESS sequence with additional editing pulses showed within subject CVs of 5.27% and 8.64%, respectively . The GABA CV was similar to the current study with a smaller voxel (18 cm3) but nearly five times the number of acquisitions. Large voxels in conjunction with a low number of acquisitions allowed acquisition of unedited (STEAM) and edited (MEGA-PRESS) data from the same voxel and kept scan times to a minimum allowing for complete data acquisition of multiple regions and multiple localization methods in this study. However, GABA CVs were comparable to those of previously published studies, which can most likely be attributed to the increased SNR at 7 T and suppression of the macromolecule signals using the MEGA-PRESS-IVS technique (8).
In summary, both short TE STEAM and MEGA-PRESS-IVS at 7 T show excellent reproducibility of twelve metabolites including GABA in the AC, and nine metabolites including GABA in the DLPFC. Although the STEAM sequence has previously been shown to be well suited to use at high magnetic field strengths (1,2,9), other pulse sequences have been demonstrated recently at 7 T such as ‘SPECIAL’ (25) or ‘SEMI-LASER’ (26) which have significantly higher SNR ratios; it is expected that these sequences may provide even better reproducibility. It is also anticipated that sequences such as these may be used in combination with spectral editing to further improve edited GABA detection and reproducibility at 7 T (27).
Acknowledgments
Grant Support: NIH T32MH067533 (SAW) K01MH077230 (LMR) and P41EB015909, R01MH096263 and R21MH082322 (PBB)
References
- 1.Tkac I, Andersen P, Adriany G, Merkle H, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at 7 T. Magn Reson Med. 2001;46(3):451–456. doi: 10.1002/mrm.1213. [DOI] [PubMed] [Google Scholar]
- 2.Tkac I, Oz G, Adriany G, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: metabolite quantification at 4T vs. 7T. Magn Reson Med. 2009;62(4):868–879. doi: 10.1002/mrm.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephenson MC, Gunner F, Napolitano A, et al. Applications of multi-nuclear magnetic resonance spectroscopy at 7T. World J Radiol. 2011;3(4):105–113. doi: 10.4329/wjr.v3.i4.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi C, Dimitrov IE, Douglas D, et al. Improvement of resolution for brain coupled metabolites by optimized (1)H MRS at 7T. NMR Biomed. 2010;23(9):1044–1052. doi: 10.1002/nbm.1529. [DOI] [PubMed] [Google Scholar]
- 5.Mekle R, Mlynarik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med. 2009;61(6):1279–1285. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- 6.Deelchand DK, Van de Moortele PF, Adriany G, et al. In vivo 1H NMR spectroscopy of the human brain at 9.4 T: initial results. J Magn Reson. 2010;206(1):74–80. doi: 10.1016/j.jmr.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frahm J, Merboldt KD, Hanicke W. Localized proton spectroscopy using stimulated echoes. J Magn Reson. 1987;72(3):502–508. doi: 10.1002/mrm.1910170113. [DOI] [PubMed] [Google Scholar]
- 8.Edden RA, Barker PB. Spatial effects in the detection of gamma-aminobutyric acid: improved sensitivity at high fields using inner volume saturation. Magn Reson Med. 2007;58(6):1276–1282. doi: 10.1002/mrm.21383. [DOI] [PubMed] [Google Scholar]
- 9.Kegeles LS, Mao X, Stanford AD, et al. Elevated Prefrontal Cortex gamma-Aminobutyric Acid and Glutamate-Glutamine Levels in Schizophrenia Measured In Vivo With Proton Magnetic Resonance Spectroscopy. Arch Gen Psychiatry. 2012;69(5):449–459. doi: 10.1001/archgenpsychiatry.2011.1519. [DOI] [PubMed] [Google Scholar]
- 10.Olbrich HM, Valerius G, Rusch N, et al. Frontolimbic glutamate alterations in first episode schizophrenia: evidence from a magnetic resonance spectroscopy study. World J Biol Psychiatry. 2008;9(1):59–63. doi: 10.1080/15622970701227811. [DOI] [PubMed] [Google Scholar]
- 11.Pouwels PJ, Frahm J. Differential distribution of NAA and NAAG in human brain as determined by quantitative localized proton MRS. NMR Biomed. 1997;10(2):73–78. doi: 10.1002/(sici)1099-1492(199704)10:2<73::aid-nbm448>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 13.Tkac I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999;41(4):649–656. doi: 10.1002/(sici)1522-2594(199904)41:4<649::aid-mrm2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 14.Gruetter R. Automatic, localized in vivo adjustment of all first- and second-order shim coils. Magn Reson Med. 1993;29(6):804–811. doi: 10.1002/mrm.1910290613. [DOI] [PubMed] [Google Scholar]
- 15.Henry PG, Dautry C, Hantraye P, Bloch G. Brain GABA editing without macromolecule contamination. Magn Reson Med. 2001;45(3):517–520. doi: 10.1002/1522-2594(200103)45:3<517::aid-mrm1068>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 16.Soher BJ, Young K, Bernstein A, Aygula Z, Maudsley AA. GAVA: spectral simulation for in vivo MRS applications. J Magn Reson. 2007;185(2):291–299. doi: 10.1016/j.jmr.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 18.Pienaar L, Turnball K. The chapman-richards generalization of von bertalanffy's growth model for basal area growth and yield in even-aged stands. Forest Sci. 1973;19:2–22. [Google Scholar]
- 19.Tkac I, Gruetter R. Methodology of H NMR Spectroscopy of the Human Brain at Very High Magnetic Fields. Appl Magn Reson. 2005;29(1):139–157. doi: 10.1007/BF03166960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norris DG. High field human imaging. J Magn Reson Imaging. 2003;18(5):519–529. doi: 10.1002/jmri.10390. [DOI] [PubMed] [Google Scholar]
- 21.Mullins PG, Chen H, Xu J, Caprihan A, Gasparovic C. Comparative reliability of proton spectroscopy techniques designed to improve detection of J-coupled metabolites. Magn Reson Med. 2008;60(4):964–969. doi: 10.1002/mrm.21696. [DOI] [PubMed] [Google Scholar]
- 22.Evans CJ, McGonigle DJ, Edden RA. Diurnal stability of gamma-aminobutyric acid concentration in visual and sensorimotor cortex. J Magn Reson Imaging. 2010;31(1):204–209. doi: 10.1002/jmri.21996. [DOI] [PubMed] [Google Scholar]
- 23.Bogner W, Gruber S, Doelken M, et al. In vivo quantification of intracerebral GABA by single-voxel(1)H-MRS-How reproducible are the results? Eur J Radiol. 2010;73(3):526–531. doi: 10.1016/j.ejrad.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 24.O'Gorman RL, Michels L, Edden RA, Murdoch JB, Martin E. In vivo detection of GABA and glutamate with MEGA-PRESS: reproducibility and gender effects. J Magn Reson Imaging. 2011;33(5):1262–1267. doi: 10.1002/jmri.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mekle R, Mlynárik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med. 2009;61(6):1279–85. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- 26.Boer VO, van Lier AL, Hoogduin JM, Wijnen JP, Luijten PR, Klomp DW. 7-T (1) H MRS with adiabatic refocusing at short TE using radiofrequency focusing with a dual-channel volume transmit coil. NMR Biomed. 2011;24(9):1038–1046. doi: 10.1002/nbm.1641. [DOI] [PubMed] [Google Scholar]
- 27.Andreychenko A, Boer VO, Arteaga de Castro CS, Luijten PR, Klomp DW. Efficient spectral editing at 7 T: GABA detection with MEGA-sLASER. Magn Reson Med. 2011 Dec 28; doi: 10.1002/mrm.24131. [DOI] [PubMed] [Google Scholar]
- 28.Geramita M, van der Veen JW, Barnett AS, Savostyanova AA, Shen J, Weinberger DR, Marenco S. Reproducibility of prefrontal γ-aminobutyric acid measurements with J-edited spectroscopy. NMR Biomed. 2011;24(9):1089–1098. doi: 10.1002/nbm.1662. [DOI] [PubMed] [Google Scholar]