Abstract
Stringent regulation of biochemical signalling pathways involves feedback and feedforward loops, which underlie robust cellular responses to external stimuli. Regulation occurs in all horizontal layers of signalling networks, primarily by proteins that mediate internalization of receptor–ligand complexes, dephosphorylation of kinases and their substrates, as well as transcriptional repression. Recent studies have unveiled the role of miRNAs (microRNAs), post-transcriptional regulators that control mRNA stability, as key modulators of signal propagation. By acting as genetic switches or fine-tuners, miRNAs can directly and multiply regulate cellular outcomes in response to diverse extracellular signals. Conversely, signalling networks temporally control stability, biogenesis and abundance of miRNAs, by regulating layers of the miRNA biogenesis pathway. In the present mini-review, we use a set of examples to illustrate the extensive interdependence between miRNAs and signalling networks.
Keywords: cancer, growth factor, microRNA (miRNA), network, tyrosine kinase
Introduction
Growth factors are compact polypeptides that transmit fate-determining signals to target cells expressing the cognate receptors. The latter are transmembrane glycoproteins whose cytoplasmic portions harbour protein kinase activity [1]. Stimulation of such receptor kinases leads to downstream activation of instructive signalling through cascades of protein kinases and other biochemical pathways, which culminate in phenotype-encoding genetic programmes. Years of studies of the biochemical mechanisms underlying linear signal transmission have not been able to reconcile a major difficulty: how can a cell transform the gradual changes in the concentration of different ligand growth factors, using only a handful of signalling pathways, into discrete (sometimes binary) outputs? Initial answers indicate that intrinsic hard-wired regulatory mechanisms shape the linear inputs into a non-linear output [2]. FFLs (feedback and feedforward loops) determine the duration, amplitude and frequency of a signal, and bring about signal amplification, binary switching or oscillations that may predominate critical alterations of the cellular steady state. In line with this emerging concept, attempts are being made by systems biologists to decipher structural and functional principles of signalling networks by integrating dynamic interactions of forward driving, as well as their regulatory elements, into functional largely autonomous modules [3]. Moreover, because feedback regulatory elements control signal magnitude, deviations from tight regulation is often manifested in pathological states, such as hyperproliferative diseases, including atherosclerosis and cancer.
In the milieu of cellular components able to exert feedback regulation, miRNAs (microRNAs) emerge as important regulatory elements common to diverse signalling networks. miRNAs are negative regulators of gene expression, capable of exerting pronounced influences upon the translation and stability of mRNAs [4]. Approximately 1000 human miRNAs have been identified to date [5]. It is estimated that each miRNA targets several hundreds of distinct mRNAs molecules [6], leading to the speculation that the majority of human mRNAs are subject to regulation by miRNAs. miRNAs are distinguished by their size of ~22 nt, and unique biogenesis pathway, a stepwise enzymatic cleavage of RNA precursors by RNase III endonucleases [7], which generates the final duplex, and incorporates it into the RISC (RNA-induced silencing complex) particle [8]. Once their maturation is complete, miRNA molecules become competent to target mRNAs for decay or for translational arrest [9,10]. Targeting of an mRNA by an miRNA is mediated by base-pairing between nucleotides 2–8 of the miRNA (numbered from the miRNA’s 5′-end; referred to as the ‘seed sequences’) and a target element in the transcript’s 3′-UTR (untranslated region) [11].
Despite great advances, the miRNA field remains largely uncharted with regard to the physiological function of these small molecules; uncovering the true function of individual miRNA molecules remains a formidable challenge. First, miRNAs are frequently encoded by families of genes with shared seed sequences [12], which complicate genetic analyses. Secondly, each miRNA binds with multiple putative targets that have disparate functions [13]. As in the case of transcription factor-binding sites, predictions based solely on seed sequence compatibility of an miRNA may not be able to establish a priori which transcript is most meaningful and thus worthy of experimental validation. Thirdly, the degree of target down-regulation imposed by miRNAs tends to be moderate: an miRNA typically causes less than 50% reduction at the protein level [14]. These considerations suggest that even though most genes might serve as targets of miRNAs, only a fraction of these interactions will prove instrumental for overt biological responses and phenotypes. The present mini-review provides a select set of examples (see Table 1) that relate the function of miRNAs to sophisticated functions in information-transfer networks, from fine-tuners that maintain output integrity, through sentinels of bi-stable cellular states, to mediators of pathological processes, such as cancer (Figure 1).
Table 1.
Examples of miRNAs involved in cancer and cell-fate decisions
| Pathway component | Target mRNA | miRNA | Relationship to cancer | Reference |
|---|---|---|---|---|
| Receptors | EGFR | miR-7 | Glioblastoma | [39] |
| ERBB2 | miR-331-3p | Prostate cancer | [40] | |
| ERBB2/ERBB3 | miR-125a/miR-125b | Breast cancer | [41] | |
| TGFB1 | miR-141 | EMT | [26] | |
| VEGFA | miR-126 | Angiogenesis | [42] | |
| Signalling molecules | NRAS/KRAS | Let-7 | Vulva development and lung cancer | [35] |
| PTEN | miR-21 | Hepatocellular cancer | [43] | |
| KRAS | miR-143 | Colorectal cancer | [44] | |
| ERK5 (MAPK7) | miR-143/miR-145 | Adipocyte differentiation and bladder cancer | [45] | |
| Ligands | IL6 | Let-7 | Genetic switch of tumour initiation | [33] |
| VEGFA | miR-34a, miR-15a, miR-16 and more | Nasopharyngeal carcinoma | [42] | |
| Transcription factors | Yan | miR-7 | Photoreceoptor differentiation | [22] |
| E2F1 | miR-330 | Prostate cancer | [46] | |
| EGR1 | miR-191 | Maintenance of cellular arrest and breast cancer | [20] |
EGFR, EGF receptor; MAPK7, mitogen-activated protein kinase 7; PTEN, phosphatase and tensin homologue deleted on chromosome 10; VEGFA, vascular endothelial growth factor A.
Figure 1. Examples of network motifs involving signalling components and miRNAs.
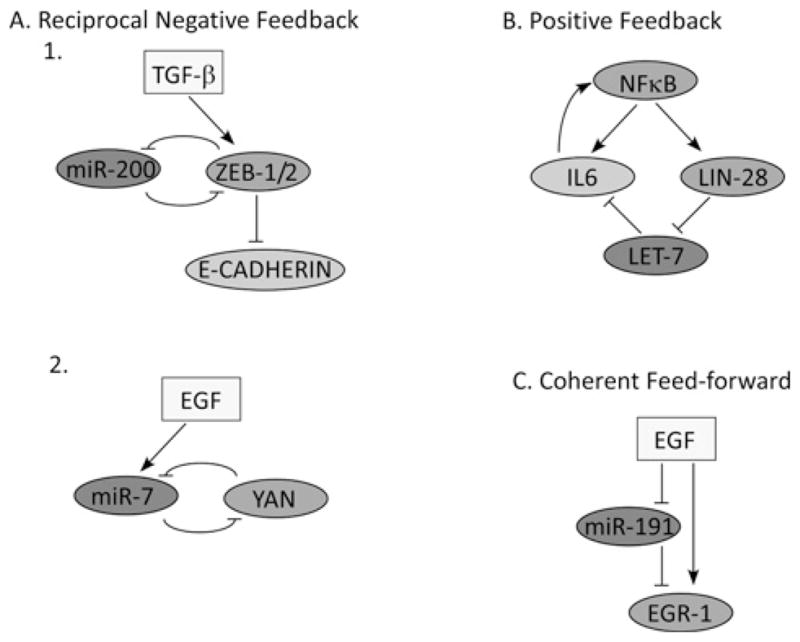
(A) Reciprocal negative regulation. 1. An epithelial phenotype is maintained by high levels of miR-200, which inhibits ZEB family repressor proteins and up-regulates E-cadherin. The switch to a mesenchymal state can be induced by TGFβ, which up-regulates ZEB proteins. This in turn maintains a mesenchymal state through repressive ZEB-1/2 binding to the promoters of E-cadherin and miR-200. 2. Progenitor cells in the eye of D. melanogaster are maintained by high levels of the transcription factor YAN. EGFR (EGF receptor) signalling promotes differentiation of progenitor cells to photoreceptor cells by reducing the levels of YAN both by direct ERK (extracellular-signal-regulated kinase)-stimulated degradation of YAN and by the induction of miR-7, which maintains persistently low levels of YAN. (B) Positive feedback. Transient activation of NF-κB increases IL-6 levels both by direct transcriptional activation of the IL6 promoter, and by induction of LIN28 which reduces Let-7 miRNA, the target of which is IL-6. In turn, IL-6 reactivates NF-κB. Thus positive feedback transforms a reversible transient signal into a self-sustained loop. (C) Coherent feedforward. EGR1 induction relies on the activation of MAPK (mitogen-activated protein kinase) and the degradation of miR-191, which targets EGR1.
Tuning of signalling networks by miRNAs
Because the outcomes of signalling events are highly dynamic, transient and non-linear, the upstream pathways and networks might benefit from the scales of quantitative fluctuations imposed by miRNAs. Especially powerful is the potential to regulate dozens or even hundreds of different transcripts by tightly controlling synthesis or turnover of the respective miRNAs, particularly during rapid changes that occur in the cellular microenvironment. Indeed, several recent studies reported tight regulation of miRNA transcription [15–17], as well as rapid and simultaneous induction of sets of miRNAs in response to external stimuli [18,19]. Regulation of mature miRNA expression can be achieved also by rapid processing of the precursors. For example, treatment of vascular smooth muscle cells with TGFβ (transforming growth factor β) promotes a rapid increase in expression of mature miR-21 through a post-transcriptional step, namely promoting the processing of primary transcripts of miR-21 (pri-miR-21) into precursor miR-21 (pre-miR-21) by the DROSHA microprocessor complex [18]. The latter forms a complex with pri-miR-21, TGFβ-specific SMAD signal transducers and the RNA helicase p68 [also known as DDX5 (DEAD box 5)]. Upon up-regulation, miR-21 down-regulates PDCD4 (programmed cell death 4), a negative regulator of smooth muscle contractile genes, thereby facilitating differentiation into contractile cells.
In contrast with miRNA induction through transcription and biogenesis, turnover of miRNAs has received only limited attention. In one recent report, stimulation of spontaneously immortalized normal mammary cells with EGF (epidermal growth factor) was shown to cause rapid turnover of a set of ~25 miRNAs, denoted ID-miRs (immediately down-regulated miRNAs) [20]. EGF stimulation within this cellular context elicits a well-orchestrated genetic programme leading to cell migration, and ID-miR turnover appears to be essential for migration onset. Interestingly, the targets of ID-miRs were shown to be the earliest genes to be induced by EGF in migrating cells, namely transcripts encoding IEGs (immediate early genes) [e.g. FOS and EGR1 (early growth response 1)]. Thus the ID-miRs, which are highly abundant in arrested cells, silence basal IEG expression to prevent untimely cellular activation (Figures 1C and 2). In line with roles as suppressors of growth factor signalling, the abundance of ID-miRs is relatively low in breast cancer and in brain tumours driven by constitutive EGF signalling. Rapid and co-ordinated decay of multiple miRNAs similarly takes place in retinal neurons, in response to exposure to light [21]. Concentrations of specific miRNAs (e.g. the miR-183/miR-96/miR-182 cluster, miR-204 and miR-211) were down-regulated during dark adaptation and up-regulated in light-adapted retinas, with both rapid decay and increased transcription being responsible for the respective changes. Adaptation of the retina to different levels of illumination is likely to be mediated by the voltage-dependent glutamate transporter Slc1a1, which serves as an miR-183/miR-96/miR-182 target in photoreceptor cells. Conceivably, simultaneous signal-induced metabolism of miRNAs plays a cardinal role in adaptations to environmental changes, such that future studies will uncover more examples.
Figure 2. Regulation of miRNA biogenesis and function by signalling networks.
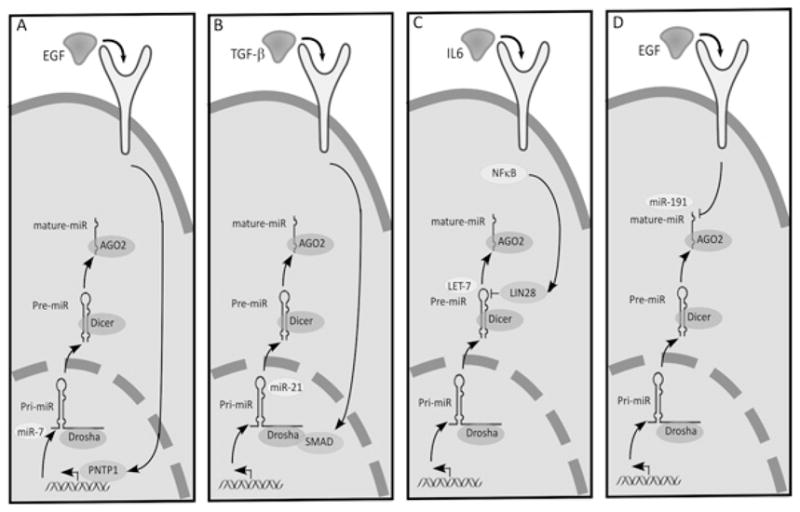
(A) EGF signalling activates the transcription factor PNTP1, which binds to and activates miR-7’s promoter. (B) TGFβ activates SMAD proteins, which translocate to the nucleus, where they associate with Drosha and RNase III to induce maturation of pri-miR-21. (C) Activation of LIN28 causes its association with precursors of the Let-7 family of miRNAs to induce their degradation. (D) EGF induces the degradation of a class of miRNAs, named ID-miRs (e.g. miR-191), which enables induction of IEGs such as c-FOS. AGO2, Argonaute 2; Pre-miR, precursor miRNA; Pri-miR, primary transcript of miRNA.
Along with mediating cellular differentiation, miRNAs can regulate binary switches that robustly select persistent cell fates. An interesting example is provided by the Drosophila melanogaster eye [22]. In resting cells, the transcription factor YAN represses miR-7 transcription, whereas miR-7 represses Yan mRNA expression in photoreceptors. Differentiation to photoreceptors is triggered by transient EGF signalling, which degrades YAN and up-regulates miR-7 in cells as they begin differentiating into photoreceptors. The resulting reciprocal negative feedback ensures mutually exclusive expression, with YAN in progenitor cells and miR-7 in photoreceptor cells (Figure 1A).
miRNAs regulating motile phenotypes and metastasis
EMTs (epithelial–mesenchymal transitions) take place both during embryonic development, when tissue remodelling and cell migration shape the future organism, and in pathological situations such as in tumour progression [23]. During EMT, epithelial cells lose the adherent and tight junctions that keep them in contact, undergo profound changes in their cytoskeletal architecture and migrate over long distances. EGF, along with TGFβ and other growth factors, enhance EMT, in part by regulating several transcription factors (e.g. Snail, Twist and ZEB proteins [24]). On their part, the latter enhance synthesis and secretion of specific matrix metalloproteinases, down-regulate protease inhibitors and repress E-cadherin (epithelial cadherin), an essential adhesive molecule that maintains epithelial integrity. Five miRNAs of the miR-200 family were shown to regulate the expression of the E-cadherin transcriptional repressors ZEB1 and ZEB2 [25], and thus to play a major part in specifying the epithelial phenotype. In analogy to YAN and miR-7, ZEB-1 and ZEB-2 act as transcriptional repressors of the miR-200 family [26]. Thus, upon induction of EMT by TGFβ, a molecular memory switch comes into operation: a transient signal engages a reciprocal negative-feedback loop that persistently enhances expression of ZEB-1 and ZEB-2, thereby locking the repression of both E-cadherin and the miR-200 family [27] (Figure 1).
miRNAs as drivers of tumour initiation
Unlike the above described examples of miRNAs involved in locking a cellular state in response to an extracellular cue, malignant transformation, like other pathogenesis, represents a multistep process that requires inactivation of tumour-suppressor genes and multiple mutations that activate oncogenes [28]. There are a handful of indications for the deregulation of the expression of miRNAs in patients’ tumours. Examples include amplified or repressed expression [29], shortening of 3′-UTR sequences of oncogenic transcripts [30], aberrations impinging at seed sequences of miRNAs [31] and genomic alterations within miRNA loci [32]. Nevertheless, lines of evidence supporting involvement of miRNAs as drivers of the initial stages of malignant transformation are still sparse. For this reason, we highlight a recently uncovered molecular module that involves the NF-κB (nuclear factor κB) signalling pathway, IL-6 (interleukin 6), LIN28 and the Let-7 miRNA [33]. The module converts, within 36 h, non-transformed human mammary cells into fully transformed cells. Importantly, a critical driving force of this switch was shown to be down-regulation of the Let-7 miRNA, such that ectopic expression of any member of the Let-7 family was able to abrogate the malignant phenotype. Down-regulation of Let-7 is achieved in this system through NF-κB-mediated transcriptional induction of LIN28B. The latter is a highly conserved RNA-binding protein, which was shown in Caenorhabditis elegans to recognize the hairpin structure of the pre-miRNAs of all Let-7 family members, and cause their co-ordinate inhibition [34] (Figure 2). One of the evolutionarily conserved targets of Let-7 is the oncogenic Ras protein, and this regulation was shown to control self-renewal of breast cancer cells [35]. In another study, epithelial de-differentiation occurred through Let-7-mediated down-regulation of a different target, HMGA2 (high-mobility group A2) [36–38], a chromatin-associated protein that can modulate transcription by altering chromatin architecture. However, in the case of NF-κB, down-regulation of Let-7 drives the transformation process through a different target, namely IL-6. Notably, IL-6 expression is induced by NF-κB, which binds to the IL6 gene promoter and increases transcription. Thus NF-κB activation both releases the Let-7-mediated repression of IL6 transcripts and, at the same time, induces the transcription of the IL6 gene. Because IL-6 transmits signals through the NF-κB pathway, this configuration creates a potent positive-feedback loop (Figure 1B). Clearly, this powerful oncogenic module implies that we understand only part of the potential complexity of miRNAs and transcription factors acting at the interface linking signal transduction and the regulation of gene expression.
Concluding remarks
The advancement of high-throughput genomic screens, along with the development of software able to predict targets of miRNAs, will eventually yield high granularity maps combining signal transduction pathways, transcriptional programmes and the relevant miRNAs. Although we are still far from this goal, identifying miRNAs affecting information-transfer events can both assign functionality to numerous miRNAs and also advance our understanding of how signals of diverse molecular nature coalesce into cell fate decisions. In similarity to the current pharmacological translation of high-content knowledge on kinase cascades and the avalanche of new kinase inhibitors to treat cancer and other diseases, it is conceivable that detailed understanding of roles played by miRNAs in signalling networks will lead to novel therapies. Achieving this important goal depends on the development of innovative pharmacological platforms, such as the locked nucleic acids technology, capable of synthesizing and delivering stable, specific and effective RNA-based drug molecules.
Acknowledgments
Y.Y. is the incumbent of the Harold and Zelda Goldenberg Professorial Chair.
Funding
Our laboratory is supported by research grants from the National Cancer Institute [grant number CA72981], the M.D. Moross Institute for Cancer Research, Kekst Family Institute for Medical Genetics, Women’s Health Research Center funded by Bennett-Pritzker Endowment Fund, Marvelle Koffler Program for Breast Cancer Research, Harry and Jeanette Weinberg Women’s Health Research Endowment, the European Commission and the German Research Foundation.
Abbreviations used
- E-cadherin
epithelial cadherin
- EGF
epidermal growth factor
- EGR1
early growth response 1
- EMT
epithelial–mesenchymal transition
- IEG
immediate early gene
- IL-6
interleukin 6
- miRNA
microRNA
- ID-miR
immediately down-regulated miRNA
- NF-κB
nuclear factor κB
- TGFβ
transforming growth factor β
- UTR
untranslated region
References
- 1.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 4.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 8.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 9.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 10.Tijsterman M, Plasterk RH. Dicers at RISC: the mechanism of RNAi. Cell. 2004;117:1–3. doi: 10.1016/s0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Li W, Jin YX. Computational identification of novel family members of microRNA genes in Arabidopsis thaliana and Oryza sativa. Acta Biochim Biophys Sin. 2005;37:75–87. [PubMed] [Google Scholar]
- 13.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 14.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 15.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 17.Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, Enerly E, Johnsen H, Buganim Y, Solomon H, et al. p53-repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avraham R, Sas-Chen A, Manor O, Steinfeld I, Shalgi R, Tarcic G, Bossel N, Zeisel A, Amit I, Zwang Y, et al. EGF decreases the abundance of microRNAs that restrain oncogenic transcription factors. Sci Signaling. 2010;3:ra43. doi: 10.1126/scisignal.2000876. [DOI] [PubMed] [Google Scholar]
- 21.Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 23.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 24.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 25.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 26.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1–SIP1 and the microRNA-200 family regulates epithelial–mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 30.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin Y, Wang E, Wu M, Shen SH. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–4541. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-κB, Lin28, Let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res. 2008;14:2334–2340. doi: 10.1158/1078-0432.CCR-07-4667. [DOI] [PubMed] [Google Scholar]
- 38.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 40.Epis MR, Giles KM, Barker A, Kendrick TS, Leedman PJ. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem. 2009;284:24696–24704. doi: 10.1074/jbc.M109.030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 42.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin. Chim. Acta. 2010;411:846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 44.Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24:2754–2759. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clape C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, Iborra F, Avances C, Villalba M, Culine S, Fajas L. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS ONE. 2009;4:e7542. doi: 10.1371/journal.pone.0007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT, Goan YG, Lu PJ. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene. 2009;28:3360–3370. doi: 10.1042/BST20110623. Received 15 June 2011. [DOI] [PubMed] [Google Scholar]


