Summary
Most mammalian pre-mRNAs are alternatively spliced in a manner that alters the resulting open reading frame. Consequently, alternative pre-mRNA splicing provides an important RNA-based layer of protein regulation and cellular function. The ubiquitous nature of alternative splicing coupled with the advent of technologies that allow global interrogation of the transcriptome have led to an increasing awareness of the possibility that widespread changes in splicing patterns may contribute to lymphocyte function during an immune response. Indeed, a few notable examples of alternative splicing have clearly been demonstrated to regulate T-cell responses to antigen. Moreover, several proteins key to the regulation of splicing in T cells have recently been identified. However, much remains to be done to truly identify the spectrum of genes that are regulated at the level of splicing in immune cells and to determine how many of these are controlled by currently known factors and pathways versus unknown mechanisms. Here we describe the proteins, pathways, and mechanisms that have been shown to regulate alternative splicing in human T cells and discuss what is and is not known about the genes regulated by such factors. Finally, we highlight unifying themes with regards to the mechanisms and consequences of alternative splicing in the adaptive immune system and give our view of important directions for future studies.
Keywords: gene regulation, alternative splicing, T-cell response, splicing factors, mRNA profiling
Introduction
The adaptive immune system is marked by its ability to detect an invading pathogen or foreign antigen and direct an appropriate effector response. T cells, for example, recognize antigen through the T-cell receptor (TCR), which triggers extensive proximal signaling and downstream signaling cascades. These signaling events ultimately lead to changes in cell morphology, proliferation, secretion of cytokines and cytotoxins, migration and homing, and a plethora of effector function (1). Studies over the past decades have focused on how transcriptional control regulates gene expression in an immune response. However, post-transcriptional mechanisms of gene regulation are increasingly emerging as important regulators of immune cell function. In particular, one mechanism of post-transcriptional gene regulation that has garnered much interest in the past few years is alternative pre-mRNA splicing.
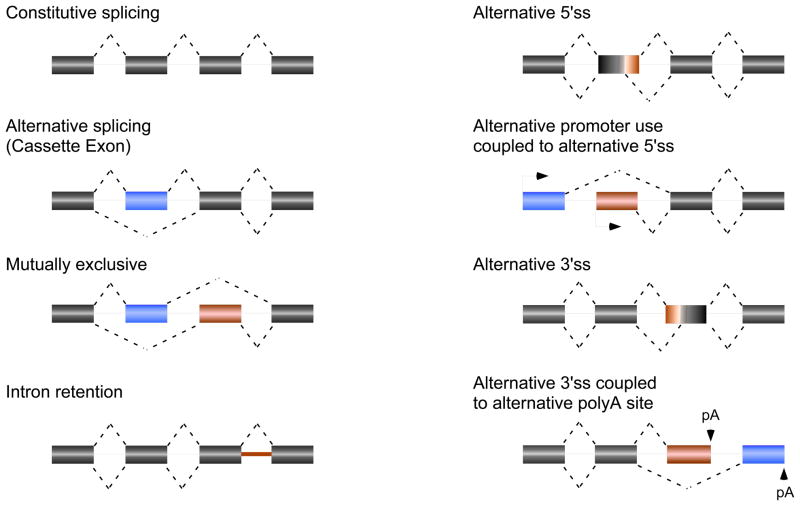
Alternative splicing (AS) is an essential and ubiquitous mechanism for regulated gene expression that goes beyond turning genes ‘on’ and ‘off’. In broad terms, AS refers to any case of variable inclusion of an exon, intron, or portion-thereof in some fraction of transcripts derived from a given gene. As such, AS results in the generation of multiple distinct mRNAs from a given gene. The most common form of AS is variable inclusion of an internal ‘cassette’ exon, although essentially all other potential patterns have been described in at least some genes (2) (Fig. 1). Variable inclusion of sequences internal to a gene (cassette exon, alternative splice site usage, or intron retention) typically leads to alterations in coding sequence, either by adding or deleting a peptide domain or by introducing premature stop codons (PTCs). By contrast, differential splicing of the 5′ or 3′ UTRs can alter the presence of regulatory motifs and miRNA binding sites, thereby influencing mRNA stability, expression, and/or translation (3, 4). Thus, regardless of pattern, AS has a profound effect on the identity, level, or expression pattern of the resulting protein.
Fig. 1. Common patterns of AS.
Rectangles represent exons and lines represent introns. Dotted lines connect exons that are joined together to form the final mRNA. Red and blue regions are those that can be differentially included in the mature mRNA. Dotted lines above and below represent the two alternate possible splicing patterns.
It is now well established that the majority of multi-exon mammalian genes (>90%) undergo AS (2, 5). Given the diversity and plasticity required for immune effector functions it seems inevitable that cells of the adaptive immune system rely on regulated pre-mRNA splicing to at least some extent to control protein expression and function. For many years, our laboratory has investigated the mechanisms by which AS is regulated in human T cells. Here we review what we and others have learned regarding the control of AS during immune responses, the functional impact of such regulation, and the questions that emerge from our current knowledge.
Breadth and impact of alternative splicing in immune responses
The accessibility of high-throughput genomics has shifted the way we think about AS from a gene-centric to a networks point of view. In light of this, a flurry of high throughput genomic studies has emerged in an attempt to identify AS programs that are coordinated in response to stimuli, perturbations to cell states, and differentiation among others. In particular, the studies listed in Table 1 have provided a quantitative and defined analysis of AS in various functional states of primary immune cells and model cell lines, and support a role for widespread regulated splicing in immune responses. Specifically, these studies have identified hundreds of potential changes in AS during T-cell activation, B-cell stimulation, and HIV (human immunodeficiency virus) infection. The described splicing changes include those predicted to impact the coding potential of the affect genes, as well as differential use of terminal exons which alters the presence of miRNA binding sites in the 3′ UTR (Table 1).
Table 1.
Studies Identifying Splicing Regulation in Immune Cells
| Cell Type | Conditions | Experimental Platform | Reference | Observations |
|---|---|---|---|---|
| JSL1 Jurkat, primary human CD4+ T cells | PMA (Jurkat), PHA (primary T cells) | RNA-seq | Martinez 2012 RNA 18:1029 | 178 AS events in response to activation of Jurkat cells, 80% validation rate (43/52) by RT-PCR. Validated in primary CD4+ T cells. AS genes enriched for functions relevant to immune response |
| primary human CD4+ T cells | HIV-1 infection | exon array | Imbeault 2012 PLoS Pathogens 8:e1002861 | 323, 129 and 107 AS events are regulated in response to HIV-1 infection of CD4+ T cells at 24, 48 and 72 h post-infection. |
| PBMCs | PHA, IL-2 & ionomycin | exon array | Whistler 2010 BMC Genomics 11:496 | 472 alternatively spliced transcripts, >70% validation rate (5/7) by RT-PCR, AS genes enriched for splicing factors |
| primary human CD2+ T, CD19+ B cells | α-CD3/28 (T cells), α-CD40, IL2 +IL10 (B-cells) | exon array | Grigoryev 2009 PLoS One 4:e7906 | Array predicts 50–70% of constitutively expressed genes in T cells and 40% in B cells are alternatively spliced. Only 4/22 predictions (18%) validate by RT-PCR |
| Primary murine T cells | α-CD3/28 | exon array | Sandberg 2008 Science 320:1643 | Quantify and validate AS of alternate terminal exons. Do not differentiate between mechanism of AS and APA |
| Primary murine T cells | naive and memory T cells purified from mice | exon array | Wu 2008 Immunity 29:863–75 | Provide quantitative measure of individual exons in numerous genes that differ in array expression between naive and memory murine T cells. No RT-PCR validation given. |
| JSL1 Jurkat, primary human CD4+ and CD8+ T cells | PMA (Jurkat), PHA (primary cells) | exon junction array | Ip RNA 2007 13: 563 | First large scale analysis of AS in T cells. Initial analysis done in Jurkat cells, validated predictions in primary CD4+ and CD8+ T cells |
One general conclusion from the profiling studies in Table 1 is that AS regulates a unique and substantial set of genes that are not altered at the level of transcription (6–8). This suggests that transcription and AS contribute independently to T-cell antigen responses, underscoring the unique role of AS in molding immune responses. Furthermore, some of the studies in Table 1 found that signal-responsive AS events are enriched in genes involved in many aspects of T-cell biology and effector functions (6, 7). Enriched activities include canonical T-cell signaling pathways, such as nuclear factor-κB (NF-κB) and c-Jun N-terminal kinase (JNK) and mitogen-activated protein kinase (MAPK) signaling, Rho-mediated changes in lymphocyte migration, and genes involved in cell proliferation (6, 7), again highlighting the potential impact of AS on immune responses.
We note that an important aspect of the papers in Table 1 was providing sufficient data to warrant the conclusions drawn. As high-throughput analyses are becoming more accessible, the need for caution in data analysis and interpretation is essential. It is widely accepted among splicing experts that a minimum threshold for confidence in AS is a 10% change (i.e. 20% inclusion to 30% inclusion of a variable exon) (2, 5). Below this differential, the ability to detect and quantify changes is not sufficient for rigorous conclusions. Indeed, in our hands, a minimum differential threshold of 20 percentage points has been required in analysis of RNA-Seq data in order for predicted changes to hold up to independent validation (6). Furthermore, the biologic significance of very minor changes in exon inclusion, even if statistically significant, is highly suspect. Thus, it is imperative that any profiling studies state the predicted differences for any AS events reported. Finally, defining the AS event beyond providing a gene name is necessary for any conclusions to be drawn, as there are often more than one potential AS event within a given gene.
Regardless of the platform and analysis pipeline used to profile mRNA expression, rigorous validation of AS predictions by independent assays such as reverse transcriptase polymerase chain reaction (RT-PCR) are of utmost importance to define threshold criteria for meaningful data and ultimately identify confirmed AS events. Importantly, validation rates differ considerably among published studies (Table 1), thus predictions from exon array or RNA-Seq cannot be assumed a priori to be accurate. However, even if the conclusions from some of the profiling experiments are overstated, the general conclusion that AS plays a functionally relevant role in immune responses has been well documented. Indeed, a literature search reveals close to one hundred genes that have now been validated to exhibit significant changes in isoform expression during an immune response, including several instances in which the AS changes are known to impact lymphocyte function (Table 2).
Table 2.
Validated Instances of Alternative Splicing during Immune Responses
| Gene | Exon a | Cell Type b | Condition c | Change | Reference |
|---|---|---|---|---|---|
| Cell Surface Receptors | |||||
| CD3-zeta | intron 8 | T | α-CD3/28 | retention | (10) |
| CD3-zeta | intron 4 | PBMC | PHA | retention | (116) |
| CD28 | exon 3 | T | α-CD3/28 | inclusion | (117) |
| CD45 | exon 4 | Jk, CD4 | various | skipping | (17) |
| CD45 | exon 5 | Jk, CD4 | various | skipping | (17) |
| CD45 | exon 6 | Jk, CD4 | various | skipping | (17) |
| CD46 | mutually exclusive | T | α-CD3/46 | inclusion of exon 14 | (118) |
| CD6 | exon 5 | T | various | skipping | (119) |
| CD8 | exon 7 | CD8 | α-CD3/28 | retention | (120) |
| CD83 | exon 3–4 | PBMC | various | inclusion | (121) |
| CD96 | exon 4 | CD4, CD8 | resting | skipping | (122) |
| CTLA-4 | exon 3 | T | α-CD3/28 | inclusion | (117) |
| Fas | exon 6 | PBMC | PHA | inclusion | (123) |
| HMMR | exon 3 | CD4, CD8 | PHA | inclusion | (8) |
| ICAM-1 | exon 2–5 | T, Thy | LPS | variable | (124) |
| PECAM1 | exon 14–15 | Jk | PMA | skipping | (125) |
| SEC16A | exon 29 | Jk, CD4 | PMA, PHA | variable d | (6) |
| SEC16A | exon 30 | Jk, CD4 | PMA, PHA | inclusion | (6) |
|
| |||||
| Cytokine-related | |||||
|
| |||||
| IL-4 | exon 2 | T | α-CD3 | inclusion | (126) |
| IL-7Rα | exon 6 | PBMC | HIV | variable | (30) |
| TIR8 | exon 8 | Jk, CD4 | PMA, PHA | inclusion | (6) |
|
| |||||
| Transcriptional regulators | |||||
|
| |||||
| ATF2 | exon 14 | Jk | PMA | inclusion | (6) |
| BRD8 | exon 20 | Jk, CD4 | PMA, PHA | skipping | (6) |
| EHMT2 | exon 10 | Jk, CD4 | PMA, PHA | inclusion | (6) |
| FOXP3 | exon 7 | CD4, Tregs | α-CD3/28 | skipping | (127) |
| GATA3 | exon 4 | CD4 | PHA | skipping | (8) |
| HIF1α | exon 14 | CD4 | PHA | inclusion | (8) |
| ILF3 | exon 9 | CD4, CD8 | PHA | inclusion | (8) |
| IRF1 | exon 7 | CD4, CD8 | PHA | variable e | (8) |
| LEF1 | exon 6 | Jk, Thy, CD4 | various | variable | (8, 45) |
| RUNX1 | exon 1–2 | CD4 | HIV-1 | skipping | (24) |
| ZNF384 | exon 5 | Jk | PMA | inclusion | (6) |
|
| |||||
| RNA-processing | |||||
|
| |||||
| AUF-1 | exon 2 | CD4, CD8 | PHA | inclusion | (8) |
| CELF2 | exon 6 | Jk, CD4, CD8 | PMA, PHA | variable d | (8, 45) |
| EIF4G2 | exon 3 | CD4, CD8 | PHA | skipping | (8) |
| FIP1L1 | exon 8 | Jk | PMA | skipping | (6) |
| HRB | exon 3 | CD4, CD8 | PHA | skipping | (8) |
| LUC7L | exon 2a | Jk, CD4 | PMA, PHA | variable e | (6) |
| RBM25 | exon 6 | PBMC | P2I | skipping | (128) |
| SAM68 | exon 3 | CD4, CD8 | PHA | skipping | (8) |
| SFRS2 | exon 2b | PBMC | P2I | skipping | (128) |
| SFRS10 | intron 1 | PBMC | P2I | removal | (128) |
| SNRPB | ND | PBMC | P2I | inclusion | (128) |
| TIA-1 | exon 5 | Jk | resting | skipping | (48) |
| U2AF | exon 11 | PBMC | P2I | inclusion | (128) |
| ZC3H14 | exon 10 | Jk, CD4 | PMA, PHA | inclusion | (6) |
|
| |||||
| Intracellular signaling/transport | |||||
|
| |||||
| AKAP9 | exon 4 | Jk, CD4 | PMA, PHA | variable d | (6) |
| CARD8 | exon 5 | Jk | PMA | skipping | (6) |
| CLK2 | exon 7 | CD4, CD8 | PHA | skipping | (8) |
| DOCK10 | mutually exclusive | T | resting | inclusion of exon 1a | (129) |
| Erk | exon 6 | CD8 | PHA | skipping | (8) |
| ERP29 | exon 2 | Jk | PMA | inclusion | (6) |
| Fyn | mutually exclusive | Jk | PMA | inclusion of exon 7b | (39) |
| Fyn | exon 9 | Jk | PMA | skipping | (6) |
| FR4 | intron 3 | CD4, Tregs | resting | retention | (130) |
| IRAK1 | exon 11 | CD4, Tregs | α-CD3/28 | skipping | (131) |
| IRAK1 | exon 12 | Thy | total | variable | (132) |
| MAP2K7 | exon 2 | Jk, CD4 | PMA, PHA | skipping | (6) |
| MAP3K7 | exon 13 | Jk, CD4 | PMA, PHA | skipping | (6) |
| MAP4K2 | exon 9 | Jk, CD4 | PMA, PHA | variable e | (8) |
| PPP1R12A | exon 15 | Jk | PMA | skipping | (6) |
| REPS1 | exon 10 | Jk | PMA | skipping | (6) |
| TRAF3 | exon 7 | Jk | PMA | skipping | (6) |
| VAV1 | exon 6 | CD4, CD8 | PHA | skipping | (8) |
|
| |||||
| Ion channels/binding | |||||
|
| |||||
| ITPR1 | exon 54 | CD4 | HIV-1 | inclusion | (24) |
| K(Ca)3.1 | mutually | Thy | Con A | inclusion E3 | (133) |
| TRIM26 | exon 2 | Jk, CD4 | PMA, PHA | variable e | (6) |
| TRIM65 | exon 3 | Jk, CD4 | PMA, PHA | inclusion | (6) |
|
| |||||
| Cytoskeletal | |||||
|
| |||||
| CCDC14 | exon 5 | Jk | PMA | skipping | (6) |
| CLASP1 | exon 9 | CD2 | α-CD3/28 | skipping | (7) |
| EVI5 | exon 18 | CD4 | HIV-1 | inclusion | (6) |
| FAM21C | exon 5 | Jk | PMA | skipping | (6) |
| FAM33A | exon 3 | Jk | PMA | skipping | (6) |
| MACF1 | exon 47 | Jk | PMA | skipping | (6) |
| MAP7D1 | exon 6 | Jk | PMA | inclusion | (6) |
| MYL6 | exon 3 | Jk | PMA | inclusion | (6) |
| SLMAP | exon 22a | Jk | PMA | skipping | (6) |
| Tau | exon 2 | Jk | PMA | inclusion | (44) |
|
| |||||
| Mitochondrial | |||||
|
| |||||
| ARMC10 | exon 15 | Jk | PMA | skipping | (6) |
| FAM36A | exon 3 | Jk, CD4 | PMA, PHA | variable d | (6) |
| TID1 | exon 11 | Th2 | α-CD3 | skipping | (134) |
| TIMM50 | exon 6 | CD2 | α-CD3/28 | inclusion | (7) |
|
| |||||
| Other | |||||
|
| |||||
| LRRC28 | exon 9 | Jk | PMA | inclusion | (6) |
| DDH1 | exon 12 | Jk | PMA | skipping | (6) |
| FKBP1A | exon 3 | CD8 | PHA | inclusion | (8) |
| GALT | exon 3 | Jk | PMA | inclusion | (6) |
| GZMB | exon 1 | CD2 | α-CD3/28 | intron removal | (7) |
| HSPA14 | exon 4 | CD2 | α-CD3/28 | skipping | (7) |
| NGLY1 | exon 9 | Jk, CD4 | PMA, PHA | skipping | (6) |
| PIGQ | exon 10 | Jk | PMA | inclusion | (6) |
| hTERT | exon 7 & 8 | T | various | inclusion | (135) |
| UGCGL1 | exon 6 | Jk | PMA | skipping | (6) |
Notes:
Exon numbers may vary depending on convention, numbers were either taken as reported in the references or deduced from ENSEMBL. Refer to the primary article for confirmation
Codes for cell types are as follows: (Jk) Jurkat, (T) Total primary T cells, (CD4) CD4+ primary T cells, (CD8) CD8+ primary T cells, (CD2) CD2+ primary T cells, (Thy) thymocytes, (PBMC) peripheral blood mononuclear cells PBMCs, CD4+ primary Th2 cells (Th2)
Codes for stimuli are as follows: (PMA) phorbol myristate acetate, (PHA) phytohaemagglutinin, (a-CD3/28) anti-CD3 and CD28 antibodies, (ConA) conclavin A, (LPS) lipopolyssacharide, (HIV-1) human immunodeficiency virus 1, (P2I) PHA+IL2+Ionomycin. “Various” is used when more than 2 stimuli have been used.
Increased skipping (Jurkat)/Increased inclusion (CD4+ T cells)
Increased inclusion (Jurkat)/Increased skipping (CD4+ T cells)
Many of the validated AS events in immune cells have been identified in work done by our group in collaboration with the lab of Dr. Benjamin Blencowe. Our microarray profiling of naive and activated T cells was the first large scale analysis of AS in T cells (8). More recently we have expanded this analysis by employing RNA-seq to identify further novel instances of AS (6). We have utilized both technologies in a Jurkat-derived T-cell line and in both cases found that 10% of profiled exons are robustly alternatively spliced upon T-cell activation with the phorbol esther PMA (phorbol myristate acetate). Our most recent analysis identified 178 cassette exons in 168 genes that exhibit robust changes in percent inclusion (20% difference) in Jurkat cells with a validation rate of >80% by RT-PCR (6). Importantly, the majority of the validated genes also exhibit differential AS between naive and activated primary T cells. In all, our work has resulted in the validated identification of over 50 AS events that occur in as many genes in response to activation of primary and cultured T cells (Table 2).
Many of the genes that we and others have validated to undergo changes in AS during immune responses include genes with known functions in immunobiology. These include cell surface receptors, kinases, phosphatases, and adapter proteins (e.g. CD3, CD28, CD8, CTLA-4, MAP4K2, MAP3K7, MAP2K7, CD45, VAV1), transcription factors and chromatin modifying enzymes (e.g. LEF1, ATF2, GATA3, RUNX1, EHMT2), and RNA binding proteins and others involved in regulating mRNA processing of many messages (e.g. CELF2, AUF1, SAM68, TIA-1) (Table 2). These studies have provided substantive evidence for the critical role that AS plays in assuring proper function of the adaptive immune response. Previously, we have provided a comprehensive review of the known functional consequences of splicing in immunobiology (9). To provide a flavor of the functional connection between AS and lymphocyte function, we highlight below a few more recent examples where regulated pre-mRNA AS of particular classes of genes has been directly linked to immune responses and/ or susceptibility to autoimmune diseases.
Cell surface receptors: CD3ζ
The T-cell receptor-CD3 complex is responsible for recognition of antigens presented on major histocompatibility complex (MHC) molecules (1). In particular, it is the CD3ζ chain that couples antigen recognition with intracellular signaling pathways. Thus, the level of CD3ζ expression determines the ability of a T cell to be activated. Strikingly, AS of exon 8 of CD3ζ is a primary control point of CD3ζ expression (10–12). The 3′ UTR of the CD3ζ gene contains several AREs (AU-rich elements) that regulate mRNA stability, as well as a sequence that enhances translation (13). Intriguingly, this 3′ UTR contains an intron that is preferentially removed in resting cells, whereas upon T-cell activation the intron is retained (10). Removal of the intron leads to a shortened 3′ UTR, which does not affect the coding region of the protein, but lacks two ARE motifs (ARE2 and ARE3) and a translation regulatory sequence (13). Thus, the stability and translation of the intron-removed form of CD3ζ is reduced as compared to the intron-retained version (13, 14), while intron retention upon T-cell activation allows for increased CD3ζ protein expression and increased T-cell signaling (11).
Defects in CD3ζ intron retention have been linked to autoimmune disease (12). T cells from patients with systemic lupus erythematosus (SLE) preferentially express the short (intron removed) form of CD3ζ (12). Consistently, there is a decrease in CD3ζ protein associated with the TCR, and T cells are impaired in their signaling capacity and production of the proliferation cytokine interleukin-2 (IL-2) (11). Strikingly, not only is expression of CD3ζ inversely correlated with severity of SLE disease, but restoring CD3ζ in cells results in a rescue in IL-2 production (15). Thus correcting the splicing defect in CD3ζ provides a potential therapeutic opportunity for the treatment of SLE.
Cell surface receptors: CD45
One of the best-documented examples of AS in response to T-cell activation is that of the transmembrane tyrosine phosphatase CD45. In hematopoietic cells, CD45 is involved in regulating proximal antigen receptor-mediated signaling, as well as integrin- and cytokine-mediated signaling (16). Several CD45 isoforms that result from regulated pre-mRNA AS have been documented (17). CD45 AS is differentially regulated in distinct cell types, developmental stages, and in response to signaling such as antigen receptor-mediated signaling (16). In peripheral T cells, three variable exons 4, 5, and 6 that encode for CD45 extracellular domains are preferentially skipped following antigen receptor-mediated signaling, resulting in increased expression of the smallest isoform (16). The regions of the CD45 protein encoded by these variable exons are heavily glycosylated and thus prevent CD45 homodimerization. Upon T-cell activation, skipping of the CD45 variable exons leads to homodimerization at the cell surface, which leads to an inactive form of the phosphatase and decreased signaling through the TCR (18, 19). Thus AS of CD45 serves as a feedback mechanism for the maintenance of T-cell homeostasis.
A single nucleotide polymorphism (SNP) in CD45 exon 4 (C77G) has been described that leads to aberrant inclusion of this exon (20, 21). Consistent with the above model, in which loss of exon 4 skipping leads to loss of T-cell attenuation, studies have shown a correlation between this polymorphism and susceptibility to multiple sclerosis (MS) and HIV infection (22, 23) as would be anticipated from a hyperactive T-cell response. Furthermore, HIV infection itself induces exon skipping in CD45 and CD4+ T cells expressing the smallest isoform of CD45 (CD45 RO), which have been found to preferentially be infected by HIV-1 (24), suggesting a HIV-induced positive feedback loop in which CD45 AS promotes infection.
Cytokines: IL-7Rα
AS of cytokine receptors such as those in the interleukin family typically results in the generation of a soluble receptor that antagonizes or synergizes with the membrane bound receptor for ligand binding (9). One such example is the IL-7 receptor α chain (IL-7Rα), in which differential skipping of exon 6 excludes the transmembrane domain resulting in a soluble form of the receptor that is secreted in the plasma (25). IL-7Rα is expressed almost solely in cells of lymphoid origin, and mice and humans deficient in the IL-7 signaling pathway develop lymphopenia (26). IL-7Rα is particularly critical in the survival of peripheral CD4+ and CD8+ and in the generation of a memory phenotype (27, 28).
Consistent with a functional importance for IL-7Rα AS in CD4+ T cells, HIV-1-infected individuals have lower concentrations of soluble IL-7Rα in their plasma (29). Moreover, similar to the example of CD45 mentioned above, there is a genetic association of a SNP located within exon 6 of IL-7Rα and multiple sclerosis (30). Notably, the SNP located within exon 6 results in increased exon skipping and decreased expression of the membrane-bound IL-7Rα mRNA in peripheral blood mononuclear cells (PBMCs) from MS patients harboring the SNP (30). By contrast, the soluble IL-7Rα mRNA was overrepresented in SNP-containing patients relative to healthy individuals (30, 31). Interestingly, AS appears to regulate IL-7 signaling at multiple layers, since a splice variant of IL-7 cytokine itself lacking exon 5 was shown to promote T-cell survival and thymocyte maturation (32).
Intracellular signaling: Fyn
Fyn is a protein tyrosine kinase (PTK) of the Src family involved in intracellular signaling of various cellular processes including T-cell development and activation (33). The gene encoding Fyn generates two isoforms by differential inclusion of mutually exclusive exon 7s denoted 7a and 7b (34). The two exon 7s encode distinct versions of the SRC homology 2 (SH2) domain and part of the kinase domain of Fyn; therefore, AS of exon 7 is predicted to impact substrate recognition and phosphorylation (see below). The FynT isoform uses exon 7b and is the primary isoform expressed in T cells. FynT more efficiently mobilizes calcium and produces IL-2 during TCR signaling than the alternate isoform FynB, which is primarily expressed in the brain (35–37). Infection with human T-cell leukemia virus type 1 (HTLV-1) induces the expression the FynB isoform in T cells, which could serve to attenuate T-cell effector functions (38). By contrast, T-cell activation induces an even further bias toward inclusion of exon 7B, which may serve as a mechanism for sustaining and enhancing TCR signaling (39).
It was demonstrated recently that FynT more efficiently interacts with and phosphorylates the RNA-binding protein Sam68 than FynB (35). The differential interaction between FynT and FynB with SAM68 was mapped to part of the distinct SH2 kinase linker encoded by exon 7 (35). Swapping of the linker region of the FynT to the FynB isoform granted it the ability to strongly interact with and phosphorylate Sam68 (35). Notably, Sam68 has previously been shown to modulate AS of Bcl-x, in which it promotes use of the alternate upstream 5′ splice site in exon 2 to generate the pro-apoptotic Bcl-x(s) form (40). Phosphorylation of Sam68 by Fyn switches the activity of Sam68 to enhancing production of the Bcl-x(L) isoform (40). Consistent with the previous observations, expression of FynT but not FynB favors the expression of the Bcl-x(L) anti-apoptotic isoform through phosphorylation of Sam68 (35). These results suggest functional importance of the regulated Fyn AS in tuning the activity of an RNA-binding protein than in turn regulates AS of downstream target genes.
Transcription factors: LEF1
Lymphocyte enhancer factor 1 (LEF1) is an HMG-box transcription factor that is broadly expressed during embryonic development, and during adulthood is expressed in a tissue-specific manner in certain lymphocyte subsets (41, 42). LEF1 promotes expression of the T-cell antigen receptor α chain (TCRα) through binding to the TCRα enhancer (43, 44). We have shown that inclusion of LEF1 exon 6 is increased during thymic development specifically during the double negative 3b (DN3b) to double positive (DP) transition (45). Exon 6 encodes part of the context-dependent regulatory domain of the LEF1 protein, which allows its TCRα enhancer activity. During the DN3a stage, the mature TCRβ is expressed at the cell surface in complex with a surrogate TCRα chain called the pre-TCRα chain (46). One of the critical consequences of pre-TCR signaling is the rearrangement and transcription of the TCRα chain to give rise to the expression of the mature α/β TCR complex by the DP stage. We found that increased TCRα transcript levels correlate with increased LEF1 exon 6 inclusion during the DN3b to DP transition (45). Similarly, activation of the JSL1 T-cell line with PMA results in increased LEF1 exon 6 inclusion with a corresponding increase in TCRα expression. Importantly, the ability of LEF1 to enhance TCRα transcription is a direct result of LEF1 exon 6 splicing, as blocking of the LEF1 exon 6 splice site results in a substantial decrease in TCRα mRNA (45). These results suggest that during pre-TCR signaling, regulated splicing of the LEF1 transcription factor contributes to the control of TCRα transcription to produce functional TCR complexes at the cell surface.
RNA processing factors: CELF2
Many RNA-binding proteins regulate their own expression by an autofeedback loop which maintains protein expression in a particular range. The splicing factor CELF2 accomplishes this by binding to one of its own introns to induce skipping of exon 6. This AS event results in a premature stop codon and loss of subsequent translation of full length protein when the levels of CELF2 are sufficient (47). Interestingly, activation of peripheral CD4+ T cells leads to an increase in CELF2 exon 6 inclusion (6, 8, 45). Although this change in CELF2 splicing has not directly been linked to functional changes in T cells, CELF2 is known to regulate the functionally relevant splicing of LEF1 (see above and below) as well as other signal-responsive genes (NMM, MJM, KWL, unpublished data). Therefore, one can readily imagine a cascade of AS events initiating with autoregulatory splicing of CELF2. Similarly, Sam68 and TIA-1 are RNA binding proteins that have been implicated in regulated AS in T cells (see below) and have themselves been validated to undergo changes in isoform expression following antigen stimulation of T cells (8, 48). It remains to be shown, however, whether these proteins truly trigger splicing cascades following T-cell activation.
Pathways and RNA-binding proteins that regulate splicing
Importance of understanding splicing mechanism
Despite the interest in AS in immune responses, we only have mechanistic insights as to how a handful of the AS events listed in Table 2 are regulated (see below). This means that we have very little understanding of what signaling pathways, trans acting splicing factors, and sequences are mediating AS programs in immune responses. Understanding the mechanisms by which splicing is regulated in normal immune responses is critical to understanding and treating diseases of the immune system. As implied above, emerging research is providing an ever growing list of mis-regulated splicing events that correlate with autoimmune diseases and other immune defects (49). Understanding how SNPs cause splicing changes, predicting the effects of newly identified SNPs, and ultimately designing therapies to correct splicing defects, all require a detailed knowledge of the mechanisms that control particular splicing decisions.
A second motivation for understanding the mechanisms that control splicing in immune cells is so that we can better predict coregulation and functional consequences of AS. AS is often thought of as a fine-tuning mechanism, because it allows for tweaking of specific functions of a protein while maintaining others. In some instances, such tweaking is sufficient to exert a major functional change in the cell (50, 51). More often, however, concerted changes in the splicing of a family of related genes are expected to be of greater functional impact than that generated by individual AS events (50, 51). For this reason, a major step in understanding the role of splicing regulation in modulating immune responses is the identification of signaling pathways, trans-acting factors and mechanistic commonalities that orchestrate networks of regulated splicing. By analogy, knowledge of the binding sites and activity of transcription factors has allowed researchers to predict what genes are expressed downstream of an event such as NF-κB activation (52). Similarly, the ultimate goal of splicing research is to gain sufficient knowledge of what regulatory factors are activated during an immune response to be able to predict from the genetic sequence what genes and gene families undergo antigen-induced changes in splicing patterns.
Basic concepts in the regulation of alternative splicing
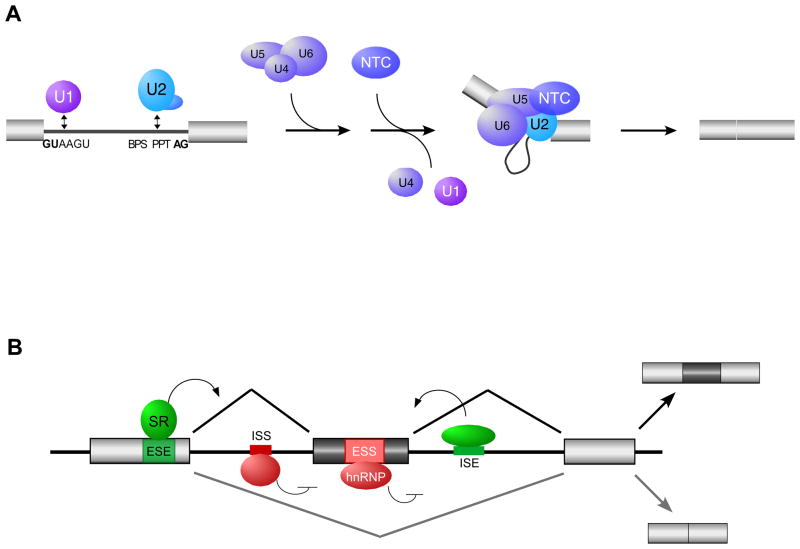
At the most basic level, the decisions of what sequences of a gene are included in the final mRNA rely on the ability of the spliceosome to recognize and process the splice sites located at the intron-exon boundaries of a pre-mRNA. The spliceosome is the enzymatic complex that catalyzes intron removal and exon joining. Instead of being a preformed enzyme, the spliceosome is built de novo on pre-mRNA transcripts through sequential interactions between the substrate and subunits of the spliceosome (Fig. 2A). The major subunits of the spliceosome are the 5 so-called snRNPs, which each contain a single non-coding RNA (snRNA) and multiple proteins (53). Exons are targeted to be spliced together by binding of the snRNPs to the splice sites, a process which is driven primarily through basepairing interactions between the snRNA component of the snRNPs and the splice site sequences (53) (Fig. 2A). Importantly, however, most mammalian splice sites only weakly basepair with the snRNAs. Therefore, auxiliary sequence elements play a crucial role in regulating splice site selection by modulating snRNP-substrate interactions (54, 55).
Fig. 2. General mechanism of splicing and its regulation.
(A) Spliceosome assembly. The 5′ end of introns are defined by the 5′ splice site (GUAAGU), and the 3′ end of introns are defined by the branch point sequence (BPS), polypirimidine tract (PPT), and the 3′ splice site AG dinucleotides. The U1, 2, 4, 5 and 6 represent the snRNPs which assemble with substrate and each other as shown. The NTC is an additional snRNA-free spliceosomal subunit. For more details see Motta-Mena et al. (58). (B) Regulation of alternative splicing. Enhancer auxiliary elements are denoted in green for exonic (ESE) or intronic (ISE) splicing enhancers. Silencer auxiliary elements are denoted in red for exonic (ESS) or intronic (ISS) splicing silencers. The activities of these auxiliary elements are often mediated through binding of SR and hnRNPs, two common families of proteins described in the text, among other RNA binding proteins.
Cis-elements that control splicing include sequences within exons such as exonic splicing enhancers (ESEs) and exonic splicing silencers (ESSs) as well as sequences within introns such as intronic splicing enhancers (ISEs) and intronic splicing silencers (ISSs) (Fig. 2B). These elements either promote (enhancers) or suppress (silencers) proper assembly of the snRNP on the substrate, typically by recruiting regulatory proteins which then interact with spliceosomal components (54, 55). Two protein families that often function as splicing regulators are the serine/arginine rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) (56, 57). When bound within exons, SR proteins tend to enhance splice site usage, whereas hnRNPs typically induce exon skipping (Fig. 2B), although there are exceptions to this generalization (54, 55, 58). Moreover, both families of proteins as well as other RNA-binding proteins can have both enhancing and silencing activity when bound to an intron. The location of binding relative to the splice sites, the presence of neighboring regulatory proteins, and the relative strength of the splice site-snRNA interactions all influence the ultimate consequence of a particular regulatory protein on the pattern of splicing. Finally, most transcripts are bound by multiple regulatory proteins such that the final decision of which alternative splice patterns will be used and how many distinct isoforms will be expressed from a single gene and pre-mRNA relies on a balance of competing regulatory factors (50, 51).
Another important aspect of splicing regulation, particularly in thinking about immune responses, is how signaling pathways can influence splicing factor activity. There are many ways by which cell signaling pathways can modulate the function of splicing regulatory factors (reviewed in 59). First of all, T-cell activation induces widespread transcriptional changes that can lead to differences in splicing factor protein expression (6–8). Similarly, AS of RNA-binding and spliceosomal proteins, such as has been identified in studies listed in Tables 1 and 2 (see CELF2 above), is likely to influence the activity of these factors and thus influence AS of downstream targets. Phosphorylation of splicing regulatory proteins can alter their activity, localization, and ability to interact with other proteins. Phosphoproteomic analyses of TCR signaling have found that proteins involved in all aspects of T-cell signaling become phosphorylated, including splicing factors and components of the spliceosome (60). Notably, TCR-induced phosphorylations within splicing proteins was preferentially observed in domains that are predicted to mediate protein-protein and/or protein-RNA interactions (60). Other mechanisms of signal-induced changes in the splicing machinery include different modes of differential protein-protein interactions, protein-RNA interactions, protein stability, and localization (59).
Despite the abundance of AS in immune responses, relatively little is know regarding the factors, mechanisms, and signaling pathways by which such changes in gene expression are controlled. The identity of signaling molecules and how they regulate the activity of splicing factors and RNA binding proteins to regulate AS have been elucidated in only a few of the validated genes listed in Table 2. Moreover, even in the cases where regulators have been characterized on individual genes, the breadth of the AS program mediated by such factors have not been well defined. Over the past decade, the primary focus of our laboratory has been to begin to identify the proteins, pathways, and mechanisms by which AS is regulated in human T cells. Below we describe the distinct proteins and mechanistic paradigms that have thus far been described by our lab and others to regulate splicing during T-cell development and function (Fig. 3). We emphasize what is known as well as what remain critical future areas of research as we seek to gain a complete understanding of how splicing shapes immune function in normal and disease states.
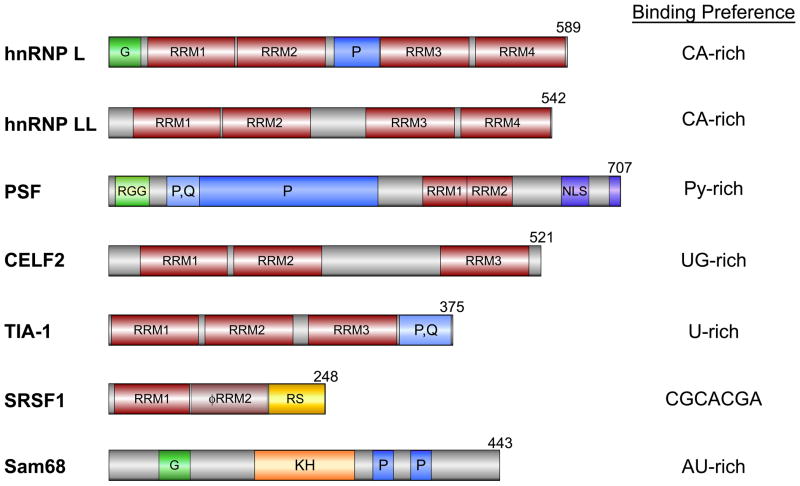
Fig. 3. Splicing factors known to regulate AS in immune responses.
Domain schematics for each factor are displayed. RRM, RNA recognition motif;φRRM, pseudo RRM; G, glycine-rich region; P, proline-rich region; RGG, arginine/glycine/glycine repeat region; P, Q, proline/glutamine rich; RS, arginine/serine-rich; KH – K homology domain. Binding preferences for each factor are specified for hnRNP L (66), hnRNP LL (64), PSF (75), CELF2 (78), TIA-1 (85), SRSF1(115), and Sam68 (97).
hnRNP L
From a mechanistic perspective, most of what we know about regulated splicing in T cells comes from decades of detailed analysis of CD45, the first gene well-documented to undergo splicing changes during the T-cell response to antigen (see above). We initially demonstrated that regulation of the three CD45 variable exons is mediated by a sequence motif, contained within each of the exons, which we designated the activation-responsive sequence [ARS (A/CYYGCA)] (39, 61). In exons 4 and 6, the ARS motif lies within a 60 nt exonic splicing silencer (ESS1) that is necessary and sufficient to confer repression of exons 4 and 6. Mutation of key residues within the ARS has demonstrated that this sequence is critical for the partial repression of exons 4 and 6 that is observed in resting T cells as well as the activation-induced hyper-repression of these exons and exon 5 upon antigen stimulation (39, 61). Furthermore, introduction of the CD45 exon 4 ESS1 element into a heterologous unregulated exon imparts basal and TCR signaling-mediated exon repression (39).
Definition of the ARS has allowed for the identification of proteins that regulate CD45 splicing through this element. Arguably, the most essential factor in the regulation of CD45 AS is hnRNP L. We first identified hnRNP L as the major ARS-binding activity in biochemical studies (62) and subsequently have confirmed hnRNP L to be the primary mediator of basal repression of the CD45 variable exons both in vitro and in cells (58, 61–64)(Fig. 4). More recently, tissue specific knockout studies in mice revealed that thymocytes depleted of hnRNPL exhibited aberrant inclusion of the CD45 exons, confirming the in vivo relevance of hnRNP L for CD45 exon repression (65).
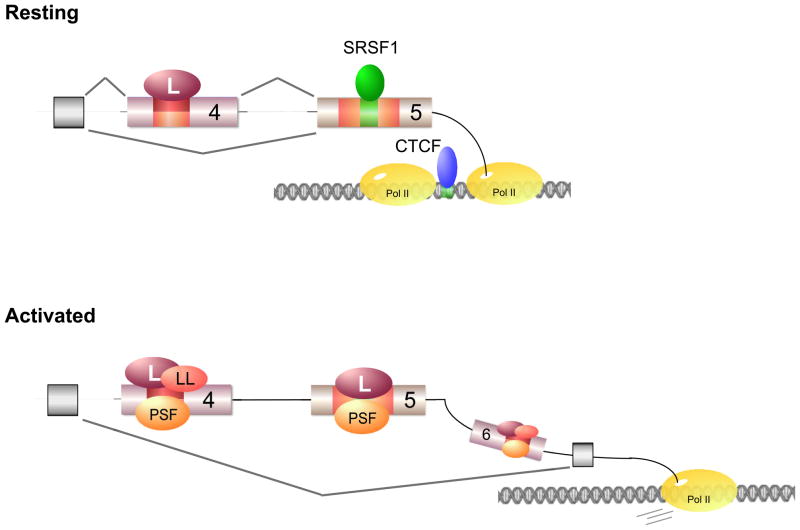
Fig. 4. Mechanism for the regulation of CD45 alternative splicing.
The CD45 ARS is depicted in orange/red and the ESE in green. In resting cells, hnRNP L (L) regulates basal levels of exon 4 repression, while SRSF1 mediates basal levels of exon 5 inclusion. Exon 5 inclusion is further regulated at the level of transcription. CTCF binds to the exon 5 ESE at the DNA level and promotes Pol II pausing around that region to promote exon 5 inclusion. Upon activation hnRNP LL and PSF join hnRNP L at the exon 4 and 6 ARSs to promote further repression, while only hnRNP L and PSF associates to exon 5.
HnRNP L is a highly abundant member of the hnRNP family of splicing factors and contains four RRMs (Fig. 3). It has both splicing silencer and enhancer activities, depending on the context in which it binds, and functions in introns or exons to regulate inclusion of cassette exons, intron retention and poly(A) site choice (58, 62, 66, 67). hnRNP L binds preferentially to CA-rich sequences, although it can associate with CA dipeptides located in a wide range of contexts (66, 68). Interestingly, the ARS itself is comprised of only a few, widely scattered, CA elements. Correspondingly, hnRNP L has only a moderate affinity for the ARS element. This use of moderate affinity is likely essential for appropriate regulation of CD45, as it allows some exon inclusion in resting T cells. Finally, the expression of hnRNP L is unchanged during T-cell responses, consistent with it largely setting a constant basal level of CD45 exon repression in both resting and activated T cells. However, hnRNP L may become hyperphosphorylated in response to antigen which may alter some of its activities (63, 69).
As discussed below, there is increasing evidence of widespread activity of hnRNP L in regulating T-cell splicing. The hnRNP L T-cell-specific knockout mice revealed that hnRNP L is critical for thymocyte development. In the absence of hnRNP L, the DN4 to DP transition is blocked causing a decrease in thymic cellularity (65). hnRNP L was also implicated in migration of single postive thymocytes to the periphery. The hnRNP L T-cell-specific knockout mice revealed that hnRNP L mediates regulation of widespread AS, as an RNA-seq comparison between hnRNP L deficient and wildtype thymocytes identified many potential hnRNPL regulated exons (65). Knowing the binding specificity of hnRNP L has also begun to allow for prediction of exons that may be regulated by this protein; however, more needs to be learned before we can do this in a truly rigorous way (see Networks section below).
hnRNP LL
Upon activation of T cells, hnRNP L is joined in binding the CD45 ARS by two additional proteins (63, 64) (Fig. 4). One of these is a paralogue of hnRNP L called hnRNP L-like (hnRNP LL), which is preferentially expressed in activated and memory T cells (Fig. 3). hnRNP LL was identified as a regulator of CD45 splicing simultaneously by our group and two others (64, 70, 71). Two of us discovered hnRNP LL through a cell-based screening approach (64, 71), while the third took advantage of mouse genetics (70).
The screen through which our laboratory identified hnRNP LL consisted of analyzing JSL1 T-cell clones that exhibited an altered ‘activation-like’ phenotype under resting conditions (64). We utilized a novel reporter construct in which CD45 exon 4 was inserted between exons encoding the synthetic transcription activator Gal4-VP16, such that expression of the active form of Gal4-VP16 was dependent on skipping of exon 4. The expression of Gal4-VP16 was, in turn, monitored by its ability to activate transcription of a reporter construct encoding GFP allowing for a readout that was highly sensitive to relatively small changes in CD45 exon 4 inclusion (72). Cells containing this dual reporter scheme were then infected with a retroviral encoded cDNA library, and resting cells exhibiting increased levels of green fluorescence protein (GFP) (indicative of increased exon 4 skipping) were isolated. Strikingly, the one commonality among several of the resulting GFP-high cell clones was aberrantly high expression of hnRNP LL (64). We confirmed the activity of hnRNP LL in the JSL1 T-cell line by demonstrating that direct overexpression of hnRNP LL resulted in increased exon 4 repression, while knockdown abolishes activation-induced repression (64). We also demonstrated that hnRNP LL mRNA and protein expression are normally upregulated in response to T-cell activation, as is direct binding of hnRNP LL to the CD45 exon 4 ESS1 (64). Similar results were obtained from the work of Oberdoerffer et al. (71), who isolated hnRNP LL in an shRNA screen for factors required for repression of exon 4 in the endogenous CD45 gene in the JSL1 cell line.
The in vivo relevance of the studies regarding hnRNP LL in human T-cell lines were supported by subsequent analysis of primary human cells (71) and by a ENU (N-ethyl-N-nitrosourea) mutagenesis study in mice (70). In the mouse study, a mutant mouse line was identified that exhibited fewer CD4+ T-helper (Th) cells. T cells from these mice, named ‘Thunder’ mice for ‘Th under normal’, were found to have altered expression of CD45 isoforms (70). Specifically, memory T cells in the Thunder mice, which normally express the smallest isoform of CD45, aberrantly express isoforms that include the variable exons (70). This altered expression of CD45 was mapped to an ENU-induced missense mutation in hnRNP LL (V136A), which abolished hnRNP LL binding to the ARS motif in CD45 by destabilizing one of the RNA binding domains (RRMs) of the protein (70).
Despite the similarity and overlapping functions between hnRNP L and hnRNP LL, these proteins have distinct binding requirements (64). Although both CD45 exons 4 and 5 contain the ARS motif and are repressed by hnRNP L, hnRNP LL does not bind to exon 5 or have a functional effect on its activation-induced repression (58, 71). By contrast, the affinity of hnRNP LL for the ARS element in exon 4 is actually higher than that of hnRNP L. However in naive T cells, hnRNP LL is expressed only at ~ 1/100th the level of hnRNP L, thus there is no significant binding or repression of exon 4 by hnRNP LL in T cells until the expression of this protein is induced by antigen stimulation (64). Notably, induction of binding of hnRNP LL to the ARS in CD45 exon 4 results in a significant increase in exon skipping over that observed from the binding of hnRNP L alone. Why association of hnRNP LL together with hnRNP L achieves more efficient repression of exon 4 than hnRNP L alone remains an open question and will require further mechanistic studies. However, it is clear that both proteins can bind simultaneously and independently to the ARS motif, and both proteins are capable of exon repression on their own (64).
A second open question with regards to hnRNP LL and T-cell function is how broadly this protein regulates splicing, and how commonly hnRNP L and hnRNP LL work in conjunction with one another. Although some targets of hnRNP LL splicing have been suggested by exon array studies in T cells (70, 71), none of these splicing events have been validated, so no conclusions can be made regarding the scope of hnRNP LL function. Studies from the Goodnow laboratory (73) have demonstrated that the regulation of CD45 by hnRNP LL is not sufficient to account for all the phenotypes of the hnRNP LL mutant mouse, implying the existence of additional physiologic targets of hnRNP LL activity. However, it is not known what such targets may be or even if all of the activity of hnRNP LL is at the level of splicing regulation rather than other steps of RNA processing that can be affected by hnRNP proteins such as mRNA stability, translation, 3′ UTR processing and even transcription.
PSF
The other factor that cooperates with hnRNP L in the signal-responsive repression of CD45 is PTB-associated splicing factor (PSF) (63, 74). PSF is not considered one of the canonical hnRNP proteins, but like the hnRNPs contains 2 RNA binding motifs as well as additional domains of low-complexity and unknown function (Fig. 3). PSF was initially identified as an essential splicing factor and subsequently has been implicated in regulating many aspects of mRNA processing and nuclear processes (75). The first direct connection of PSF to AS was our identification of PSF as one of the factors necessary for activation-induced repression of all three of the CD45 variable exons (63). We initially implicated PSF in the regulation of CD45 through biochemical purification of proteins bound to the ESS1 element in CD45 exon 4. Similar to hnRNP LL, PSF associates with the ARS element specifically in activated T cells (63) (Fig. 4). This binding of PSF to the ESS1 is independent of hnRNP L or hnRNP LL and involves sequences outside of the core part of the ARS motif (63, 64). However, PSF is required for maximal activation-induced repression of the CD45 variable exons as indicated by the fact that mutation of the PSF-recognition element increases CD45 exon inclusion in activated cells without affecting binding of hnRNP L or LL (58, 63, 64).
Unlike hnRNP LL whose transcription and protein expression are increased following activation, PSF mRNA and protein expression remain constant during an immune response (63, 64, 74). This finding suggested that the preferential binding of PSF to the CD45 pre-mRNA in activated T cells is controlled at the post-translational level. Interestingly, PSF is a direct substrate of glycogen synthase kinase 3 (GSK3) in naive T cells, in which GSK3 is active and phosphorylates PSF at threonine 687 (74). Phosphorylated PSF interacts with a protein called TRAP150, which prevents PSF from binding the CD45 pre-mRNA. Upon engagement of the TCR, GSK3 is inactivated leading to reduced PSF phosphorylation. The loss of T687 phosphorylation on PSF releases it from TRAP150, allowing PSF to bind CD45 splicing regulatory elements and repress exon inclusion (74). Conceptually, this is one of the first direct links between a signaling molecule and a splicing factor in T cells. Moreover, the accumulation of unphosphorylated PSF following T-cell activation requires de novo protein synthesis, which explains the observed kinetics of activation-induced CD45 exon repression.
The mechanism by which TRAP150 regulates PSF is not by a simple phospho-site recognition, since deletion of part of the C-terminus of PSF which includes T687, does not prevent its interaction with TRAP150 (74). This observation supports a model in which phosphorylation of PSF drives a conformational change that regulates the interaction between PSF and TRAP150. PSF has been implicated in the regulation of other alternatively spliced transcripts, and we predict the PSF/TRAP150 complex is a means of regulation for a broader set of alternatively spliced pre-mRNAs. PSF may also have additional activities in T cells as a T-cell-specific knockdown of PSF in mice results in thymic apoptosis (76). While the extent of knockdown in these studies was not sufficient to induce changes in AS, we did observe altered stability of several mRNAs including those encoding histones (76).
CELF2
Another hnRNP-related protein involved in regulated splicing in immune cells is CELF2 (45). CELF2 is a member of the CELF family of RNA processing factors, which have been extensively documented to regulate splicing during heart and skeletal muscle development (77). The CELF proteins all contain multiple RRMs and typically bind UG-rich sequences to promote or repress exon recognition (78) (Fig. 3). We have recently determined CELF2 to function as a signal-responsive splicing regulator during T-cell development as well as in response to activation of mature T cells (45).
Biochemical assays initially identified CELF2 as binding to two UG-rich ISEs that control inclusion of LEF1 variable exon 6 (see above, 45). The concentration of CELF2 protein and its corresponding binding to the LEF1 ISEs increase during the transition from DN and DP cells in thymic development, consistent with increased inclusion of LEF1 exon 6 following pre-TCR signaling. A similar change in CELF2 expression, function, and LEF1 splicing is also observed upon activation of JSL1 T cells. Knockdown of CELF2 in the JSL1 cells results in loss of LEF1 exon 6 inclusion with a concomitant decrease in TCRα mRNA, consistent with the functional consequence of regulated LEF1 splicing (see above) and confirming the role of CELF2 in the regulation of LEF1 (45, 78).
The studies demonstrating CELF2 regulation of LEF1 emphasize the biological importance of CELF2 as a lymphocyte splicing factor in addition to its activities as a brain and muscle-specific regulator. Preliminary data from our laboratory further suggests that CELF2 regulates other activation-induced splicing events in T cells (NM, MJM, KWL, unpublished data). Interestingly, the related factor CELF1 has been shown to regulate cytoplasmic mRNA stability during immune responses (79). Although CELF1 has no activity in LEF1 splicing, it is expressed in the nucleus of T cells and may regulate additional splicing events.
TIA-1
T-cell-restricted intracellular antigen 1 (TIA-1) was first identified by monoclonal antibodies that reacted with intracellular antigens in T cells but not in B-cells (80). Cloning of TIA-1 revealed it to be an RNA binding protein with three RNA-recognition motifs (Fig. 3). Interestingly, this protein is homologous to a splicing factor in the yeast S. cerivisae, Nam8, which is known to bind uridine-rich sequences in introns (81–83). Further studies confirmed that TIA-1 functions as a splicing factor as well as exhibiting other RNA-processing functions such as silencing translation of TNF-α (84, 85).
TIA-1 is the primary splicing factor responsible for regulation of FAS alternative splicing. As shown in Table 2, Fas exon 6, which encodes the transmembrane domain of the protein, is inducibly included upon T-cell activation (86). Skipping of Fas exon 6 produces a soluble version of the receptor, which can inhibit Fas signaling and therefore control apoptosis (87, 88). Knockdown of TIA-1 or the related TIAR (TIA-1 related protein) results in increased exon 6 skipping (89). A uridine-rich sequence located downstream of the exon 6 5′ splice site was identified as an ISE required for the ability of TIA-1 to enhance exon 6 inclusion (89).
In an interesting case of positive feedback regulation, TIA-1 is phosphorylated by Fas-activated serine/threonine kinase (FASTK), which, as its name implies, is a kinase that becomes activated in Jurkat T cells in response to Fas engagement with ligand (90). Strikingly, knockdown in Jurkat cells or overexpression in HeLA cells of FASTK phenocopies modulation of TIA-1 expression with regards to Fas exon 6 inclusion (48). In addition, FASTK has little effect on Fas splicing in cells in which TIA-1 and TIAR were depleted or the uridine-rich ISE was mutated (48). Together, these results suggest that phosphorylation of TIA-1 by FASTK promotes the ability of TIA-1 to enhance exon 6 inclusion. Moreover, these studies propose that Fas signaling can use AS to enhance itself in a feedforward manner in which initial Fas signaling activates FASTK and TIA-1 to increase expression of membrane-bound receptors. Whether this pathway is involved in the T-cell activation-induced exon 6 inclusion remains unanswered. Furthermore, it is unknown whether the many other targets of TIA-1 function that have been identified are likewise alternatively spliced in response to Fas or TCR signaling (91–93).
SRSF1
Alternative splicing factor/splicing factor 2 (ASF/SF2 now called SRSF1) is a member of the ubiquitous SR family of splicing factors that promote constitutive and alternative splicing (56). Like all SR proteins, SRSF1 contains N-terminal RRM type RNA binding motifs and a C-terminal domain rich in Arg-Ser dipeptides (56) (Fig. 3). The C-terminal Arg-Ser (RS) domain provides the SR protein family their name and is thought to function primarily as a protein-protein interaction domain that promotes recruitment of the snRNPs and enhancement of exon inclusion (56). The splicing of most exons is controlled in at least some manner by SR proteins. Therefore it is not surprising that, where investigated, SR proteins have activities in controlling splicing in immune cells.
One example of SR protein function in immune-related AS comes, not surprisingly, from studies of CD45. Specifically, we implicated SRSF1 in regulation of CD45 exon 5. Like the other CD45 variable exons, CD45 exon 5 contains the two copies of the ARS motif; however, they are separated by a strong ESE (61). Although hnRNP L and PSF mediate exon 5 repression through binding to the ARS, the ESE is also important for regulation and specifically for promoting the relatively high level of inclusion of this exon (58, 61). We identified SRSF1 as a protein that binds to the exon 5 ESE and promotes exon inclusion in an ESE-dependent manner (58). Furthermore, we demonstrated that hnRNP L counters the SRSF1 enhancer activity, by directly competing for binding exon 5. Therefore, although it is now apparent that the ESE also functions at the level of transcription (see CTCF below), we conclude that the balance of hnRNP L and SRSF1 contributes to the degree of CD45 exon 5 inclusion, consistent with the notion that the balance of hnRNP and SR activities plays a fundamental role in many splicing decisions. We also note that several other SR proteins, including SRSF2 (SC35), SRSF3 (SRp20), and SRSF7 (9G8) have also been correlated to changes in CD45 AS (94, 95), although the mechanism of activity and even whether these are direct or indirect effects has not been determined.
SRSF1 also mediates the activation-induced CD3ζ AS highlighted in Table 2 (10). As described above, following T-cell activation there is a shift toward the CD3ζ isoform that retains an intron within the 3′ UTR, thus promoting protein expression. SRSF1 regulates CD3ζ AS by binding to the ARE2 element within the 3′ UTR and promoting intro retention at the 3′ UTR (10). Consistent with the intron-retained version of the CD3ζ message being the functional version of this mRNA, depletion of SRSF1 resulted in decreased CD3ζ protein expression while overexpression resulted in increased intron retention and CD3ζ protein production (10). Furthermore, during T-cell activation, increased levels of SRSF1 correlate with an increase in the intron-retained CD3ζ 3′ UTR. SRSF1 protein levels has been further linked to CD3ζ protein levels in T cells from SLE patients (10). Interestingly, several SR proteins in addition to SRSF1 show altered expression (both increased and decreased) in response to antigen stimulation of T cells. Given the abundant functional role of SR proteins, such changes in SR protein expression are likely to affect widespread changes in splicing efficiency and AS patterns (96).
SAM68
Sam68 is a member of the signal transduction and RNA (STAR) family of proteins that contain a KH type RNA-binding domain as well as binding sites for signaling proteins (97) (Fig. 3). STAR family proteins typically have activities in mRNA stability and translation; however, Sam68 has been shown to regulate several AS events including the T-cell activation-induced regulation of CD44 splicing (98). CD44 is a cell surface adhesion molecule involved in T-cell homing, and some of its splice variants have been correlated with tumor malignancy (reviewed in 9). The CD44 gene contains 10 variable exons that are differentially included in T cells in various combinations. Whereas naive T cells express the shortest CD44 isoform that lacks all of the variable exons, antigen stimulation induces the inclusion of various combinations of the additional 10 variable exons (99, 100). AS of exon v5 is the most investigated of the CD44 variable exons. Deletion experiments of CD44 exon v5 identified both an ESS and an ESE that regulate its inclusion (100). Activation-induced inclusion of CD44 exon v5 in T cells was also shown to be dependent on the Ras-Raf-MEK-ERK signaling pathway (101). The Ras signaling cascade leads to interactions between Sam68 and the SR-related protein SRm160 to form a complex which binds the ESE and promotes exon inclusion (98, 102). Furthermore, Sam68 can also interact with the Brm subunit of SWI/SNF a complex involved in chromatin-remodeling (103). Upon activation by the ERK pathway Sam68 binds to CD44 exon v5 and then interacts with or signals to Brm, to promote Pol II stalling (98, 102). As part of a growing recognition of the mechanistic interplay between transcription and splicing, it is known that slowing Pol II elongation rates can promote inclusion of variable exons, by providing the spliceosome time to recognize and assemble on weak splice sites (104). Consistent with this model, recruitment of Brm to CD44 through Sam68 promotes inclusion of the variable v5 exon of this gene through transcriptional pausing (103). Thus, Sam68 regulation of CD44 is an example of how an RNA-binding protein can influence AS by modulating transcription rates.
CTCF
Another excellent example of AS mediated by regulation of transcription comes from recent studies on the DNA-binding protein CCCTC-binding factor (CTCF) (105). CTCF binds DNA and has classically been thought of as an insulator that binds in intergenic chromosomal regions (106). However, recently, CTCF was shown to bind extensively within genes, where it correlates with sites of Pol II pausing. Remarkably, CTCF binding sites are also enriched in or near alternative exons, raising the possibility that binding of CTCF regulates elongation of Pol II to favor exon inclusion. Proof of this model was provided for CD45 exon 5 (105). Specifically, it was shown that B-cell lines with high expression of CD45 exon 5 (CD45RB high) exhibited greater CTCF binding to exon 5 than cells that were CD45RB low. Surprisingly, the binding site for CTCF was shown to encompass the ESE previously identified as essential to high inclusion of exon 5 (58, 61) (Fig. 4). In cells and in vitro, CTCF binding to exon 5 promoted Pol II pausing upstream of CTCF binding sites (105). Importantly, CTCF depletion results in loss of CD45 exon 5 inclusion, as does mutation of the CTCF binding site, providing a direct causal link between CTCF binding and AS (105).
CTCF-promoted exon 5 inclusion is independent of hnRNP LL, which does not influence CD45 exon 5 skipping in the transition from naive to activated T cells (58, 71, 105). Instead, CTCF binding to exon 5 is inhibited by DNA methylation, and this correlates with loss of exon inclusion (105). Notably, memory T cells, which express reduced amounts of CD45 exon 5, have increased DNA methylation and decreased CTCF binding at exon 5 relative to naive T cells (that show high levels of exon 5 inclusion) (105). RNA-Seq of CTCF-depleted B-cell lines combined with CTCF ChIP-seq revealed genome-wide effects of CTCF-regulated AS (105). Furthermore, comparison of the CTCF-binding sites identified by ChIP-seq in B cells with publicly available studies in primary CD4+ T cells revealed conservation of CTCF binding sites. Therefore, regulation of CTCF binding is likely to contribute broadly to the regulation of AS,
Splicing networks and the importance of identifying regulatory features
Although the detailed dissection of the regulation of individual genes is essential, to ultimately understand the full functional consequence of AS in immune responses we need to have a broader understanding of the networks of genes that are coordinately regulated to achieve a concerted functional outcome. This is particularly true given the concept of AS as a fine-tuning mechanism discussed above. While the RNA-binding proteins discussed above are good candidates for directing programs of coordinated splicing, predicting targets of these proteins is not simple. While some of the known splicing factors have stringent binding specificity, others bind more promiscuously to pre-mRNAs (Fig. 3). Furthermore, combinatorial control of splicing by multiple RNA-binding proteins that bind to pre-mRNAs independently or in complex with one another, adds many layers of control, as seen in the example of CD45 regulation (Fig. 4). Such modular binding and/or function of proteins allows tight control of the extent to which a given AS event is expressed (58, 107, 108) but precludes straightforward prediction of functional targets.
Global mRNA profiling during various immune responses or in cells depleted of various regulatory proteins is an increasingly common method to identify coregulated AS events. In the case of protein depletion, such profiling studies combined with transcriptome-wide binding maps, generated by approaches such as CLIP-seq, are an important first step toward differentiating between direct and indirect effects of various regulatory proteins. Such genome-wide analyses also aid in defining binding signatures that are predictive of particular splicing patterns. However, even when predicted events are validated, profiling studies are simply a starting place for deeper understanding. Detailed analysis of individual genes and/or biochemical analysis of particular regulatory factors is ultimately required to truly understand the mechanisms by which genes are regulated and how specific combinations of proteins lead to changes in splicing during immune responses.
The most thorough comparison of factor binding with functional effects on splicing in lymphocytes was the recent analysis of CTCF function in B cells described above (105). RNA-seq and microarray analysis of mRNAs following depletion of a splicing factor in T cells has also been applied to hnRNP L and hnRNP LL (65, 70, 71). For hnRNP L, mRNA from thymocytes of hnRNP L depleted versus wildtype mice was analyzed by RNA-seq (65). This study detected no major changes in gene transcription upon hnRNP L depletion, suggesting that the observed differences in mRNA profiles were due solely to AS (65). The investigators found that hnRNP L target genes were enriched in microtubule and cytoskeletal functions. Although upwards of 200 AS events were predicted by RNA-seq, only 4 events were validated (65). Nevertheless, this study implies that hnRNP L regulates a broader network of genes in thymocytes.
For hnRNP LL, exon array analysis of resting and activated hnRNP LL-depleted T cells suggested several hnRNP LL dependent AS events (71), as did analysis of naive and memory T cells from hnRNP LL mutant mice (70). Both studies propose that hnRNP LL regulates a set of AS events upon T-cell activation. Although, in neither case were any of the array predictions confirmed by RT-PCR, so the validity of the predictions remains unknown. Two other regulatory proteins known to exhibit increased activity during T-cell response are PSF and CELF2 (see above, 45, 74). Both of these proteins have been shown in other cell types to regulate the splicing of multiple genes (47, 78, 109–111). These results suggest that PSF and CELF2 also regulate a program of genes in response to T-cell activation. Consistent with this prediction, several exons that we have identified as undergoing activation-responsive AS in T cells are flanked by sequence motifs that match the binding consensus for CELF2 (6). Preliminary data from our laboratory indicate that CELF2 regulates at least some of these activation-induced splicing events in T cells (NM, MJM, KWL, unpublished data), although a comprehensive analysis has not been completed. For PSF, the binding site is less well defined and thus less predictive (75). Therefore, genes regulated by PSF will likely have to be identified experimentally through profiling experiments such as those described above.
Of the other proteins thus far implicated in T-cell splicing, various studies have shown that TIA-1 preferentially binds U-rich elements and that alternative exons with U-rich intronic sequences downstream of 5′ splice sites are frequently regulated by TIA-1 (91, 93). Whether some of the TIA-1-regulated AS events identified in other cell types are also regulated in T cells or whether there are others remains an open question. Similarly, Sam68 and SRSF1 have been implicated in the regulation of many AS events including during the processes of adipogenesis, neuronal development, apoptosis, and heart development (40, 112–114). However, thus far there are no studies inquiring the broad scope of either Sam68 or SRSF1 in AS regulation in immune responses.
In addition to defining the scope of genes that are substrates of various regulatory proteins in lymphocytes, much remains to be learned about the connections between signal transduction and splicing factors during immune responses. Several canonical signaling pathways are activated downstream of the TCR and costimulatory receptors (1). Future studies will be needed to discover signaling axes involved in the coordinated regulation of AS. Attributing regulatory control of splicing to signaling pathways will provide a higher resolution understanding of how signal transduction is coupled to splicing. Signaling pathways that modulate the activity of splicing factors in T cells remain largely undescribed. For the regulatory proteins described above, we know only a few of the upstream signaling molecules that regulate their activity such as GSK3 for PSF, ERK for Sam68, and FASTK for TIA-1 (Fig. 5). Importantly, a single signaling pathway may regulate the activity of multiple factors, and various pathways may impinge on the same factor. For example, a particularly interesting question with regards to PSF is whether all activities are regulated by its phosphorylation by GSK3 and/or interaction with TRAP150 described above. As the cellular phenotype of PSF knockdown is distinct from the TRAP150-inhibited condition (76), it is strongly predicted that PSF has both GSK3/TRAP-dependent and -independent activities.
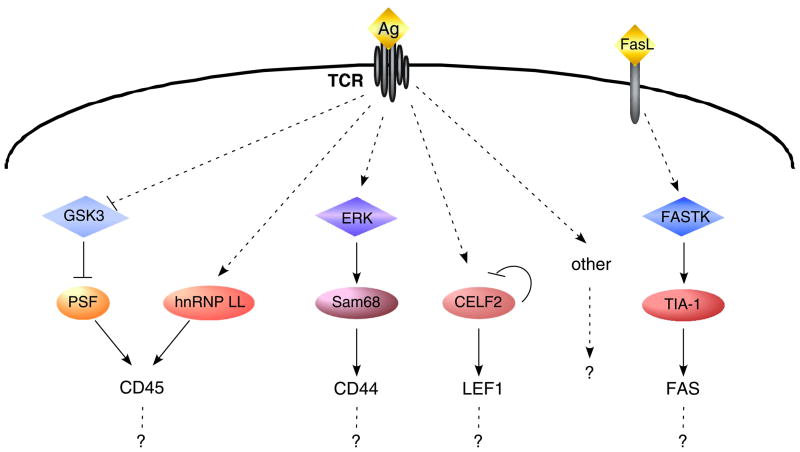
Fig. 5. Networks of signaling pathways that regulate AS in T cells.
Summary of several known kinases (blue/purple diamonds) and RNA binding factors (orange/red ovals) that have been shown to be regulated in response to antigen (Ag) stimulation or Fas Ligand (FasL) signaling in T cells, and the genes (text) that they regulate. Solid arrows and lines indicate direct activities, dotted arrows/lines indicate known relationships that are likely indirect. ‘other’ indicates the fact that there are likely many other signaling pathways that are also involved in mediating antigen-induced changes in AS that are yet to be discovered. Question marks are meant to emphasize that although each signaling pathway so far has only been linked to regulation of 1–2 genes, each branch likely regulates a program of co-regulated genes. Cross-coordination between branches is also anticipated, as observed for CD45. See text for detailed description of each pathway.
To begin to define networks of AS, we have compared the responsiveness of specific signal-induced AS events to various stimuli in cultured and primary T cells (6). We classified distinct networks of signal-induced AS events by their responsiveness to ionomycin relative to PMA. Strikingly, we found the different networks identified in this way are enriched for distinct sequence features (6). Among the enriched motifs we identified the ARS motif as present in several exons that respond to PMA signaling alone in addition to CD45 (6). Some of the additional motifs identified in this study may likewise regulate specific networks of genes distinct from CD45; however, individual experiments will be required to test the functionality of these motifs. Mapping regulatory motifs also serves to predict splicing outcome of other AS events that contain particular motifs. For examples, our initial definition of the ARS motif allowed us to predict T-cell activation responsive exons in the Tau and Fyn genes that we subsequently validated (39).
Finally, motif mapping is important to enhance our predictive capacity of disease-causing SNPs. For example, our group has made important contributions to understanding how the SNP in CD45 exon 4 (C77G) affects AS. Despite the link between the C77G SNP and susceptibility to MS and HIV infection (see above), the molecular consequences of this disease relevant SNP had remained unexplored. The C77G disrupts the ESS1 and confers aberrantly high exon 4 inclusion without affecting the protein coding capacity (20, 21). We recently found that the C77G polymorphism weakly affects the binding of hnRNP L to exon 4 and dramatically abrogates the binding of two additional hnRNP proteins, hnRNP E2 and hnRNP K (108). Even though hnRNP K and hnRNP E2 make little contribution to CD45 exon splicing under normal conditions, upon depletion of hnRNP L, these proteins play a compensatory role (62, 63, 108). The C77G mutation both reduces the binding of hnRNP L and prevents the compensatory function of hnRNP K and E2, to result in a significant overall loss of splicing repression. Therefore, a thorough biochemical understanding of the essential regulatory element controlling exon 4 expression and the proteins that associate with this sequence permitted a mechanistic explanation for the phenotype of the C77G SNP. Such a detailed appreciation of the molecular basis of disease is the first step toward predicting the consequences of similar polymorphisms and designing approaches to correct or override such genetically encoded defects.
Conclusions and implications for future studies
Many lines of evidence suggest broad regulation of AS during immune responses. Several cases have been described in which AS serves to modulate lymphocyte activity. In addition, some of the signaling pathways, splicing factors, and auxiliary elements that drive AS in immune effector functions have been characterized. However, many more genes and regulatory factors have yet to be investigated, and even the full spectrum of AS regulation in lymphocytes – in different cell types and during different phases of an immune response – remains to be rigorously explored. It is imperative that research continues to thoroughly map the factors, mechanisms, networks, and consequences of AS in the immune system. Given the potential for AS to broadly shape cellular function and the growing number of instances in which aberrant splicing is known to contribute to defects in immune function, future studies will undoubtedly continue to uncover new and exciting connections between AS and immune responses and suggest novel approaches to the treatment of human disease.
Acknowledgments
KWL and the work from the Lynch laboratory described herein are supported by R01 GM084034 and GM067719. NMM has been supported by GM084034-S1.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 6.Martinez NM, et al. Alternative splicing networks regulated by signaling in human T cells. RNA. 2012;18:1029–1040. doi: 10.1261/rna.032243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grigoryev YA, et al. Genome-wide analysis of immune activation in human T and B cells reveals distinct classes of alternatively spliced genes. PLoS ONE. 2009;4:e7906. doi: 10.1371/journal.pone.0007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch KW. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol. 2004;4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- 10.Moulton VR, Tsokos GC. Alternative splicing factor/splicing factor 2 regulates the expression of the zeta subunit of the human T cell receptor-associated CD3 complex. J Biol Chem. 2010;285:12490–12496. doi: 10.1074/jbc.M109.091660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuzaka K, et al. TCR zeta mRNA with an alternatively spliced 3′-untranslated region detected in systemic lupus erythematosus patients leads to the down-regulation of TCR zeta and TCR/CD3 complex. J Immunol. 2003;171:2496–2503. doi: 10.4049/jimmunol.171.5.2496. [DOI] [PubMed] [Google Scholar]
- 12.Nambiar MP, et al. Polymorphisms/mutations of TCR-zeta-chain promoter and 3′ untranslated region and selective expression of TCR zeta-chain with an alternatively spliced 3′ untranslated region in patients with systemic lupus erythematosus. J Autoimmun. 2001;16:133–142. doi: 10.1006/jaut.2000.0475. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhury B, et al. Stability and translation of TCR zeta mRNA are regulated by the adenosine-uridine-rich elements in splice-deleted 3′ untranslated region of zeta-chain. J Immunol. 2006;177:8248–8257. doi: 10.4049/jimmunol.177.11.8248. [DOI] [PubMed] [Google Scholar]
- 14.Chowdhury B, et al. Decreased stability and translation of T cell receptor zeta mRNA with an alternatively spliced 3′-untranslated region contribute to zeta chain down-regulation in patients with systemic lupus erythematosus. J Biol Chem. 2005;280:18959–18966. doi: 10.1074/jbc.M501048200. [DOI] [PubMed] [Google Scholar]
- 15.Nambiar MP, et al. Reconstitution of deficient T cell receptor zeta chain restores T cell signaling and augments T cell receptor/CD3-induced interleukin-2 production in patients with systemic lupus erythematosus. Arthritis Rheum. 2003;48:1948–1955. doi: 10.1002/art.11072. [DOI] [PubMed] [Google Scholar]
- 16.Hermiston ML, Xu Z, Weiss A. CD45: a critical regulator of signaling thresholds in immune cells. Annu Rev Immunol. 2003;21:107–137. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- 17.Lynch KW, Weiss A. A model system for the activation-induced alternative-splicing of CD45 implicates protein kinase C and Ras. Mol Cell Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dornan S, et al. Differential association of CD45 isoforms with CD4 and CD8 regulates the actions of specific pools of p56lck tyrosine kinase in T cell antigen receptor signal transduction. J Biol Chem. 2002;277:1912–1918. doi: 10.1074/jbc.M108386200. [DOI] [PubMed] [Google Scholar]
- 19.Xu Z, Weiss A. Negative regulation of CD45 by differential homodimerization of the alternatively spliced isoforms. Nat Immunol. 2002;3:764–771. doi: 10.1038/ni822. [DOI] [PubMed] [Google Scholar]
- 20.Lynch KW, Weiss A. A CD45 polymorphism associated with Muliple Sclerosis disrupts an exonic splicng silencer. J Biol Chem. 2001;276:24341–24347. doi: 10.1074/jbc.M102175200. [DOI] [PubMed] [Google Scholar]
- 21.Zilch CF, Walker AM, Timon M, Goff LK, Wallace DL, Beverley PCL. A point mutation within CD45 exon A is the cause of variant CD45RA splicing in humans. Eur J Immunol. 1998;28:22–29. doi: 10.1002/(SICI)1521-4141(199801)28:01<22::AID-IMMU22>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 22.Tchilian EZ, et al. A point mutation in CD45 may be associated with an increased risk of HIV-1 infection. AIDS. 2001;15:1892–1894. doi: 10.1097/00002030-200109280-00024. [DOI] [PubMed] [Google Scholar]
- 23.Jacobsen M, et al. A point mutation in PTPRC is associated with the development of multiple sclerosis. Nat Genet. 2000;26:495–499. doi: 10.1038/82659. [DOI] [PubMed] [Google Scholar]
- 24.Imbeault M, Giguere K, Ouellet M, Tremblay MJ. Exon level transcriptomic profiling of HIV-1-infected CD4(+) T cells reveals virus-induced genes and host environment favorable for viral replication. PLoS Pathog. 2012;8:e1002861. doi: 10.1371/journal.ppat.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodwin RG, et al. Cloning of the human and murine interleukin-7 receptors: demonstration of a soluble form and homology to a new receptor superfamily. Cell. 1990;60:941–951. doi: 10.1016/0092-8674(90)90342-c. [DOI] [PubMed] [Google Scholar]
- 26.Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol. 2007;7:144–154. doi: 10.1038/nri2023. [DOI] [PubMed] [Google Scholar]
- 27.Schluns KS, Kieper WC, Jameson SC, Lefrancois L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol. 2000;1:426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- 28.Kondrack RM, Harbertson J, Tan JT, McBreen ME, Surh CD, Bradley LM. Interleukin 7 regulates the survival and generation of memory CD4 cells. J Exp Med. 2003;198:1797–1806. doi: 10.1084/jem.20030735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose T, Lambotte O, Pallier C, Delfraissy JF, Colle JH. Identification and biochemical characterization of human plasma soluble IL-7R: lower concentrations in HIV-1-infected patients. J Immunol. 2009;182:7389–7397. doi: 10.4049/jimmunol.0900190. [DOI] [PubMed] [Google Scholar]
- 30.Gregory SG, et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 31.McKay FC, et al. Haplotypes of the interleukin 7 receptor alpha gene are correlated with altered expression in whole blood cells in multiple sclerosis. Genes Immun. 2008;9:1–6. doi: 10.1038/sj.gene.6364436. [DOI] [PubMed] [Google Scholar]
- 32.Vudattu NK, Magalhaes I, Hoehn H, Pan D, Maeurer MJ. Expression analysis and functional activity of interleukin-7 splice variants. Genes Immun. 2009;10:132–140. doi: 10.1038/gene.2008.90. [DOI] [PubMed] [Google Scholar]
- 33.Palacios EH, Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23:7990–8000. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
- 34.Cooke MP, Perlmutter RM. Expression of a novel form of the fyn proto-oncogene in hematopoietic cells. New Biol. 1989;1:66–74. [PubMed] [Google Scholar]
- 35.Brignatz C, et al. Alternative splicing modulates autoinhibition and SH3 accessibility in the Src kinase Fyn. Mol Cell Biol. 2009;29:6438–6448. doi: 10.1128/MCB.00398-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidson D, Chow LM, Fournel M, Veillette A. Differential regulation of T cell antigen responsiveness by isoforms of the src-related tyrosine protein kinase p59fyn. The J Exp Med. 1992;175:1483–1492. doi: 10.1084/jem.175.6.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davidson D, Viallet J, Veillette A. Unique catalytic properties dictate the enhanced function of p59fynT, the hemopoietic cell-specific isoform of the Fyn tyrosine protein kinase, in T cells. Mol Cell Biol. 1994;14:4554–4564. doi: 10.1128/mcb.14.7.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weil R, et al. Altered expression of tyrosine kinases of the Src and Syk families in human T-cell leukemia virus type 1-infected T-cell lines. J Virol. 1999;73:3709–3717. doi: 10.1128/jvi.73.5.3709-3717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothrock C, Cannon B, Hahm B, Lynch KW. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- 40.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Genderen C, et al. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 42.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 43.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected] Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 44.Waterman ML, Fischer WH, Jones KA. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 45.Mallory MJ, Jackson J, Weber B, Chi A, Heyd F, Lynch KW. Signal- and development-dependent alternative splicing of LEF1 in T cells is controlled by CELF2. Mol Cell Biol. 2011;31:2184–2195. doi: 10.1128/MCB.05170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leiden JM, Thompson CB. Transcriptional regulation of T-cell genes during T-cell development. Curr Opin Immunol. 1994;6:231–237. doi: 10.1016/0952-7915(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 47.Dembowski JA, Grabowski PJ. The CUGBP2 splicing factor regulates an ensemble of branchpoints from perimeter binding sites with implications for autoregulation. PLoS Genet. 2009;5:e1000595. doi: 10.1371/journal.pgen.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Izquierdo JM, Valcarcel J. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J Biol Chem. 2007;282:1539–1543. doi: 10.1074/jbc.C600198200. [DOI] [PubMed] [Google Scholar]
- 49.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 53.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 54.House AE, Lynch KW. Regulation of Alternative Splicing: More than just the ABCs. J Biol Chem. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- 55.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 57.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette J-F, Revil T, Chabot B. hnRNP Proteins and Splicing Control. Austin: Landes Biosciences; 2007. [DOI] [PubMed] [Google Scholar]
- 58.Motta-Mena LB, Heyd F, Lynch KW. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Molecular cell. 2010;29:223–234. doi: 10.1016/j.molcel.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heyd F, Lynch KW. Degrade, move, regroup: signaling control of splicing proteins. Trends Biochem Sci. 2011;36:397–404. doi: 10.1016/j.tibs.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mayya V, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 61.Tong A, Nguyen J, Lynch KW. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J Biol Chem. 2005;280:38297–38304. doi: 10.1074/jbc.M508123200. [DOI] [PubMed] [Google Scholar]
- 62.Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol. 2007;27:6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Topp JD, Jackson J, Melton AA, Lynch KW. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA. 2008;14:2038–2049. doi: 10.1261/rna.1212008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaudreau MC, Heyd F, Bastien R, Wilhelm B, Moroy T. Alternative splicing controlled by heterogeneous nuclear ribonucleoprotein L regulates development, proliferation, and migration of thymic pre-T cells. J Immunol. 2012;188:5377–5388. doi: 10.4049/jimmunol.1103142. [DOI] [PubMed] [Google Scholar]
- 66.Hui J, et al. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hui J, Stangl K, Lane WS, Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol. 2003;10:33–37. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- 69.Liu G, et al. A conserved serine of heterogeneous nuclear ribonucleoprotein L (hnRNPL) mediates depolarization-regulated alternative splicing of potassium channels. J Biol Chem. 2012;287:22709–22716. doi: 10.1074/jbc.M112.357343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu Z, et al. Memory T cell RNA rearrangement programmed by heterogeneous nuclear ribonucleoprotein hnRNPLL. Immunity. 2008;29:863–875. doi: 10.1016/j.immuni.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levinson N, et al. Use of transcriptional synergy to augment sensitivity of a splicing reporter assay. RNA. 2006;12:925–930. doi: 10.1261/rna.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu Z, Yates AL, Hoyne GF, Goodnow CC. Consequences of increased CD45RA and RC isoforms for TCR signaling and peripheral T cell deficiency resulting from heterogeneous nuclear ribonucleoprotein L-like mutation. J Immunol. 185:231–238. doi: 10.4049/jimmunol.0903625. [DOI] [PubMed] [Google Scholar]
- 74.Heyd F, Lynch KW. Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol Cell. 2010;40:126–137. doi: 10.1016/j.molcel.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 76.Heyd F, Lynch KW. PSF controls expression of histone variants and cellular viability in thymocytes. Biochem Biophys Res Commun. 2011;414:743–749. doi: 10.1016/j.bbrc.2011.09.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dasgupta T, Ladd AN. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip Rev RNA. 2011;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beisang D, Rattenbacher B, Vlasova-St Louis IA, Bohjanen PR. Regulation of CUG-binding protein 1 (CUGBP1) binding to target transcripts upon T cell activation. J Biol Chem. 2012;287:950–960. doi: 10.1074/jbc.M111.291658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anderson P, et al. A monoclonal antibody reactive with a 15-kDa cytoplasmic granule-associated protein defines a subpopulation of CD8+ T lymphocytes. J Immunol. 1990;144:574–582. [PubMed] [Google Scholar]
- 81.Puig O, Gottschalk A, Fabrizio P, Seraphin B. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 1999;13:569–580. doi: 10.1101/gad.13.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Forch P, et al. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 83.Gottschalk A, et al. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- 84.Piecyk M, et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Del Gatto-Konczak F, et al. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol Cell Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu C, Cheng J, Mountz JD. Differential expression of human Fas mRNA species upon peripheral blood mononuclear cell activation. Biochem J. 1995;310:957–963. doi: 10.1042/bj3100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheng J, et al. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994;263:1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- 88.Cascino I, Fiucci G, Papoff G, Ruberti G. Three functional soluble forms of the human apoptosis-inducing Fas molecule are produced by alternative splicing. J Immunol. 1995;154:2706–2713. [PubMed] [Google Scholar]
- 89.Izquierdo JM, et al. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 90.Tian Q, Taupin J, Elledge S, Robertson M, Anderson P. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J Exp Med. 1995;182:865–874. doi: 10.1084/jem.182.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 92.Aznarez I, et al. A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008;18:1247–1258. doi: 10.1101/gr.073155.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Z, et al. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010;8:e1000530. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.ten Dam GB, et al. Regulation of alternative splicing of CD45 by antagonistic effects of SR protein splicing factors. J Immunol. 2000;164:5287–5295. doi: 10.4049/jimmunol.164.10.5287. [DOI] [PubMed] [Google Scholar]
- 95.Wang H-Y, Xu X, Ding J-H, Bermingham JR, Fu X-D. SC35 plays a role in T cell development and alternative splicing of CD45. Mol Cell. 2001;7:331–342. doi: 10.1016/s1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 96.Screaton GR, et al. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochim Biophys Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 98.Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 99.Arch R, et al. Participation in normal immune responses of a metastasis-inducing splice variant of CD44. Science. 1992;257:682–685. doi: 10.1126/science.1496383. [DOI] [PubMed] [Google Scholar]
- 100.Konig H, Ponta H, Herrlich P. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 1998;17:2904–2913. doi: 10.1093/emboj/17.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weg-Remers S, Ponta H, Herrlich P, Konig H. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 2001;20:4194–4203. doi: 10.1093/emboj/20.15.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng C, Yaffe MB, Sharp PA. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev. 2006;20:1715–1720. doi: 10.1101/gad.1430906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struc Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 104.Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struc Mol Biol. 2006;13:5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- 105.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Singh P, Lee DH, Szabo PE. More than insulator: multiple roles of CTCF at the H19-Igf2 imprinted domain. Front Genet. 2012;3:214. doi: 10.3389/fgene.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Preussner M, et al. HnRNP L and L-like cooperate in multiple-exon regulation of CD45 alternative splicing. Nucleic Acids Res. 40:5666–5678. doi: 10.1093/nar/gks221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Motta-Mena LB, Smith SA, Mallory MJ, Jackson J, Wang J, Lynch KW. A disease-associated polymorphism alters splicing of the human CD45 phosphatase gene by disrupting combinatorial repression by heterogeneous nuclear ribonucleoproteins (hnRNPs) J Biol Chem. 2011;286:20043–20053. doi: 10.1074/jbc.M111.218727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ray P, Kar A, Fushimi K, Havlioglu N, Chen X, Wu JY. PSF suppresses tau exon 10 inclusion by interacting with a stem-loop structure downstream of exon 10. J Mol Neurosci. 45:453–466. doi: 10.1007/s12031-011-9634-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim KK, Kim YC, Adelstein RS, Kawamoto S. Fox-3 and PSF interact to activate neural cell-specific alternative splicing. Nucleic Acids Res. 2011;39:3064–3078. doi: 10.1093/nar/gkq1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ladd AN, Taffet G, Hartley C, Kearney DL, Cooper TA. Cardiac tissue-specific repression of CELF activity disrupts alternative splicing and causes cardiomyopathy. Mol Cell Biol. 2005;25:6267–6278. doi: 10.1128/MCB.25.14.6267-6278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huot ME, et al. The Sam68 STAR RNA-binding protein regulates mTOR alternative splicing during adipogenesis. Mol Cell. 2012;46:187–199. doi: 10.1016/j.molcel.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 113.Iijima T, et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–1614. doi: 10.1016/j.cell.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu X, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 115.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 116.Atkinson TP, Hall CG, Goldsmith J, Kirkham PM. Splice variant in TCRzeta links T cell receptor signaling to a G-protein-related signaling pathway. Biochem Biophys Res Commun. 2003;310:761–766. doi: 10.1016/j.bbrc.2003.09.073. [DOI] [PubMed] [Google Scholar]
- 117.Magistrelli G, et al. A soluble form of CTLA-4 generated by alternative splicing is expressed by nonstimulated human T cells. Eur J Immunol. 1999;29:3596–3602. doi: 10.1002/(SICI)1521-4141(199911)29:11<3596::AID-IMMU3596>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 118.Astier AL, Meiffren G, Freeman S, Hafler DA. Alterations in CD46-mediated Tr1 regulatory T cells in patients with multiple sclerosis. J Clin Invest. 2006;116:3252–3257. doi: 10.1172/JCI29251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Castro MA, et al. Extracellular isoforms of CD6 generated by alternative splicing regulate targeting of CD6 to the immunological synapse. J Immunol. 2007;178:4351–4361. doi: 10.4049/jimmunol.178.7.4351. [DOI] [PubMed] [Google Scholar]
- 120.Thakral D, Dobbins J, Devine L, Kavathas PB. Differential expression of the human CD8beta splice variants and regulation of the M-2 isoform by ubiquitination. J Immunol. 2008;180:7431–7442. doi: 10.4049/jimmunol.180.11.7431. [DOI] [PubMed] [Google Scholar]
- 121.Dudziak D, Nimmerjahn F, Bornkamm GW, Laux G. Alternative splicing generates putative soluble CD83 proteins that inhibit T cell proliferation. J Immunol. 2005;174:6672–6676. doi: 10.4049/jimmunol.174.11.6672. [DOI] [PubMed] [Google Scholar]
- 122.Meyer D, et al. CD96 interaction with CD155 via its first Ig-like domain is modulated by alternative splicing or mutations in distal Ig-like domains. J Biol Chem. 2009;284:2235–2244. doi: 10.1074/jbc.M807698200. [DOI] [PubMed] [Google Scholar]
- 123.Liu C, Cheng J, Mountz JD. Differential expression of human Fas mRNA species upon peripheral blood mononuclear cell activation. Biochemistry J. 1995;310:957–963. doi: 10.1042/bj3100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.King PD, Sandberg ET, Selvakumar A, Fang P, Beaudet AL, Dupont B. Novel isoforms of murine intercellular adhesion molecule-1 generated by alternative RNA splicing. J Immunol. 1995;154:6080–6093. [PubMed] [Google Scholar]
- 125.Wang Y, Su X, Sorenson CM, Sheibani N. Modulation of PECAM-1 expression and alternative splicing during differentiation and activation of hematopoietic cells. J Cell Biochem. 2003;88:1012–1024. doi: 10.1002/jcb.10451. [DOI] [PubMed] [Google Scholar]
- 126.Alms WJ, Atamas SP, Yurovsky VV, White B. Generation of a variant of human interleukin-4 by alternative splicing. Mol Immunol. 1996;33:361–370. doi: 10.1016/0161-5890(95)00154-9. [DOI] [PubMed] [Google Scholar]
- 127.Kaur G, Goodall JC, Jarvis LB, Hill Gaston JS. Characterisation of Foxp3 splice variants in human CD4+ and CD8+ T cells--identification of Foxp3Delta7 in human regulatory T cells. Mol Immunol. 2010;48:321–332. doi: 10.1016/j.molimm.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 128.Whistler T, Chiang CF, Lonergan W, Hollier M, Unger ER. Implementation of exon arrays: alternative splicing during T-cell proliferation as determined by whole genome analysis. BMC Genomics. 2010;11:496. doi: 10.1186/1471-2164-11-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Alcaraz-Garcia MJ, et al. Human and mouse DOCK10 splicing isoforms with alternative first coding exon usage are differentially expressed in T and B lymphocytes. Hum Immunol. 2011;72:531–537. doi: 10.1016/j.humimm.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 130.Jia Z, et al. A novel splice variant of FR4 predominantly expressed in CD4+CD25+ regulatory T cells. Immunol Invest. 2009;38:718–729. doi: 10.3109/08820130903171003. [DOI] [PubMed] [Google Scholar]
- 131.Birzele F, et al. Next-generation insights into regulatory T cells: expression profiling and FoxP3 occupancy in Human. Nucleic Acids Res. 39:7946–7960. doi: 10.1093/nar/gkr444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jensen LE, Whitehead AS. IRAK1b, a novel alternative splice variant of interleukin-1 receptor-associated kinase (IRAK), mediates interleukin-1 signaling and has prolonged stability. J Biol Chem. 2001;276:29037–29044. doi: 10.1074/jbc.M103815200. [DOI] [PubMed] [Google Scholar]
- 133.Ohya S, Fujimori T, Kimura T, Yamamura H, Imaizumi Y. Novel spliced variants of large-conductance Ca(2+)-activated K(+)-channel beta2-subunit in human and rodent pancreas. J Pharmacol Sci. 2010;114:198–205. doi: 10.1254/jphs.10159fp. [DOI] [PubMed] [Google Scholar]
- 134.Syken J, Macian F, Agarwal S, Rao A, Munger K. TID1, a mammalian homologue of the drosophila tumor suppressor lethal(2) tumorous imaginal discs, regulates activation-induced cell death in Th2 cells. Oncogene. 2003;22:4636–4641. doi: 10.1038/sj.onc.1206569. [DOI] [PubMed] [Google Scholar]
- 135.Jalink M, Ge Z, Liu C, Bjorkholm M, Gruber A, Xu D. Human normal T lymphocytes and lymphoid cell lines do express alternative splicing variants of human telomerase reverse transcriptase (hTERT) mRNA. Biochem Biophys Res Commun. 2007;353:999–1003. doi: 10.1016/j.bbrc.2006.12.149. [DOI] [PubMed] [Google Scholar]