Summary
The important role of microRNAs in directing immune responses has become increasingly clear. Here, we highlight discoveries uncovering the role of specific microRNAs in regulating the development and function of natural killer (NK) cells. Furthermore, we discuss the impact of NK cells on the entire immune system during global and specific microRNA ablation in the settings of inflammation, infection, and immune dysregulation.
Keywords: natural killer, NK, microRNA, development, cytotoxicity
General introduction
Natural killer (NK) cells are critical mediators of host immunity to malignancy and infection. Humans (and mice) lacking functional NK cells exhibit increased susceptibility to a variety of viral pathogens, especially herpesviruses including cytomegalovirus, Epstein-Barr virus, varicella zoster virus, and herpes simplex virus (1–6). Similarly, NK cells mediate potent anti-tumor responses in mice (7–10) and, as evidenced in a growing number of clinical studies, in humans (7, 11–19).
NK cell function is controlled by germ-line encoded surface receptors, which can be broadly categorized as either activating or inhibitory. Activating receptors, such as NKG2D and Ly49H in mice or KIR2DL and CD94-NKG2C in humans, recognize self- or virally encoded molecules expressed on infected or malignant cells and transduce signals that drive cytotoxic responses (20–23). In contrast, many of the inhibitory receptors bind to self-major histocompatibility complex (MHC) class I molecules expressed on healthy cells and are important for restraining NK cell function (22, 24–26). In addition, mature NK cells express receptors for and are highly responsive to pro-inflammatory and pro-survival cytokines such as interleukin-12 (IL-12), IL-18, IL-2, Type I interferons (IFNs), and IL-15, which can activate NK cells alone or synergize with signals downstream of activating receptors (20, 27–33).
Productive NK cell activation leads to cytotoxicity, cytokine secretion, and in specific cases, the robust proliferation of antigen-specific effector cells (34). NK cell-mediated cytotoxicity, which involves the targeted release of lytic molecules such as perforin and granzyme B, results in apoptosis of virus-infected, malignant, or diseased host cells. In addition, NK cell-derived chemokines and cytokines, most notably IFN-γ, can exert potent regulatory effects on targets cells, as well as recruit and activate other cells of the immune system, including macrophages and dendritic cells, and T and B lymphocytes, which mediate further anti-viral and anti-tumor immunity (35, 36).
In addition to cytotoxicity and cytokine secretion, certain stimuli (e.g. specific viral proteins) are able to trigger robust clonal proliferation in NK cells. This response shares many features with CD8+ T-cell responses, including antigen-specificity and – dependency, defined proliferation and contraction phases, and the development of a small pool of long-lived ‘memory’ cells, which exhibit heightened functionality upon secondary antigen exposure (34, 37–40). For example, certain mouse strains (such as C57BL/6) harbor a subset of NK cells that express the Ly49H activating receptor (6, 41, 42). This receptor can recognize the m157 glycoprotein encoded by the mouse cytomegalovirus (MCMV). During MCMV infection in C57BL/6 mice, binding of the Ly49H receptor to the m157 viral protein expressed on infected cells triggers a robust, clonal expansion of Ly49H+ NK cells (43–45). A similar clonal-like expansion has been observed for human NK cells expressing the activating receptor, CD94-NKG2C, during human cytomegalovirus (HCMV) infection, suggesting a conserved mechanism of antigen-driven NK cell expansion (46).
MicroRNAs (miRNAs) comprise a large family of small, non-coding regulatory RNAs that act as post-transcriptional repressors of protein-coding target messenger RNA (mRNA) species (47). miRNAs are initially generated as long primary transcripts (pri-mRNAs) that fold into a hairpin secondary structure (48). Pri-mRNAs are then processed by a nuclear enzyme complex containing the RNase III-like enzyme, Drosha, and its double-stranded RNA (dsRNA)-binding co-factor, DiGeorge syndrome critical region 8 protein (Dcgr8), into 70–80 nucleotide dsRNA precursors (known as pre-miRNAs) (49–51). Following export to the cytoplasm via the nuclear transporter Exportin V (52, 53), pre-mRNAs undergo further enzymatic trimming by a second RNase III-like enzyme, Dicer, yielding 16–25 nucleotide length RNA duplexes that contain the mature miRNA (54). One strand of the mature miRNA duplex interacts with an Argonaute family protein (55), of which there are four known in mammals (Ago1-Ago4), to form a miRNA-induced silencing complex (miRISC) (47). Regulation is achieved by binding of the miRNA (as part of the larger miRISC) to imperfect complementary ‘seed’ sequences often located in the 3′-untranslated region (3′-UTR) of the target mRNA. Formation of the miRNA:mRNA complex promotes degradation and/or prevents translation of the target mRNA, thus silencing protein expression of targeted genes (47, 56–58).
miRNAs in higher vertebrates are often highly conserved and can each target hundreds of distinct protein-coding targets (59–62). Although complete elimination of any one target is rarely achieved by miRNAs in mammalian cells, simultaneous downmodulation of many genes provides a mechanism by which a single miRNA, or several miRNAs in concert, can drastically modify the global expression profile of a cell (62, 63). For this reason, miRNAs are important regulators of diverse aspects of cellular development and function. Here we summarize recent findings on the role of miRNAs in NK cell biology in the context of development, inflammation, tumor surveillance, and viral infection.
miRNAs in NK cell development and maturation
Like T and B cells, NK cells develop from pluripotent common lymphoid progenitor (CLP) cells in a multi-stage process that occurs primarily in the bone marrow in adults (although peripheral sites of development have been described) (34, 64–67). Discrete developmental stages can be identified by the sequential acquisition of distinct surface receptors. Very early NK cell progenitors (pre-NK cells) in mice express CD122, the shared γ-chain of the IL-2 and IL-15 receptors. Studies in IL-15- and IL-15R-deficient mice, as well as in vitro models of human and mouse NK cell differentiation, have shown that IL-15 is required for the survival of NK cells past the progenitor stage (68–71). As development progresses, pre-NKs acquire expression of the NK cell lineage-defining receptors that will ultimately control their activation and effector function; examples include NK1.1 (in C57BL/6 and SJL mouse strains), CD94-NKG2A, NKG2D, CD49b, and natural cytotoxicity receptors, along with the Ly49 receptors in mice and NKG2C, NKG2A, and KIR in humans. These immature NK cells then acquire the final vestiges of cytotoxic and pro-inflammatory functionality: expression of pre-formed transcripts for granzyme B, perforin, and IFN-γ, the translation of which is rapidly initiated upon NK cell activation in the periphery. These functional but still immature NK cells emigrate from the bone marrow and undergo further differentiation and maturation in peripheral tissues such as the spleen and liver. In mice, terminal maturation in the periphery is associated with downregulation of CD27 and upregulation of CD11b expression, as well as acquisition of full cytolytic and cytokine secretion potential.
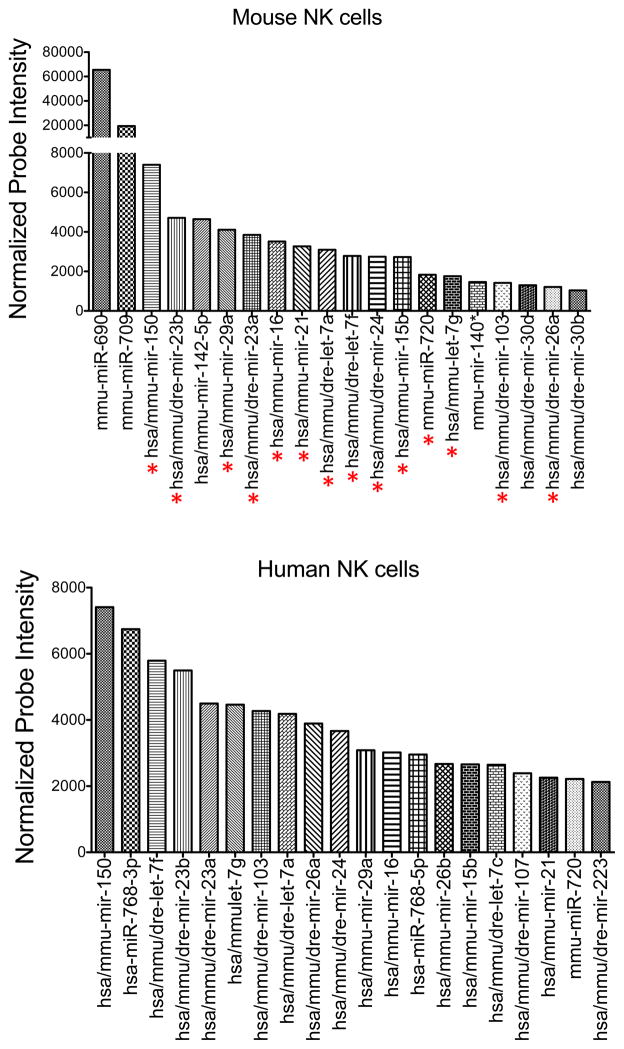
Recent work in animals lacking specific or global miRNAs, in combination with comprehensive miRNA expression profiling studies, has allowed investigators to explore the mechanisms by which miRNAs regulate NK cell development. Microarray studies revealed the expression of nearly 200 unique miRNAs in human and mouse primary NK cells (72). Among these, 80% of those identified in human NK cells could be found in their mouse counterparts, and 59% of the miRNAs present in mouse NK cells were also found in human NK cells, indicating significant interspecies overlap (72). Among the most highly expressed miRNAs in both mouse and human NK cells were miR-150, miR-23b, miR-29a, miR-23a, miR-16, miR-21, let-7a, let-7f, miR-24, miR-15b, miR-720, let-7g, miR-103, and mir-26a (Fig. 1). Similar results were generated in separate studies that used next generation sequencing to identify >400 miRNAs in human (73) and >300 in mouse NK cells (74). In the latter study, the top 10 most highly expressed in NK cells accounted for ~65% of the total miRNA pool (74).
Fig. 1. Top 20 most highly expressed miRNAs in resting mouse and human NK cells by microarray.
Total RNA was extracted from sorted mouse splenic NK cells (NK1.1+ TCRβ−) and human peripheral blood NK cells (CD56+CD3−). Expression of individual miRNAs was assessed by microarray. After normalization, 170 oligonucleotide probes gave mean fluorescence values above background. Shown are the normalized expression values (in arbitrary units) of the top 20 most highly expressed miRNAs in mouse (top) and human (bottom) NK cells. miRNAs common to both groups are indicated with a red asterisk below the top graph.
Early studies addressing the importance of miRNAs in NK cell development took advantage of the universal role of Dicer and Dgcr8 in miRNA biogenesis. To circumvent the embryonic requirement for Dicer and Dgcr8, Bezman et al. (72) studied the impact of global miRNA-deficiency on NK cell development using genetically modified mice expressing either loxP-flanked (‘floxed’) Dicer1 or Dgcr8 alleles and a chimeric Cre recombinase that could be specifically activated by exogenous tamoxifen treatment. In this system, drug-induced deletion of Dicer1 or Dgcr8 led to a significant decrease in the number of splenic and liver NK cells, and this defect was shown to stem from impaired survival and proliferation in these cells (72). In addition, both Dicer- and Dcgr8-deficient mice harbored a relative deficit of mature (CD27loCD11bhi) NK cells and a relative surplus of immature (CD27hiCD11blo) NK cells, highlighting a role for miRNAs in NK cell maturation (72). Although Dicer is required for the biogenesis of other small RNAs (including siRNA, shRNA, and snoRNAs), the fact that deletion of Dcgr8, which is involved only in miRNA synthesis, similarly impaired NK cell development suggested that loss of miRNAs was specifically responsible for the observed defect in both animal cohorts (72). Sullivan et al. (75) further showed that the requirement for miRNAs in NK cell development was cell-intrinsic, because deletion of Dicer1 only in developing lymphocytes (via transgenic expression of a human CD2 promoter-driven Cre cassette) also led to a reduced frequency and impaired maturation of NK cells.
Although global miRNA deficiency impairs NK cell homeostasis and maturation, recent work by Thomas et al. (76) suggests that an overabundance of miRNAs may similarly have a negative impact on NK cell development. Mice lacking Eri1, a 3′-to-5′ exoribonuclease that suppresses small RNA function in mammalian cells, have a global and sequence-independent increase in miRNA abundance. Interestingly, these mice also exhibited a substantial reduction and aberrant maturation of peripheral NK cells, defined by delayed acquisition of Ly49 receptors in the bone marrow and a reduction of cells expressing Ly49D and Ly49H in the periphery (76). Using mixed bone marrow chimeric animals, the requirement for Eri1 in developing NK cells was found to be cell-intrinsic. Thus, either elevation or reduction of the cellular levels of miRNA expression has a negative impact on NK cell homeostasis.
Although informative, modulation of global miRNA levels, as occurs in Dicer-, Dcgr8-, and Eri1-deficient mice, does not allow for delineation of which miRNAs regulate NK cell development and their individual mechanisms of action. Rather, experimental modulation of individual miRNAs has been necessary to understand their specific roles in the context of NK cell biology. MiR-150, for example, is important for B and T-cell development and function (77–80) and, given its high expression in both mouse and human resting NK cells (72) (Fig. 1), it was identified as a potential regulator of NK cell function. Indeed, genetic deletion of miR-150 led to a significant reduction in the number of peripheral, mature NK cells in mice (81). This reduction was attributed to a developmental blockade in NK cell maturation, as evidenced by the accumulation of immature NK cells in both the bone marrow and the peripheral organs of miR-150−/− animals. Reciprocally, transgene-mediated overexpression of miR-150 increased the number and frequency of NK cells in mice and led to the development of NK cells with a more mature phenotype (81). The developmental phenotype of miR-150−/− NK cells was attributed in part to increased expression of c-Myb, a known miR-150 target (78) and a direct transcriptional activator of Myc (82, 83) and Bcl-2 (84), genes with important pro-proliferative and pro-survival functions, respectively, in developing lymphocytes. miR-150−/− NK cells exhibited elevated levels of c-Myb and its downstream targets, c-Myc and Bcl-2, supporting this hypothesis (81). Moreover, mice lacking one Myb allele exhibited a phenotype that closely mirrored that of miR-150 overexpression, i.e. an increase in the number and frequency of mature NK cells (81). Thus, miR-150, on its own, is critical for NK cell development and maturation in part because of its function as a regulator of c-Myb expression (81).
Another miRNA, miR-181, was recently reported to be upregulated in developing human NK cells. Consistent with its known role in T and B-cell development (85, 86), knockdown of miR-181 in human CD34+ hematopoietic progenitor cells dramatically blocked their ability to develop into NK cells in vitro (87). Mechanistically, miR-181 was found to be required for the repression of nemo-like kinase (NLK) (87), a known inhibitor of Notch signaling [which is important for NK cell development (88, 89)]. Knockdown of miR-181 led to an increase in NLK protein, which correlated with impaired NK cell development, whereas either NLK deficiency or miR-181 overexpression enhanced development (87). The signals that regulate miR-181 expression in developing NK cells are not fully understood. However, miR-181a was recently shown to be induced by and act downstream of the T-cell receptor (TCR) in mouse T cells (85). In this context, miR-181a acted as a modulator of TCR signal strength by repressing the protein tyrosine phosphatases, Ptpn22, Shp2, Dusp5, and Dusp6, and thereby boosting basal TCR signals. Given that all of these phosphatases are expressed in mouse NK cells (90, 91), it has been suggested that activating NK cell receptors may similarly utilize miR-181 to modulate signal strength downstream of receptor engagement in NK cells (72). Thus, both global and specific ablation of miRNAs results in the dysregulation of NK cell development.
miRNAs in NK cell homeostasis and cancer
In addition to acting as regulators of developmental processes, studies in NK cell lymphomas have implicated miRNAs as important repressors of oncogenic transformation during homeostasis. Overexpression of c-Myc, a classic oncogene implicated in the pathogenesis of a wide variety of cancers, was recently shown to repress a number of anti-proliferative miRNAs during tumorigenesis (92). In keeping with a putative tumor suppressor function, a number of miRNAs (e.g. miR-150, miR-342-5p, miR-22*, miR-181a-2*, miR-101, miR-26a, miR-26b, miR-28-5, miR-363, and miR-146) have been found to be downregulated in human NK/T-cell lymphomas (NKTLs) (93–95). Moreover, downregulation of at least one of these, miR-146a, was shown to independently predict poor clinical outcomes in NKTL patients (93). Additional evidence that miRNAs may function as tumor suppressors comes from experimental data showing that ectopic re-expression of many of these down-regulated miRNAs, including miR-146a and miR-150, was sufficient to reduce the growth rate and/or increase the incidence of apoptosis in NK cell lines (93–95). Computational prediction of the mRNAs targeted by these miRNAs revealed an enrichment of genes involved in cell cycle regulation, p53 signaling, and mitogen-activated protein kinase (MAPK) signaling, pathways with critical regulatory roles in cellular transformation. Several of the predicted targets, including STMN1, IRF4, PRDM11, and BCL2, were verified experimentally to be overexpressed in NKTL tumors (94).
Specific examples of miRNAs that regulate oncogenic potential in NK cells include miR-150, miR-146a, and miR-30b. As mentioned above, re-expression of miR-150 in mir-150lo NKTL cell lines can promote apoptosis and reduce cellular proliferation, as well as dampen phosphoinositide-3-kinase-(PI3K)-AKT activation (95), a pathway that has been implicated in NK/T-cell lymphomagenesis (96). These activities were linked to the ability of miR-150 to repress DKC1, a protein involved in telomere maintenance (97), and AKT2, a PI3K-AKT-associated kinase with putative oncogenic functions in human hepatocellular carcinomas and colorectal cancers (98, 99). Re-expression of miR-150 in human NKTL cell lines reduced DKC1 and AKT2 levels, lowered telomerase activity, and increased levels of tumor suppressor proteins such as Bim and p53, thereby promoting cellular senescence and apoptosis, respectively (95). Like miR-150, re-introduction of miR-146a in miR-146lo NKTL cell lines was shown to suppress cellular proliferation and promote apoptosis (93). This was attributed to the ability of miR-146a to repress TRAF6, an activator of NFκB signaling, and thereby reduce expression of the anti-apoptotic factor, BCL-2, which is upregulated downstream of the NFκB pathway (93). And lastly, miR-30b was shown in NKTL cells to directly target the 3′-UTR of BLIMP1 (94), an important lineage-specification factor associated with B-cell lymphomas in patients. Thus, miRNAs are important for promoting cell cycle regulation, cellular senescence, and apoptosis, processes that result in immortalization and aberrant proliferation during miRNA dysregulation.
Although some may act as tumor suppressors, other miRNAs may promote cell proliferation and survival, features that can also be dysregulated in transformed cells to exacerbate oncogenic traits. miR-21 and miR-155, for example, have been shown to be over-expressed in NKTL cells and knockdown of either resulted in increased expression of phosphatase and tensin homolog (PTEN), programmed cell death 4 (PDCD4), and SHIP1, factors that dampen NK cell proliferation and function (96). In addition to promoting the expression of negative regulatory factors, knockdown of miR-21 or miR-155 decreased expression of the pro-proliferative factor, pAKT. Conversely, overexpression of miR-21 or miR-155 had the opposite effect on the expression levels of these genes (96).
Thus, miRNAs regulate processes associated with basal homeostasis, cell turnover, and survival at steady-state (i.e. in the absence of inflammation, infection, or disease), and dysregulated miRNA expression in immune cells, including NK cells, can result in diseases such as cancer (Fig. 2). miRNAs will undoubtedly be discovered to play specific roles in NK cell homeostasis. In addition to promoting transformation of NK cells themselves, miRNA-specific loss of NK cell function could lead to diminished tumor immunosurveillance by this arm of the immune system, resulting in miRNA-unrelated but NK cell-related transformation and disease.
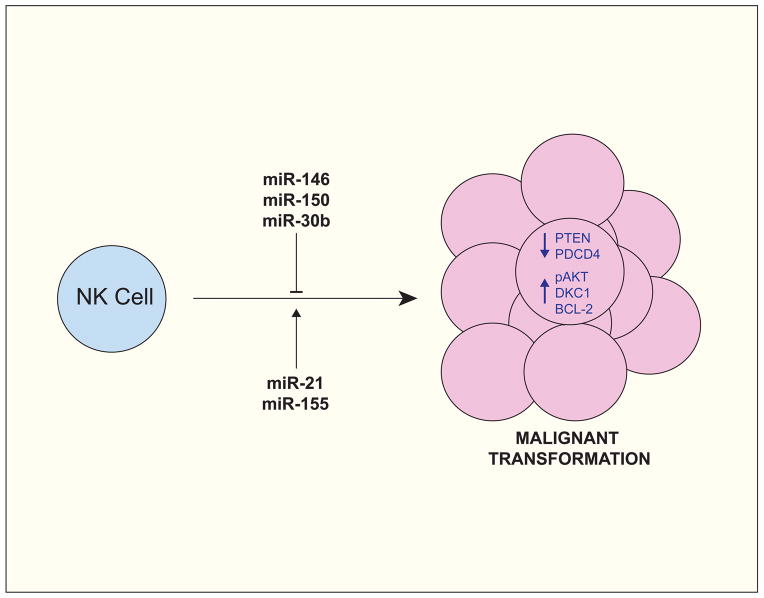
Fig. 2. miRNAs have oncogene and tumor suppressor activity in NK cells.
miR-150 and miR-21 are often over-expressed in Natural Killer/T cell Lymphomas (NKTLs) and have been shown to promote oncogenic transformation by repressing tumor suppressor factors (e.g. PTEN and PDCD4) and by promoting pro-proliferative pathways (e.g. the PI3K-AKT cascade). In contrast, miR-146, miR-150, and miR-30b, which are often under-expressed in NKTLs, have been attributed with tumor suppressor activity, owing in part to their ability to repress pro-proliferative and pro-survival factors, such as AKT2, DKC1, and, indirectly, BCL-2.
miRNAs in NK cell effector responses
Evidence that miRNAs regulate not only NK cell development but also effector function is rooted in early studies evaluating the effects of disrupting Dicer or Dcgr8 in mouse NK cells. Although these studies reported conflicting details of exactly how miRNAs regulate NK cell function, they nevertheless agree that global loss of miRNAs has a significant impact on NK cell activation and effector function. Bezman et al. (72) showed that tamoxifen-induced global deletion of either Dicer1 or Dcgr8 in adult mice impaired the ability of NK cells to degranulate or produce IFN-γ in response to activation through activating receptors, including NK1.1, NKp46, and Ly49H. Interestingly, this defect was not observed when the cells were stimulated with the proinflammatory cytokines, IL-12 and IL-18 (72). In addition to cytokine production, miRNA deficiency in these animals also impaired the ability of antigen-specific Ly49H+ NK cells to expand in response to MCMV challenge in vivo. This defect was attributed to a requirement for miRNAs in the survival of activated NK cells (72). Contradictory results were obtained in an independent study by Sullivan et al. (75), where it was shown that deletion of Dicer1 in developing NK cells, using human CD2 promoter-controlled Cre expression, actually enhanced IFN-γ production and degranulation by NK cells following activation via pro-inflammatory cytokines, stimulation through the NK1.1 receptor, or incubation with YAC-1 target cells, an NK-sensitive tumor cell line. Similarly, Dicer-deficient NK cells in this study produced significantly higher levels of IFN-γ in response to MCMV infection in vivo. The conflicting results of these two studies are likely due to differences in the systems used to delete Dicer in NK cells. In contrast to miRNA ablation, increasing global levels of miRNAs through deletion of Eri1 (a negative regulator of miRNAs) had no impact on NK cell degranulation, IFN-γ production, or cytotoxicity against target cells in vitro (76). However, Eri1−/− NK cells were limited in their ability in proliferate in response MCMV infection in vivo, and this defect correlated with a substantial increase in viral titers in Eri1−/− mice. Collectively, the studies in Dicer-, Dcgr8-, and Eri1-deficient animals highlight a critical role for miRNAs in regulating NK cell activation and effector function.
Beyond global perturbation of miRNA levels, studies on individual miRNAs have further refined our understanding of how specific miRNAs regulate NK cell activation. Several groups have identified unique miRNAs that regulate IFN-γ production in resting and activated NK cells. For example, either cytokine-induced or transgene-mediated upregulation of miR-155 enhanced IFN-γ production by activated human NK cells, whereas miR-155 knockdown impaired this activity (73, 100). Mechanistically, miR-155 promoted NK cell responses by targeting and suppressing SHIP-1, a potent negative regulator of NK cell effector function. Similar defects in IFN-γ production were observed in NK cells from mice lacking Bic, the gene encoding miR-155, suggesting an evolutionarily conserved role for this miRNA (100). Like miR-155, miR-150 is also required for optimal IFN-γ production by mouse NK cells. Compared to wildtype NK cells, fewer miR150−/− NK cells produce IFN-γ after stimulation in vitro, whereas transgenic overexpression of miR-150 in mice was shown to enhance IFN-γ production by activated NK cells (81).
Beyond IFN-γ production, miRNAs also regulate other NK cell effector responses, including cytotoxicity and the secretion of other proinflammatory cytokines, such as TNFα. In human NK cells, miR-30c-1* was recently shown to act downstream of the activating NK cell receptor, CD226, to promote cytotoxic functionality and TNF-α production (101). This function was attributed to the ability of miR-30c-1* to target HMBOX1, a transcription factor shown to inhibit NK cell activation.
Rather than promote effector responses, some miRNAs may function to repress the translation of mRNA for effector molecules, such as granzyme B and IFNγ, which are present at high levels in resting NK cells but remain untranslated in the absence of activation stimuli. For example, mir-29 and members of the miR-15/16 family (i.e. miR-15a/15b/16) restrict IFN-γ production by NK cells by directly targeting the IFN-γ 3′-UTR (75, 102). Consistent with the presumed role of these miRNAs in restraining NK cell function until activating signals are received, stimulation of mouse NK cells in vitro led to a reduction in miR-29 or miR-15/16 expression and a concomitant increase in IFN-γ production. Ma et al. (102) further showed that NK cells in mice expressing a miR-29 ‘sponge’ construct (which competes with endogenous miR-29 targets and reduces the amount of ‘free’ miR-29) produced exaggerated levels of IFN-γ following activation in vitro or infection with Listeria monocytogenes in vivo. This latter response correlated with enhanced clearance of bacterial burden, indicating that miR-29 may be manipulated during infection to suppress protective immune responses. miRNAs also regulate untimely cytotoxic activity in resting NK cells. miR-223, for example, directly targets and represses the 3′-UTR of granzyme B at steady state, whereas stimulation with IL-15 causes a decrease in miR-223 expression and a corresponding increase in granzyme B levels (74). Comparable roles for miR-27a*, miR-30e, and miR-378 have also been described. miR-27a* was found to directly target the 3′-UTR of perforin and granzyme B transcripts in human NK cells and miR-27a* knockdown substantially increased NK cell lysis of target cells in vitro (103). Similarly, miR-378 and miR-30e, two miRNAs abundantly expressed in resting human NK cells, were found to target granzyme B and perforin transcripts, respectively (104). Like miR-223, cytokine stimulation reduced miR-378 and miR-30e expression and thereby augmented activation-induced cytotoxicity by human NK cells.
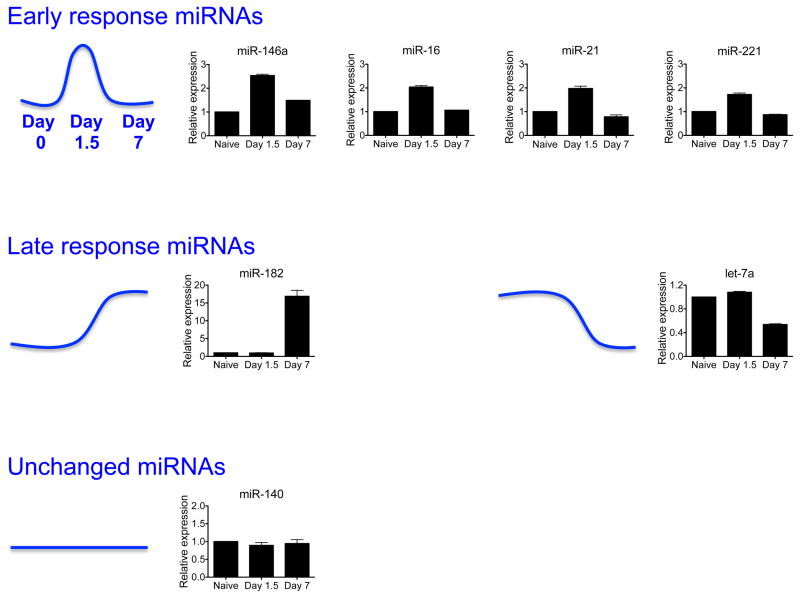
To further understand how miRNAs might regulate effector responses in antigen-specific NK cells responding to viral infection, we sorted Ly49H+ NK cells from MCMV-infected animals on days 0, 1.5, and 7 post-infection and used quantitative real-time PCR (qRT-PCR) to assess expression of 7 miRNAs with known or suspected roles in NK cell biology (miR-146a, miR-16, miR-21, miR-221, miR-182, let-7a, and miR-140), most of which have been discussed above. The observed expression patterns could be broadly grouped into three categories (Fig. 3): (i) miRNAs transiently upregulated only at day 1.5 post-infection, or early response miRNAs (e.g. miR-146a, miR-16, miR-21, and miR-221); (ii) miRNAs upregulated (e.g. miR-182) or downregulated (e.g. let-7a) only on day 7 post-infection, or late response miRNAs; and (iii) miRNAs whose expression remained unchanged at the assessed time points. Early response miRNAs would be predicted to act downstream of signals from proinflammatory cytokines, which are particularly abundant in the first 24–72 h post-infection (27). As such, these miRNAs might be predicted to regulate ‘innate’ NK cell functions, such as IFN-γ secretion and cytotoxicity, or shape the clonal proliferation of antigen-specific Ly49H+ NK cells that is initiated during this time frame. In keeping with this hypothesis, roles for miR-16 in controlling IFN-γ production (75) and for miR-21 in promoting proliferation (96) in NK cells have already been discussed. Future studies will be necessary to more fully understand how and if the other early response miRNAs identified in our study regulate NK cell function.
Fig. 3. miRNA profile of NK cells during MCMV infection.
Expression of select miRNAs in splenic Ly49H+ NK cells sorted from animals infected with Smith strain MCMV, as assessed by qRT-PCR. Levels of mature miRNAs were normalized to sno-202. Data are shown as the fold change at day 1.5 and day 7 versus day 0 (naïve). miRNAs transiently upregulated on day 1.5 post-infection (i.e. Early response miRNAs) include miR-146a, miR-16, miR-21, and miR-221. miRNAs upregulated or downregulated on day 7 post-infection (i.e. Late response miRNAs) include miR-182 and let-7a. miR-140 levels remained constant on the assessed time points.
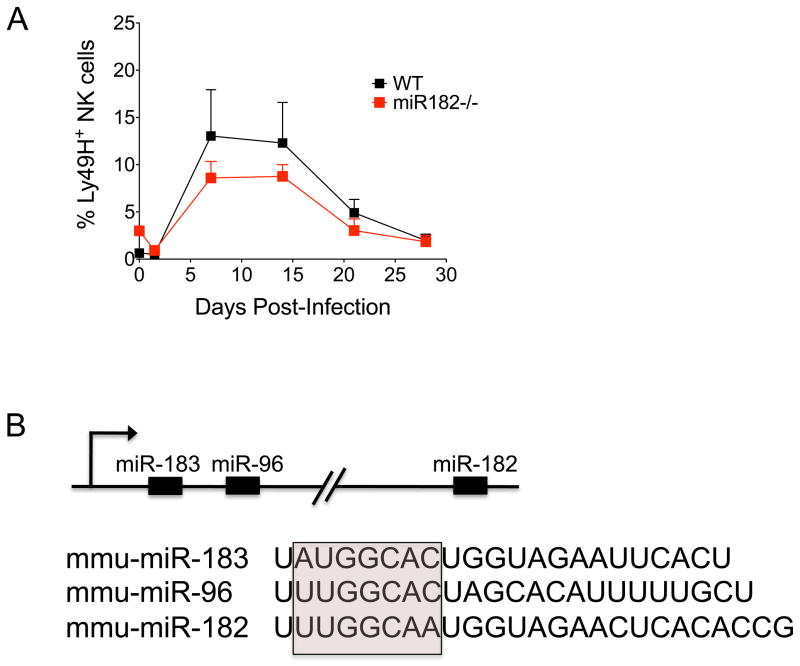
Given the delayed expression of late response miRNAs, it is less likely that they regulate cytotoxicity or IFN-γ production. Instead, these miRNAs may be important instructors of the antigen-driven clonal expansion (or contraction) of Ly49H+ NK cells, which peaks at day 7 post-infection, then slowly declines over the following weeks until a small pool of long-lasting ‘memory’ NK cells remains. One of these miRNAs, miR-182, has been shown to act downstream of the IL-2 receptor (IL-2R) in T-helper cells to promote clonal expansion by regulating Foxo1 transcripts (105). Given that NK cells also require IL-2R signaling for their development and maturation, we used miR-182−/− mice to investigate the extent to which miR-182 was necessary for NK cell expansion during MCMV infection. Normal NK cell numbers were found in miR-182−/− mice (data not shown), indicating that miR-182 was not required for NK cell maturation or homeostasis at steady state (i.e. in the absence of inflammation). miR-182−/− NK cells also exhibited normal expression of markers associated with maturation and activation, including CD27, CD11b, KLRG1, and CD69 expression (data not shown), further underscoring unperturbed NK cell development in these animals. To address the role of miR-182 in NK cell expansion during infection, we co-transferred equal numbers of wildtype (WT) and miR-182−/− Ly49H+ NK cells into Ly49H-deficient hosts and evaluated their ability to expand following MCMV challenge. We observed a similar expansion of WT and miR-182−/− Ly49H+ NK cells at day 7 and equal percentages of memory NK cells at day 28 post-infection (Fig. 4A). The absence of a defect in NK cell expansion and memory formation in miR-182−/− animals may be due to functional compensation by other miRNAs. Indeed, other miR-183 ~ miR-182 family members, which along with miR-182 are all expressed from a single pri-miR transcript, all share a nearly identical seed sequence (Fig. 4B), indicating that these miRNAs may target an overlapping suite of mRNAs. Additional studies would be necessary to address whether deletion of the whole miR-183~182 cluster would impact NK cell responses during MCMV infection. Alternatively, upregulation of miR-182 on day 7 post-infection may be an inconsequential after-effect of the extensive changes in gene expression that occur during MCMV infection. However, the observation that the expression of miR-183 ~ 182 cluster miRNAs was also significantly elevated in primary human fibroblasts infected with human CMV suggests that there might be a common mechanism connecting miR-183 ~ 182 family members and CMV (106).
Fig. 4. miR-182 is dispensable for NK cell responses to MCMV infection.
(A) Equal numbers of WT (CD45.1+) and miR-182−/− (CD45.2+) Ly49H+ NK cells were co-transferred into Ly49H-deficient hosts. Following infection with MCMV, the percentage of adoptively transferred WT (black) and miR-182−/−(red) Ly49H+ NK cells within the total NK cell population in blood was assessed at the indicated time points post-infection. Error bars show SEM (n = 3). Data are representative of three independent experiments. (B) Schematic representation of the miR-183 ~ 182 cluster. miR-183, miR-96, and miR-182 share sequence homology, particularly within the seed sequence used to bind target mRNAs (pink box).
In summary, an increasing number of studies indicate that specific miRNAs can regulate both early and late responses of NK cells against viral infection, and disruption of many of these individual miRNAs can result in susceptibility to infection. Future studies, in particular those using conditional ablation of individual miRNAs in genetically engineered mice (107), will undoubtedly identify additional miRNAs with key regulatory roles in NK cell responses to viral infection.
miRNAs as common regulators of NK and CD8+ T-cell programs
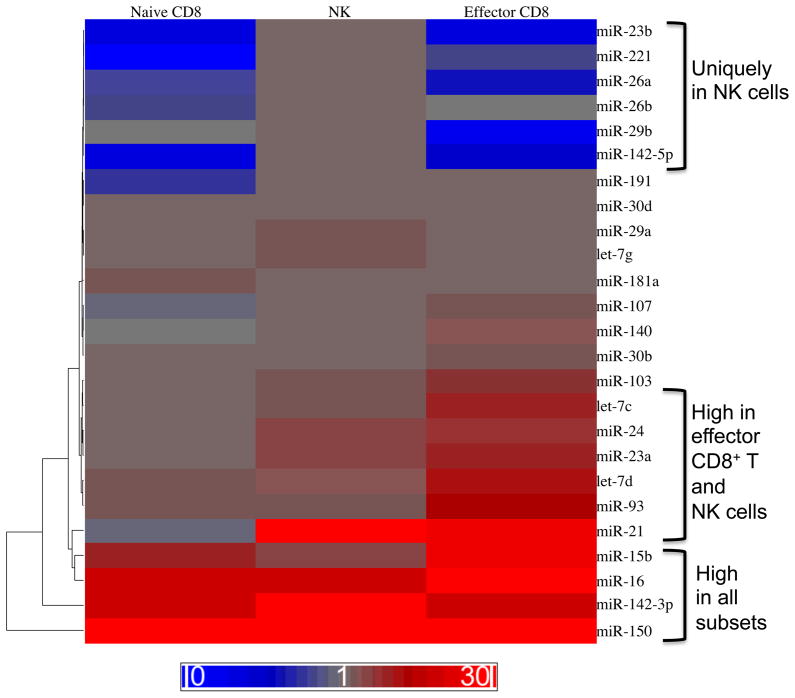
NK cells have long been appreciated to share many common features with CD8+ T cells, most notably their shared ability to kill infected or malignant target cells through direct cytotoxic mechanisms. Developmental parallels also exist between NK and CD8+ T cells, including shared origin from CLP cells, a reliance on common γ-chain-dependent cytokines (e.g. IL-15 and IL-7) for survival during development and homeostasis, and analogous education processes during development (i.e. ‘licensing’ or ‘disarming’ for NK cells and thymic selection for T cells) (34). Activation is similarly dependent on engagement of activating receptors (i.e. the TCR for CD8+ T cells and receptors such as NKG2D or Ly49H for NK cells) and pro-inflammatory cytokines (e.g. IL-12, IL-18, and Type I interferons). Moreover, it is now appreciated that antigen-specific engagement of certain activating receptors on NK cells can drive a strong proliferative response and the generation of long-lived ‘memory’ cells with enhanced functionality in response to antigen re-exposure. The kinetics and nature of this response is highly reminiscent of antigen-specific CD8+ T-cell responses against pathogens (34). Given these parallels, specific miRNAs may function in both cell types to regulate common pathways of development, homeostasis, and effector function. Consistent with this hypothesis, both NK cells and naïve CD8+ T cells are reduced in frequency and number in Dicer- and Dgcr8-deficient mice (72, 75, 108, 109). Moreover, a survey of published miRNA profiling studies reveals substantial overlap in the specific miRNAs expressed by NK cells and CD8+ T cells. Examples include miR-142-3p, mi-R150, miR-16, miR-23a, miR-15b, miR-29a, miR-30b, miR-146, and miR-26a (72–74, 110, 111). To more precisely compare miRNAs expressed by naive NK cells and naive and effector CD8+ T cells, we used qRT-PCR to evaluate the relative expression of 35 unique miRNAs in the three cell populations. Several broad patterns of expression were especially notable: (i) miRNAs highly expressed in all three cell subsets, (ii) miRNAs with moderate-to-high expression in NK cells and effector CD8+ T cells, and (iii) miRNAs uniquely upregulated in NK cells as compared to either CD8+ T-cell population (Fig. 5).
Fig. 5. Common miRNA expression programs between NK and CD8+ T-cell populations.
miRNAs expressed in resting NK cells as compared to resting (naive) CD8+ T cells or effector CD8+ T cells. NK1.1+ TCRβ− NK cells were sorted from C57BL/6 mice. Naive CD8+ T cells were sorted from CD45.1+CD45.2+ P14 mice bearing the DbGP33-specific TCR. Effector CD8+ T cells were sorted from chimeric mice generated by adoptive transfer of P14 T cells into C57BL/6 (CD45.2+) recipients that were infected intraperitoneally with 2 × 105 plaque forming units (PFU) of LCMV Armstrong. Levels of mature miRNAs were measured by qRT-PCR and normalized to sno202. Fold expression relative to sno-202 is depicted as a heat map. Three patterns
Among those highly expressed in all three subsets (e.g. miR-150, miR-142-3p, miR-16, miR-15b, miR-93, and let-7d), several have described roles in lymphocyte biology. For example, miR-150 promotes NK development (as described above) (81) and, at least in the context of overexpression, represses invariant natural killer T (iNKT) (81) and B-cell development (79). Similarly, miR-16 and miR-15b, which repress IFN-γ transcript in NK cells (75), have also been identified as tumor suppressor factors in B-cell leukemias, where they repress expression of oncogenes such as BCL2, MCL1, CCND1, and WNT3a (112). Although no role has yet to be described for miR-142-3p in NK or CD8+ T-cells, it is known to regulate the cyclic AMP (cAMP) pathway in CD4+ T cells (113). Future work will provide insight into whether these miRNAs indeed have overlapping roles in CD8+ T-cells and NK cells, perhaps as regulators of development or homeostasis.
In terms of the miRNAs highly expressed only by effector CD8+ T cells and NK cells, including miR-21, miR-23a, miR-24, let-7c, and miR-103, it is tempting to speculate that these may function to control effector functions common to both cell types, such as IFN-γ secretion and cytotoxicity. Studies in mice have hinted at such a role for miR-21, which is often upregulated in B and T cells from human systemic lupus erythematosus patients and from genetically defective mouse strains with lupus-like disease symptoms (114–116). Indeed, silencing of miR-21 in ‘lupus-prone’ mice was shown to reverse symptoms of autoimmunity, including splenomegaly and aberrant PDCD4 expression and disease-associated alteration in the CD4+/CD8+ T-cell ratio, suggesting that miR-21 may function as an important regulator of autoimmune lymphocyte responses in lupus (114). Altogether, the evidence strongly suggests that parallel pathways of regulation are at work in NK and CD8+ T cells, where individual miRNA expression is modulated to enhance or repress specific cellular processes and functions.
Conclusions and future directions
Mounting evidence underscores the critical role of miRNAs as regulators of development, maturation, proliferation, differentiation, and activation of NK cells (Fig. 6). Within the context of complex transcriptional networks, the control of gene expression has been historically attributed to transcription factors. However, a growing number of studies have shown that miRNAs are equally critical regulators of cellular mRNA and protein levels during both homeostasis and disease. Whereas transcription factors function in many respects as ‘on-off’ switches of gene expression, miRNAs act more like rheostats, finely tuning the level of translatable mRNAs in response to specific developmental or stimulatory cues and changing cellular requirements. This rapid and reversible mechanism of action makes them well-suited to control NK and T-cell effector responses, such as cytokine production and cytotoxicity, which must be rapidly mounted in the face of viral or malignant assaults but then rapidly disengaged with equal potency to prevent pathogenic destruction of healthy, bystander cells.
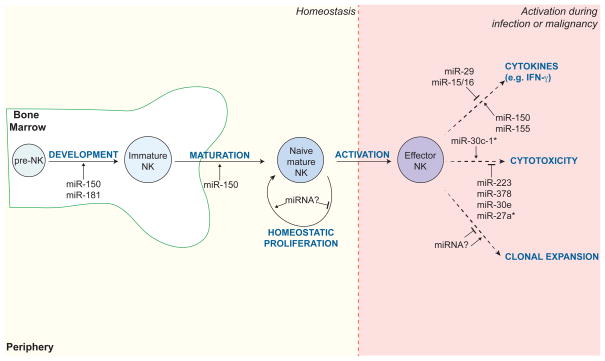
Fig. 6. miRNAs regulate the development, maturation, and effector function of NK cells.
A summary of individual miRNAs known to regulate NK cell development and function at steady state and in the context of activation (e.g. during infection or malignancy). Examples include miR-150 and miR-181, which are required for NK cell development and maturation in the bone marrow; miR-150, miR-155, miR-29, and miR-15/16, which modulate cytokine production by activated NK cells; and miR-30c-1*, miR-223, miR-378, miR-30e, miR-27a*, which control expression of cytotoxic molecules, such as granzyme B and perforin.
Future studies are required to appreciate the full spectrum of NK cell activities controlled by miRNAs, as well as to understand the activating and developmental signals that orchestrate their expression during homeostasis, infection, and cancer. High throughput and genome-wide technologies that match individual miRNAs to their cognate target transcripts, such as Ago-CLIP (117, 118), will be particularly useful in dissecting the specific cellular pathways controlled by individual miRNAs in NK cells in vivo.
Acknowledgments
A.M.B. is supported by a NIH T32 award (CA009149). J.C.S. is supported by the Searle Scholars Program, the Cancer Research Institute, and grants from the NIH (AI085034 and AI100874). L.L.L. is an American Cancer Society Professor and funded by NIH grant AI068129. An American Cancer Society Postdoctoral Fellow grant supported N.A.B. We thank members of the Sun and Lanier labs for their insightful discussions and suggestions.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Orange JS. Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect. 2002;4:1545–1558. doi: 10.1016/s1286-4579(02)00038-2. [DOI] [PubMed] [Google Scholar]
- 2.Orange JS, et al. Deficient natural killer cell cytotoxicity in patients with IKK-gamma/NEMO mutations. J Clin Invest. 2002;109:1501–1509. doi: 10.1172/JCI14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nandakumar S, Woolard SN, Yuan D, Rouse BT, Kumaraguru U. Natural killer cells as novel helpers in anti-herpes simplex virus immune response. J Virol. 2008;82:10820–10831. doi: 10.1128/JVI.00365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shanley JD. In vivo administration of monoclonal antibody to the NK 1. 1 antigen of natural killer cells: effect on acute murine cytomegalovirus infection. J Med Virol. 1990;30:58–60. doi: 10.1002/jmv.1890300113. [DOI] [PubMed] [Google Scholar]
- 5.Ghiasi H, Cai S, Perng GC, Nesburn AB, Wechsler SL. The role of natural killer cells in protection of mice against death and corneal scarring following ocular HSV-1 infection. Antiviral Res. 2000;45:33–45. doi: 10.1016/s0166-3542(99)00075-3. [DOI] [PubMed] [Google Scholar]
- 6.Scalzo AA, et al. The effect of the Cmv-1 resistance gene, which is linked to the natural killer cell gene complex, is mediated by natural killer cells. J Immunol. 1992;149:581–589. [PubMed] [Google Scholar]
- 7.Ruggeri L, et al. Innate immunity against hematological malignancies. Cytotherapy. 2002;4:343–346. doi: 10.1080/146532402760271127. [DOI] [PubMed] [Google Scholar]
- 8.Ruggeri L, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 9.Street SE, et al. Innate immune surveillance of spontaneous B cell lymphomas by natural killer cells and gammadelta T cells. J Exp Med. 2004;199:879–884. doi: 10.1084/jem.20031981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Iizuka K, Aguila HL, Weissman IL, Yokoyama WM. In vivo natural killer cell activities revealed by natural killer cell-deficient mice. Proc Natl Acad Sci USA. 2000;97:2731–2736. doi: 10.1073/pnas.050588297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruggeri L, Mancusi A, Capanni M, Martelli MF, Velardi A. Exploitation of alloreactive NK cells in adoptive immunotherapy of cancer. Curr Opin Immunol. 2005;17:211–217. doi: 10.1016/j.coi.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Ruggeri L, Mancusi A, Perruccio K, Burchielli E, Martelli MF, Velardi A. Natural killer cell alloreactivity for leukemia therapy. J Immunother. 2005;28:175–182. doi: 10.1097/01.cji.0000161395.88959.1f. [DOI] [PubMed] [Google Scholar]
- 13.Hsu KC, Dupont B. Natural killer cell receptors: regulating innate immune responses to hematologic malignancy. Semin Hematol. 2005;42:91–103. doi: 10.1053/j.seminhematol.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Hsu KC, et al. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood. 2005;105:4878–4884. doi: 10.1182/blood-2004-12-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pegram HJ, et al. Alloreactive natural killer cells in hematopoietic stem cell transplantation. Leukemia Res. 2011;35:14–21. doi: 10.1016/j.leukres.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 16.Gillgrass A, Ashkar A. Stimulating natural killer cells to protect against cancer: recent developments. Exp Rev Clin Immunol. 2011;7:367–382. doi: 10.1586/eci.10.102. [DOI] [PubMed] [Google Scholar]
- 17.Venstrom JM, et al. KIR and HLA genotypes are associated with disease progression and survival following autologous hematopoietic stem cell transplantation for high-risk neuroblastoma. Clin Cancer Res. 2009;15:7330–7334. doi: 10.1158/1078-0432.CCR-09-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giebel S, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003;102:814–819. doi: 10.1182/blood-2003-01-0091. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Martinez A, et al. KIR-HLA receptor-ligand mismatch associated with a graft-versus-tumor effect in haploidentical stem cell transplantation for pediatric metastatic solid tumors. Ped Blood Cancer. 2009;53:120–124. doi: 10.1002/pbc.21955. [DOI] [PubMed] [Google Scholar]
- 20.Vidal SM, Khakoo SI, Biron CA. Natural killer cell responses during viral infections: flexibility and conditioning of innate immunity by experience. Curr Opin Virol. 2011;1:497–512. doi: 10.1016/j.coviro.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pyzik M, et al. Distinct MHC class I-dependent NK cell-activating receptors control cytomegalovirus infection in different mouse strains. J Exp Med. 2011;208:1105–1117. doi: 10.1084/jem.20101831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 23.Bubic I, et al. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J Virol. 2004;78:7536–7544. doi: 10.1128/JVI.78.14.7536-7544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 25.Karlhofer FM, Ribaudo RK, Yokoyama WM. The interaction of Ly-49 with H-2Dd globally inactivates natural killer cell cytolytic activity. Trans Assoc Am Phys. 1992;105:72–85. [PubMed] [Google Scholar]
- 26.Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell. 2010;142:847–856. doi: 10.1016/j.cell.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orange JS, Biron CA. Characterization of early IL-12, IFN-alphabeta, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J Immunol. 1996;156:4746–4756. [PubMed] [Google Scholar]
- 28.Orange JS, Biron CA. An absolute and restricted requirement for IL-12 in natural killer cell IFN-gamma production and antiviral defense. Studies of natural killer and T cell responses in contrasting viral infections. J Immunol. 1996;156:1138–1142. [PubMed] [Google Scholar]
- 29.Nguyen KB, et al. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J Immunol. 2002;169:4279–4287. doi: 10.4049/jimmunol.169.8.4279. [DOI] [PubMed] [Google Scholar]
- 30.Sun JC, Madera S, Bezman NA, Beilke JN, Kaplan MH, Lanier LL. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J Exp Med. 2012;209:947–954. doi: 10.1084/jem.20111760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romee R, et al. Cytokine activation induces human memory-like NK cells. Blood. 2012;120:4751–4760. doi: 10.1182/blood-2012-04-419283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young HA, Ortaldo J. Cytokines as critical co-stimulatory molecules in modulating the immune response of natural killer cells. Cell Res. 2006;16:20–24. doi: 10.1038/sj.cr.7310004. [DOI] [PubMed] [Google Scholar]
- 33.Ortaldo JR, et al. Regulation of ITAM-positive receptors: role of IL-12 and IL-18. Blood. 2006;107:1468–1475. doi: 10.1182/blood-2005-04-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat Rev Immunol. 2011;11:645–657. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malhotra A, Shanker A. NK cells: immune cross-talk and therapeutic implications. Immunotherapy. 2011;3:1143–1166. doi: 10.2217/imt.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcenaro E, Carlomagno S, Pesce S, Moretta A, Sivori S. NK/DC crosstalk in anti-viral response. Adv Exp Med Biol. 2012;946:295–308. doi: 10.1007/978-1-4614-0106-3_17. [DOI] [PubMed] [Google Scholar]
- 37.Sun JC, Lopez-Verges S, Kim CC, DeRisi JL, Lanier LL. NK cells and immune “memory”. J Immunol. 2011;186:1891–1897. doi: 10.4049/jimmunol.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paust S, von Andrian UH. Natural killer cell memory. Nat Immunol. 2011;12:500–508. doi: 10.1038/ni.2032. [DOI] [PubMed] [Google Scholar]
- 39.Paust S, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paust S, Senman B, von Andrian UH. Adaptive immune responses mediated by natural killer cells. Immunol Rev. 2010;235:286–296. doi: 10.1111/j.0105-2896.2010.00906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown MG, et al. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 42.Lee SH, et al. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat Genet. 2001;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- 43.Smith HR, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci USA. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 45.Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM. Specific and nonspecific NK cell activation during virus infection. Nat Immunol. 2001;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- 46.Lopez-Verges S, et al. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci USA. 2011;108:14725–14732. doi: 10.1073/pnas.1110900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 48.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 49.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 50.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 51.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 52.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 53.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr Topics Microbiol Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- 55.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 56.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 57.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 58.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 59.Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA discovery. nat. Genet. 2006;38 (Suppl):S2–7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 60.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 61.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev. 2006;214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 65.Freud AG, et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med. 2006;203:1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 67.Di Santo JP, Vosshenrich CA. Bone marrow versus thymic pathways of natural killer cell development. Immunol Rev. 2006;214:35–46. doi: 10.1111/j.1600-065X.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- 68.Kennedy MK, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lodolce JP, et al. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9:669–676. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 70.Ma A, Koka R, Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 71.Vosshenrich CA, et al. Roles for common cytokine receptor gamma-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J Immunol. 2005;174:1213–1221. doi: 10.4049/jimmunol.174.3.1213. [DOI] [PubMed] [Google Scholar]
- 72.Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DG, Lanier LL. Distinct requirements of microRNAs in NK cell activation, survival, and function. J Immunol. 2010;185:3835–3846. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu X, et al. Identification of microRNA transcriptome involved in human natural killer cell activation. Immunol Lett. 2012;143:208–217. doi: 10.1016/j.imlet.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 74.Fehniger TA, et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010;20:1590–1604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sullivan RP, et al. MicroRNA-deficient NK cells exhibit decreased survival but enhanced function. J Immunol. 2012;188:3019–3030. doi: 10.4049/jimmunol.1102294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thomas MF, et al. Eri1 regulates microRNA homeostasis and mouse lymphocyte development and antiviral function. Blood. 2012;120:130–142. doi: 10.1182/blood-2011-11-394072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghisi M, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood. 2011;117:7053–7062. doi: 10.1182/blood-2010-12-326629. [DOI] [PubMed] [Google Scholar]
- 78.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 79.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Almanza G, Fernandez A, Volinia S, Cortez-Gonzalez X, Croce CM, Zanetti M. Selected microRNAs define cell fate determination of murine central memory CD8 T cells. PloS One. 2010;5:e11243. doi: 10.1371/journal.pone.0011243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bezman NA, Chakraborty T, Bender T, Lanier LL. miR-150 regulates the development of NK and iNKT cells. J Exp Med. 2011;208:2717–2731. doi: 10.1084/jem.20111386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zobel A, Kalkbrenner F, Guehmann S, Nawrath M, Vorbrueggen G, Moelling K. Interaction of the v-and c-Myb proteins with regulatory sequences of the human c-myc gene. Oncogene. 1991;6:1397–1407. [PubMed] [Google Scholar]
- 83.Nakagoshi H, Kanei-Ishii C, Sawazaki T, Mizuguchi G, Ishii S. Transcriptional activation of the c-myc gene by the c-myb and B-myb gene products. Oncogene. 1992;7:1233–1240. [PubMed] [Google Scholar]
- 84.Heckman CA, Mehew JW, Ying GG, Introna M, Golay J, Boxer LM. A-Myb up-regulates Bcl-2 through a Cdx binding site in t(14;18) lymphoma cells. J Biol Chem. 2000;275:6499–6508. doi: 10.1074/jbc.275.9.6499. [DOI] [PubMed] [Google Scholar]
- 85.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 86.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 87.Cichocki F, et al. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187:6171–6175. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bachanova V, et al. Activated notch supports development of cytokine producing NK cells which are hyporesponsive and fail to acquire NK cell effector functions. Biol Blood Marrow Tran. 2009;15:183–194. doi: 10.1016/j.bbmt.2008.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lehar SM, Dooley J, Farr AG, Bevan MJ. Notch ligands Delta 1 and Jagged1 transmit distinct signals to T-cell precursors. Blood. 2005;105:1440–1447. doi: 10.1182/blood-2004-08-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bezman NA, et al. Molecular definition of the identity and activation of natural killer cells. Nat Immunol. 2012;13:1000–1009. doi: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heng TS, Painter MW Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 92.Chang TC, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paik JH, et al. MicroRNA-146a downregulates NFkappaB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin Cancer Res. 2011;17:4761–4771. doi: 10.1158/1078-0432.CCR-11-0494. [DOI] [PubMed] [Google Scholar]
- 94.Ng SB, et al. Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood. 2011;118:4919–4929. doi: 10.1182/blood-2011-07-364224. [DOI] [PubMed] [Google Scholar]
- 95.Watanabe A, et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25:1324–1334. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- 96.Yamanaka Y, et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114:3265–3275. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 97.Batista LF, et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature. 2011;474:399–402. doi: 10.1038/nature10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roy HK, et al. AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis. 2002;23:201–205. doi: 10.1093/carcin/23.1.201. [DOI] [PubMed] [Google Scholar]
- 99.Xu X, et al. Akt2 expression correlates with prognosis of human hepatocellular carcinoma. Oncology Rep. 2004;11:25–32. [PubMed] [Google Scholar]
- 100.Trotta R, et al. miR-155 regulates IFN-gamma production in natural killer cells. Blood. 2012;119:3478–3485. doi: 10.1182/blood-2011-12-398099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gong J, et al. miR-30c-1* promotes natural killer cell cytotoxicity against human hepatoma cells by targeting the transcription factor HMBOX1. Cancer Sci. 2012;103:645–652. doi: 10.1111/j.1349-7006.2012.02207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ma F, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. nat. Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 103.Kim TD, et al. Human microRNA-27a* targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood. 2011;118:5476–5486. doi: 10.1182/blood-2011-04-347526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang P, et al. Identification of resting and type I IFN-activated human NK cell miRNomes reveals microRNA-378 and microRNA-30e as negative regulators of NK cell cytotoxicity. J Immunol. 2012;189:211–221. doi: 10.4049/jimmunol.1200609. [DOI] [PubMed] [Google Scholar]
- 105.Stittrich AB, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 106.Stark TJ, Arnold JD, Spector DH, Yeo GW. High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J Virol. 2012;86:226–235. doi: 10.1128/JVI.05903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park CY, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cobb BS, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu H, et al. miRNA profiling of naive, effector and memory CD8 T cells. PloS One. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 113.Huang B, et al. miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 2009;10:180–185. doi: 10.1038/embor.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garchow BG, et al. Silencing of microRNA-21 in vivo ameliorates autoimmune splenomegaly in lupus mice. EMBO Mol Med. 2011;3:605–615. doi: 10.1002/emmm.201100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pan W, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 116.Te JL, et al. Identification of unique microRNA signature associated with lupus nephritis. PloS One. 2010;5:e10344. doi: 10.1371/journal.pone.0010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]