Abstract
Sialic acids are structurally unique nine-carbon keto sugars occupying the interface between the host and commensal or pathogenic microorganisms. An important function of host sialic acid is to regulate innate immunity, and microbes have evolved various strategies for subverting this process by decorating their surfaces with sialylated oligosaccharides that mimic those of the host. These subversive strategies include a de novo synthetic pathway and at least two truncated pathways that depend on scavenging host-derived intermediates. A fourth strategy involves modification of sialidases so that instead of transferring sialic acid to water (hydrolysis), a second active site is created for binding alternative acceptors. Sialic acids also are excellent sources of carbon, nitrogen, energy, and precursors of cell wall biosynthesis. The catabolic strategies for exploiting host sialic acids as nutritional sources are as diverse as the biosynthetic mechanisms, including examples of horizontal gene transfer and multiple transport systems. Finally, as compounds coating the surfaces of virtually every vertebrate cell, sialic acids provide information about the host environment that, at least in Escherichia coli, is interpreted by the global regulator encoded by nanR. In addition to regulating the catabolism of sialic acids through the nan operon, NanR controls at least two other operons of unknown function and appears to participate in the regulation of type 1 fimbrial phase variation. Sialic acid is, therefore, a host molecule to be copied (molecular mimicry), eaten (nutrition), and interpreted (cell signaling) by diverse metabolic machinery in all major groups of mammalian pathogens and commensals.
INTRODUCTION
“Sialic acids are not only the most interesting molecules in the world, but also the most important” (151).
The whimsically hyperbolic quote above, credited to Eric Sixmister, captures the excitement that many different researchers have experienced over the past 40 years while working on a class of tertiary gene products with manifold function in both eukaryotic and prokaryotic species (151). Sialic acid (less commonly called neuraminic acid) is the designation given to a family of over 40 naturally occurring nine-carbon keto sugars acids derived from the parent compound 2-keto-3-deoxy-5-acetamido-d-glycero-d-galacto-nonulosonic acid (N-acetylneuraminic acid [Neu5Ac]). The sialic acids and related nonulosonates are unique in nature by representing the only nine-carbon sugars found in prokaryotes. In eukaryotes, sialic acids have evolved to mediate a diverse range of cell-cell and cell-molecule interactions, including (i) stabilizing glycoconjugates and cell membranes due to charge-charge repulsion, (ii) mediating cell-cell regulation and acting as chemical messengers, (iii) regulating transmembrane receptor function, (iv) affecting membrane transport, (v) controlling the half-lives of circulating glycoproteins and cells, and (vi) contributing to the permselectivity of the glomerular endothelium and slit diaphragm (118). The relative biological importance of these processes to complex metazoan animals is demonstrated by early embryonic death of homozygous mouse mutants with a defect in sialic acid synthesis (123). Some bacteria have evolved a de novo pathway for sialic acid biosynthesis that differs from the eukaryotic method, whereas other microbes use truncated synthetic pathways utilizing sialyl precursors scavenged from animal hosts (148). Many commensal and pathogenic bacteria also use environmental (host) sialic acids as sources of carbon, nitrogen, energy, and amino sugars for cell wall synthesis (98). Microbial sialic acid metabolism has now been firmly established as a virulence determinant in a range of infectious diseases. In this review we argue that unraveling the molecular mechanisms of sialometabolism and its regulation provides a unifying theme for investigating the host-pathogen interactions in a wide range of invasive (extraintestinal) infectious diseases.
The name sialic acid comes from the Greek word sialon (saliva), consistent with the discovery of these carbohydrates in bovine submaxillary mucin and brain matter (neuroamine) by the German biochemists Blix in 1936 (21) and Klenk in 1941 (74), respectively. Gottschalk (55) was the first to propose a coherent structure of Neu5Ac and also provided a survey of the early history of sialobiology, as the field has since come to be known (110). The excellent short review by Hans Faillard on the early history of sialic acid research also is highly recommended (46). Any of the more recent monographs or review articles by Roland Schauer and his colleagues should be consulted for the methodological details for working with sialic acids (e.g., 118-120). Here we focus on the metabolism and signaling functions of microbial sialic acids after briefly discussing their structure and evolution.
SIALIC ACID STRUCTURES
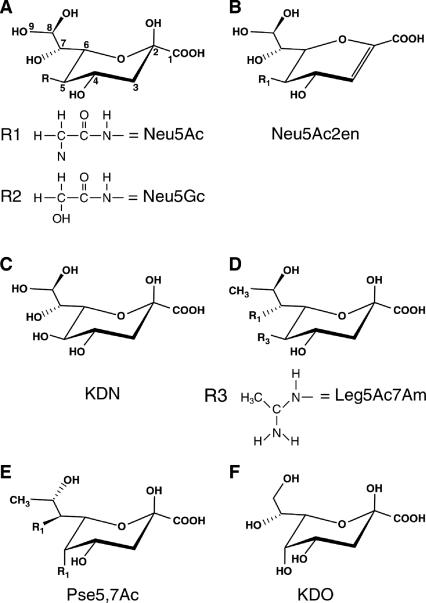
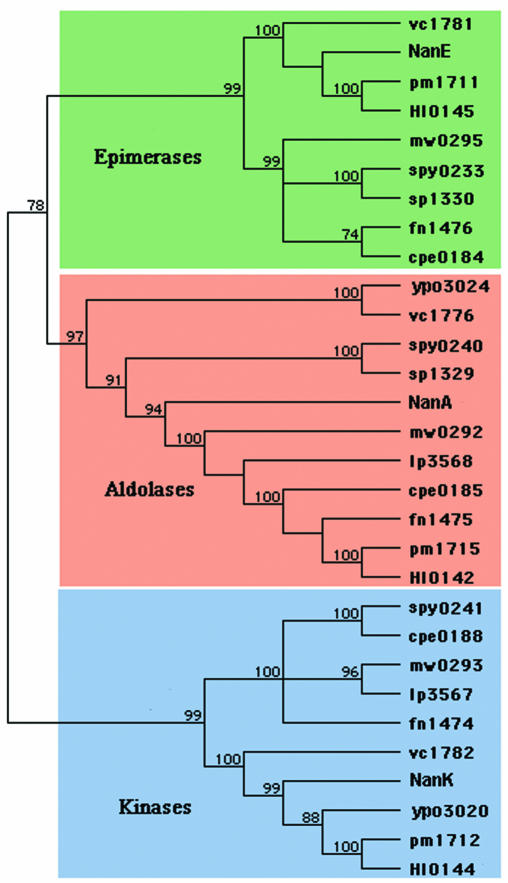
The two most common naturally occurring sialic acids differ from each other only at position five (C-5) of the respective carbon rings (Fig. 1A). The stereochemistry of Neu5Ac around C-5 was determined in 1960 by Comb and Roseman (33), using a reversible enzyme (sialate aldolase) that cleaves sialic acid under physiological conditions to the previously unknown hexosamine N-acetylmannosamine (ManNAc), the 2-epimer of N-acetylglucosamine (GlcNAc), and pyruvate. It was during research on sialic acid structure and biosynthesis that Werner Kundig, working in Roseman's laboratory, discovered ManNAc phosphorylation (77). This discovery led to the elucidation of the group translocation transporters known today as the sugar-phosphotransferase system (PTS). The PTS was the first of many phospho-relay systems to be described in biology.
FIG. 1.
Structures of sialic acids and related keto sugars. Note that the numbers in panel A indicate the relative carbon atoms in Neu5Ac and each of the succeeding ulosonic acids.
In addition to the N-acetyl and N-glycolyl groups of sialic acids shown in Fig. 1A, other common modifications to the ring or exocyclic hydroxyls include one to three acetyl groups added at C-4 or to the C-7 to C-9 hydroxyls of the glycerol side chain. Although less usual a modification than acetylation, sialic acids also may be modified with lactyl, sulfhydryl, or methyl groups. Indeed, greater than 20% of the sialic acid bound to glycoproteins of the human red blood cell membrane may be forms other than Neu5Ac, including the previously unreported unacetylated C-5 derivative of Neu5Ac, neuraminic acid (24). Although N-glycolylneuraminic acid (Neu5Gc) (Fig. 1A) is the second (after Neu5Ac) most common sialic acid of most vertebrates (mammals, birds, amphibians, reptiles, and fish), it has never been detected in bacteria. Interestingly, the mammalian structural gene encoding the hydroxylase responsible for adding the N-glycolyl group in Neu5Gc is inactive in humans, with the mutation (a deletion in one of the exons) occurring after divergence of the human lineage from that of the great apes. The absence of endogenous Neu5Gc in humans was the first biochemical Homo-Pan distinction noted, with profound implications for human physical and cultural evolution, especially infectious disease resistance (147). The comprehensive review article by Angata and Varki (7) should be consulted for details about the biological distinctions between Neu5Ac and Neu5Gc and the Homo-Pan divergence.
The carboxylate group of sialic acids is deprotonated at physiological pH (pKa of 2.6) and confers the net negative charge that dominates the physiochemical properties of the family. Glycoketosidic linkages between sialic acids and sugars such as galactose, GlcNAc, or other sialyl residues (Sia) are synthesized by sialyltransferases linking the C-2 (reducing) donor hydroxyl to an appropriate acceptor hydroxyl group as follows (110): CMP-Sia + HO-acceptor→CMP + Sia-O-acceptor, where CMP-Sia is the obligate sialyl donor for all known sialyltransferases. In solution, free sialic acid exists predominantly in its β conformation, as shown for Neu5Ac, with the carbohydrate's C-2 hydroxyl positioned axial to the ring (Fig. 1A). This form is in thermodynamic equilibrium with the minor α isomer (151). Both forms are interconverted by mutarotation as the ring opens to the straight-chain conformation followed by ring closure. The β configuration of the sialic acid substrate is maintained when coupled (activated) with CTP by CMP-sialic acid synthetase, which transfers the anomeric oxygen of β-sialic acid to the α-phosphate of CTP (3). Chemical reactivity of the acceptor hydroxyl is relatively low; therefore, all carbohydrate donors must be activated by first being coupled to a trinucleotide phosphate with removal of one or two phosphates. Sialic acid (and the octulosonate 2-keto-3-deoxy-d-manno-octulosonic acid [keto-deoxy octonate {KDO}] [Fig. 1F]) synthetases are unique enzymes in carbohydrate chemistry because they eliminate pyrophosphate instead of inorganic phosphate during the coupling reaction. Backside SN2 attack of the donor β-linked hydroxyl group by an acceptor nonreducing hydroxyl yields α-linked sialylated products (Sia-O-acceptor). Thus, all sialyltransferases operate by an inversion-of-configuration mechanism involving an activated CMP-Sia donor substrate and a suitable acceptor molecule. Although sialic acids may be linked to structures composed entirely of sugar (oligosaccharides and some polysaccharides), they are typically part of more complex structures involving lipids (glycolipids) or proteins (glycoproteins), which are collectively referred to as sialoglycoconjugates.
Dehydration (double-bond formation) at the sialic acid reducing end leads to formation of the planar structure shown for N-acetyl-2,3-didehydro-2-deoxyneuraminic acid (dideoxy-Neu5Ac [Neu5Ac2en]) in Fig. 1B. The flattened Neu5Ac2en ring mimics the transition state during hydrolysis of sialoglycoconjugates (Sia-O-acceptors) by glycosylhydrolases designated sialidases (synonymous with neuraminidases). Neu5Ac2en served as the lead compound for synthesis of the first rationally designed sialidase inhibitor, now marketed for clinical use as Relenza (zanamivir), an anti-influenza virus inhibitor that prevents spread of the virus during an infection and thus ameliorates flu symptoms without preventing infection (159). In another proposed use of sialidase inhibitors, a patent has been issued to the U.S. Army Medical Research and Materiel Command for the use of Neu5Ac2en to treat a variety of diseases involving inflammatory cells and mediators, including human immunodeficiency virus infection and AIDS (143a). Interestingly, Neu5Ac2en is formed spontaneously from CMP-Neu5Ac under physiological conditions (116) and as a by-product of sialidase action in vivo (16). Therefore, it seems that nature may have already co-opted Neu5Ac2en as a sialidase inhibitor. It is also tempting to speculate that Neu5Ac2en may be a component of the innate (antibody-independent) immune system, guarding us from certain pathogens that use sialidase for adhesion and cellular invasion. Alternatively (but not mutually exclusively), Neu5Ac2en could be part of an endogenous mammalian sialidase regulatory system, as suggested by one recent study (8).
2-Keto-3-deoxy-d-glycero-d-galacto-nonulosonic acid (keto-deoxy neuraminic acid [KDN]), 5,7-diamino-3,5,7,9-tetradeoxy-d-glycero-d-galacto-nonulosonic acid (legionaminic acid [Leg5Ac7Am]), and 5,7-diamino-3,5,7,9-tetradeoxy-l-glycero-l-manno-nonulosonic acid (pseudaminic acid [Pse5,7Ac]) (Fig. 1C to E, respectively) are sialic acid-like nonulosonates found in eukaryotes and certain, generally nonpathogenic or opportunistic bacterial species such as Legionella pneumophila, Pseudomonas aeruginosa, and Campylobacter jejuni (53, 75, 76, 126, 137, 142). It is speculated that because neither Leg5Ac7Am nor Pse5,7Ac is found in eukaryotes, these sugars would offer no protection to the bacteria, as the host would recognize the carbohydrates as foreign by mounting an effective immune response (7). Angata and Varki (7) further speculated that polymers of Leg5Ac7Am or other sialic acids might serve to protect free-living bacteria from bacteriophage infection by blocking underlying receptors. However, it is difficult to reconcile this idea with the rapid modular evolution of phage genomes (95) and the presence of polysialic acids (PSA) as bona fide virulence factors and receptors for a variety of highly lytic phages (97). In other words, blocking of potential underlying receptors with new receptor species would seem to be an inefficient mechanism for dealing with the problem of phage infection. Thus, evocation of the “Red Queen effect” (145), named after the Red Queen's quote from Alice Through the Looking Glass, “it takes all the running you can do, to keep in the same place [and if] you want to get somewhere else, you must run twice as fast as that,” to account for sialate structural diversity as a mechanism to thwart microbial infection (exogenous evolution) (7) may not be applicable for understanding bacterium-phage interactions. We may need to follow another white rabbit in order to understand how the diversity of microbial sialic acid structures has arisen.
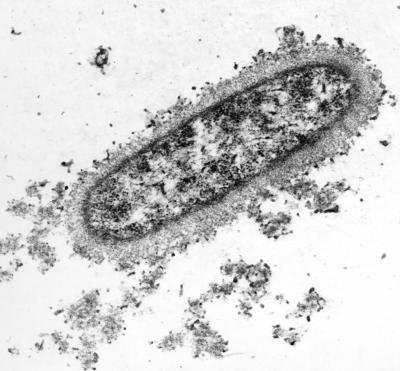
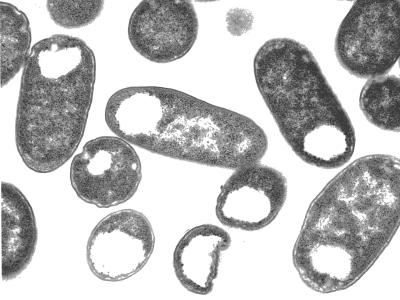
Another function of PSA and of microbial polysaccharides in general may be to conserve water. The dehydration steps prior to transmission electron microscopy of encapsulated bacteria produce amorphous blebs composed of collapsed polysaccharides (153). To visualize all but the most copious of capsules (which can be detected by particle exclusion under the light microscope), antipolysaccharide antibodies are necessary to cross-link sugar chains and stabilize the highly hydrated capsule structure (Fig. 2). Thus, bacteria, with their large surface area-to-volume ratios, are especially susceptible to desiccation. Sporulation is obviously one mechanism to combat environmental desert-like conditions, but carrying around a polysaccharide “canteen” might confer a similar survival advantage to encapsulated microorganisms. For example, mucoid strains of Escherichia coli, Acinetobacter calcoacetious, and Erwinia stewartii were found to be significantly more resistant to desiccation than corresponding nonmucoid variants (91).
FIG. 2.
Encapsulated E. coli K1. The PSA capsule of E. coli K1 was stabilized with monospecific antibody, fixed, thin sectioned, and examined by transmission electron microscopy (30, 153). Note the microcapsule surrounding the outer surface of the bacterium and the dense, granular cytoplasm.
An additional mechanism driving mammalian sialic acid structural diversification may occur within the host (endogenous evolution). It was recently shown that complex metazoans express a variety of sialic acid-binding immunoglobulin-like lectins (siglecs) that preferentially bind different sialylated acceptors (37, 38). A diverse population of sialic acids may thus facilitate cell-cell and cell-molecule interactions through direct or indirect cell signaling involving siglec-sialoglycoconjugate interactions. However, by using highly sensitive analytical methods, it has recently become apparent that the mammalian red blood cell surface may contain more Sia derivatives than there are endogenous lectins (24), suggesting that not all of the potentially vast informational content of sialoglycoconjugates may have biological significance. It will be instructive to look at the Sia diversity of red blood cell progenitors to determine whether Sia complexity alters during erythropoiesis and whether similar diversity exists in all animal tissues.
In conclusion, we suggest that the exact reason for sialic acid structural diversity in bacteria and their hosts is uncertain. However, the fact that such diversity exists is undeniable, and the number of new structures is expanding as analytical methods continue to improve. Given their usual terminal positions on host cell glycoconjugates, sialic acids are among the most prevalent molecules at the host-microbe interface. The analogy of sialic acid structural evolution seen as an arms race between and within species is thus likely to be an apt one (42).
EVOLUTION OF SIALIC ACID METABOLISM
The evolution of sialic acid metabolism is intriguing, because until recently these sugars were thought to exist only in “higher” animals (starfish to humans) and a few pathogenic bacterial species. Warren (162) used a combination of chemical assays to determine the phylogenetic distribution of sialic acids to arrive at this conclusion for multicellular organisms. His results indicated that synthesis of sialic acids was limited to complex metazoans of the deuterostome (literally “two mouths” during embryogenesis) lineage (162). Thus, neither plants, protostomes (insects, arachnids, crustaceans, gastropods, shellfish, cephalopods, and nematodes), nor fungi had detectable levels of sialic acids, suggesting that the evolution of the sialic acid biosynthetic pathway occurred near the divergence of Coelomata (protostomes and deuterostomes) roughly five hundred million years ago. The few, mostly pathogenic, bacterial species that were known to synthesize sialic acids (12) were thought to represent either a relatively recent and minor parallel evolution of the pathway or acquisition of biosynthetic genes from eukaryotes by horizontal transfer (108). Although these original observations remain largely uncontested, more sensitive analytical procedures and the burgeoning of complete genomic DNA sequences have considerably broadened our view of sialic acid distribution.
As described in the previous section on sialic acid structure, the identification of sialic acid-like molecules in a variety of bacterial species indicates that a rich diversity of nonulosonates exists in prokaryotes. The occurrence of sialic acid in certain larval stages during insect development further indicates that the basic biosynthetic machinery was intact prior to the divergence of the Coelomata (112). Adding to this discovery of increased distribution of sialic acids is the bewildering range of unicellular fungi in which sialic acids have been detected, with many apparently producing sialic acid by a de novo biosynthetic pathway (1). Sialic acids still have not been detected in plants or the Archaea, despite evidence of potential biosynthetic genes in the latter species (7). On the basis of a molecular phylogenetic approach, Angata and Varki (7) suggested that the evolution of both sialic acid and the related eight-carbon sugar KDO (Fig. 1F) diverged from enzymes involved in shikimic acid biosynthesis for aromatic amino acid production. In one intriguing scenario, the genes for sialic acid biosynthesis were transformed horizontally to a Coelomata ancestor (scenario 2 in reference 7). This suggestion is consistent with the observation that 20% of the 40 or so genes thought to have undergone horizontal transfer between bacteria and humans (117) are involved in sialic acid metabolism (7). Further analyses of these data provided strong evidence for horizontal transfer between the prokaryotic aldolase gene nanA and its orthologues in humans and other vertebrates, although the direction of this transfer could not be determined (4). These putative vertebrate aldolases strongly clustered with NanA from Yersinia pestis and Vibrio cholerae (4). Further understanding of sialic acid evolution should be aided greatly by investigating the genes and gene products involved in fungal sialic acid metabolism, a research area that currently remains almost completely unexplored.
Regardless of the exact evolutionary details of sialic acid metabolism, the widespread use of these sugars for regulating immunity and the central nervous system (5, 73, 119, 120, 125), two organs unique to deuterostomes, is consistent with the apotheosis of sialic acids in this lineage. In the context of organ system function, it will be interesting to determine what role sialic acids play in insect physiology or development. While the occurrence of sialic acid synthesis has been increasingly detected in prokaryotes and unicellular eukaryotes (see above), many more microorganisms have the metabolic machinery to degrade sialic acids as sources of carbon, nitrogen, and energy and as precursors to cell wall biosynthesis (148). The preponderance of sialic acids in animals also makes the sialic acids attractive as potential signals that could be exploited for microbial environmental (host) sensing. A description of sialic acid catabolism logically follows.
SIALIC ACID CATABOLISM
Given the preponderance of sialic acids in complex animals (the sugars are common in deuterostomes but scarce elsewhere), microorganisms that degrade sialic acid are by definition expected to closely associate as commensals or pathogens with their animal hosts. The emergence of microbial sialic acid catabolism is hypothesized to be central to a variety to host-microbe interactions.
Discovery of Sialic Acid Catabolism
Although sialic acid aldolase (lyase) was one of the first enzymes of sialometabolism to be extensively investigated at the biochemical level, it was unclear whether the aldolase functioned in catabolism, in biosynthesis, or in both processes (33). The first indication that at least some bacteria could metabolize exogenous sialic acid as a potential carbon or energy source by a lyase-dependent mechanism came from an analysis of sialic acid uptake and degradation in Clostridium perfringens (89). However, no mutants with defects in sialic acid catabolism were isolated, and the putative degradative pathway remained uncharacterized until the mid 1980s.
E. coli K1 synthesizes a homopolymer of sialic acid known as PSA. While working on the genetics of PSA biosynthesis, Vimr and Troy (156, 157) noted that only about 10% of radiolabeled sialic acid added to the growth medium was incorporated into PSA despite quantitative uptake of the label into cells. On the assumption that the bacteria expressed an efficient transporter and degradative system for sialic acid dissimilation, mutants that failed to use sialic acid as the sole carbon source were isolated. The genetic defects in two of these nan (for N-acylneuraminate) mutants were subsequently located in genes for sialate transport (nanT) and the aldolase (nanA) (156). The nanAT genes were part of an operon that responded to apparent induction based on sialic acid availability. Upon completion of the E. coli K-12 genomic DNA sequencing project (19), the nan operon was seen to potentially include, in addition to nanAT, open reading frames yhcHIJ and a putative upstream regulator, yhcK (98). The results of the genetic and physiological studies indicated that exogenous sialic acid is transported by a secondary transporter (NanT) of the major facilitator superfamily and degraded intracellularly by NanA to yield pyruvate and the amino sugar ManNAc (156, 157). The products of yhcI (renamed nanK) and yhcJ (renamed nanE) were suggested to function by first phosphorylating ManNAc and then epimerizing the ManNAc-6-P generated by the kinase reaction to GlcNAc-6-P (98). Recent biochemical analyses confirmed that NanK is an ATP-dependent kinase specific for ManNAc and that NanE is a reversible 2-epimerase (104). The upstream regulatory open reading frame yhcK was renamed nanR and shown to repress the nan operon in the absence of sialic acid (98). The function of yhcH is unknown, and a null mutation in this open reading frame conferred no detectable growth phenotype on sialic acid (70). The GlcNAc-6-P produced by NanE was shown to enter the amino sugar degradative pathway encoded by nagBA (98), converting GlcNAc-6-P ultimately to fructose-6-P by GlcNAc-6-P deacetylase (NagA) and glucosamine-6-P deaminase (NagB). Sialic acid thus can serve as the sole carbon or nitrogen source in E. coli and as a source of amino sugars (GlcNAc and glucosamine) for cell wall biosynthesis (82, 98). NanA-catalyzed degradation of sialic acid yields a triose phosphate (pyruvate) and hexosamine (ManNAc), which makes sialic acid an attractive nutritional source for microbes that associate with vertebrates.
Diversity of nan Systems
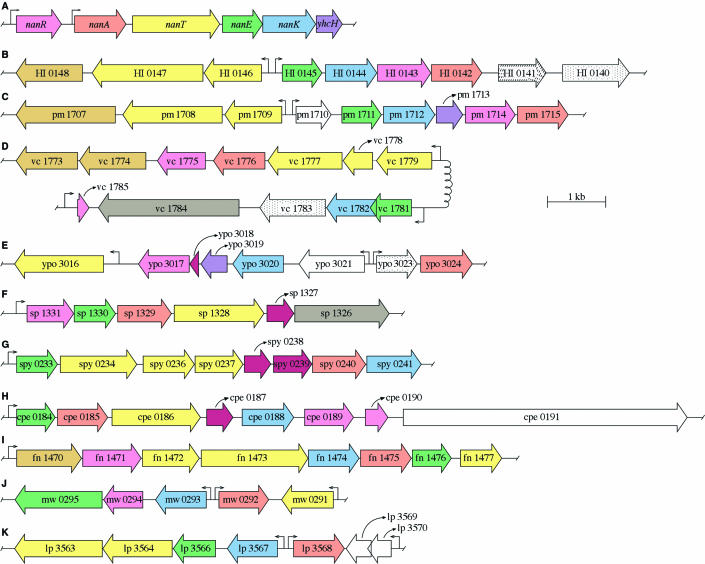
The availability of more than 264 complete bacterial genomic DNA sequences facilitated detection of complete nan systems in diverse species. A complete nan system was defined as one that minimally includes orthologues of genes encoding NanA, NanE, and NanK. This criterion held for each of the organisms shown in Fig. 3 except the following: Streptococcus pneumoniae (Fig. 3F), which contains only a weak kinase candidate (sp1331); Y. pestis, which lacks strong kinase and epimerase candidates (Fig. 3E); and Lactobacillus plantarum, which lacks an obvious epimerase candidate (Fig. 3K). Although it is possible that some organisms epimerize ManNAc to GlcNAc directly, as in the yeast Saccharomyces cerevisiae (18), Fig. 3 indicates that most bacteria that degrade sialic acids likely phosphorylate the ManNAc released by NanA prior to epimerization, as shown for E. coli (98, 104).
FIG.3.
Genetic organization of nan systems in gram-positive and gram-negative bacteria. The criteria for assigning functions to the various open reading frames are described in the text. Accession numbers (National Center for Biotechnology Information) are given in parentheses in the following. (A) E. coli (NC000913); (B) H. influenzae Rd (NC000907); (C) P. multocida Pm70 (NC002663); (D) V. cholerae N16961 (NC002505); (E) Y. pestis C092 (NC003143); (F) S. pneumoniae TIGR4 (NC003028); (G) S. pyogenes MGAS8232 (NC003485); (H) C. perfringens Str.13 (NC003366), (I) F. nucleatum ATCC25586 (NC003454), (J) S. aureus MW2 (NC003923); (K) L. plantarum WCFS1 (NC004567). Open reading frames are identified by the gene locus designation assigned to them in the database. Colored arrows indicate the known or probable gene products as follows: transcriptional regulators (magenta); aldolases (orange); transporters (yellow); epimerases (green); kinases (blue); sialidases (grey), and unknown (plum). Slashes (nagB) or dots (nagA) represent the nagBA homologues. Bronze-yellow open reading frames encode putative polypeptides with no known function but are associated with TRAP transporters as described in the text. Bent arrows indicate known or potential transcriptional start sites.
As shown in Fig. 3, most nan systems, like that of E. coli (Fig. 3A), do not encode closely linked orthologues of NagA or NagB. However, the Haemophilus influenzae nan system is an exception, as nagBA orthologues are closely linked to or part of the same nan transcriptional unit (Fig. 3B). The most recognizable feature of the operons shown in Fig. 3 is the lack of overall synteny and subsequently complex genetic organizations of the various nan systems. This conclusion is true for both the gram-negative (Fig. 3A to E) and gram-positive (Fig. 3F to K) species investigated. The lack of synteny between closely related species such as E. coli (Fig. 3A) and H. influenzae (Fig. 3B) suggests that the evolution of nan in different species was driven by the need to adapt to an animal host, in which sialic acids represent an abundant nutritional source that is generally absent in nonvertebrate environments. If this hypothesis is correct, the organizations of the different nan systems may represent each organism's individual solution to sialic acid catabolism as established during the evolution of the various host-microbe interactions.
The nanATEK-yhcH system in E. coli is derepressed over 200-fold by mutations in nanR or during growth on sialic acid as the sole carbon source, suggesting that E. coli may not usually exist in a sialic acid-rich natural environment (98). Tight nan regulation appears to result from tandem binding of two or more NanR homodimers to an operator that overlaps the nanA promoter, thus blocking RNA polymerase binding. Homologues of NanR are present in close relatives of E. coli, such as Salmonella enterica and Shigella spp., but not in any of the other species shown in Fig. 3 (70). However, most of these species may encode their own linked nan regulators (shown for various open reading frames by the magenta color), which is further evidence for the individual evolution of the various nan systems. Unraveling the genetic rationale for such apparent regulatory diversity offers great promise for better understanding the details of a variety of host-bacterium interactions.
In addition to NanAEK and NagBA, a complete nan system also should include a permease of some type for sialic acid uptake. As in the previous case of NanR, we found few obvious orthologues of NanT in the bacterial nan systems shown in Fig. 3. However, the Y. pestis nan system (Fig. 3E) includes an open reading frame (ypo3016) encoding a putative transporter with homology to NanT, while other transporters of this general type were detected in the gram-positive species S. pneumoniae (Fig. 3F), C. perfringens (Fig. 3H), Staphylococcus aureus (Fig. 3J), and L. plantarum (Fig. 3K), suggesting that these organisms rely on a mechanism of sialic acid uptake similar to that of E. coli, namely, a proton symporter or antiporter (secondary transporter of the major facilitator superfamily) (93, 146).
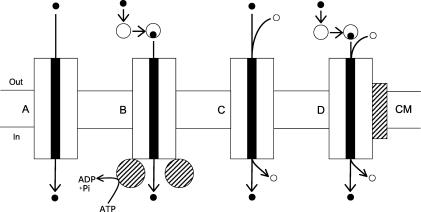
Bacterial transporters can be divided into four major types (a fifth type, not shown, includes group translocation catalyzed by the PTS): facilitated diffusion (Fig. 4A), ATP-binding cassette (ABC) transporters (Fig. 4B), secondary (ion- or proton-coupled) symporters or antiporters (Fig. 4C), and tripartite ATP-independent periplasmic (TRAP) transporters (Fig. 4D). The systems analyzed in Fig. 3 indicate that TRAP transporters are either closely linked or part of the nan operons of H. influenzae (Fig. 3B), Pasteurella multocida (Fig. 3C), V. cholerae (Fig. 3D), and Fusobacterium nucleatum (Fig. 3I). TRAP transporters include a periplasmic binding component that is thought to increase uptake affinity, a membrane-spanning polypeptide analogous to secondary transporters, and a second membrane polypeptide of unknown function (49, 72, 101). The substrates of TRAP transporters appear to be carboxylic acids such as malate, succinate, or fumarate, among which sialic acid may be considered a monocarboxylate (Fig. 1A). In some cases the two membrane TRAP transporter components may be fused into a single polypeptide (72), as in HI0147, pm1708, or fn1473 (Fig. 3B, C, and I, respectively). These TRAP transporter systems invariably include linked genes (shown in bronze-yellow) of unknown function that in at least one case were shown to be dispensable for dicarboxylic acid uptake (49). A bovine isolate of P. multocida that was attenuated for systemic pasteurellosis was isolated with an insertion mutation in pm1709 (50), which is predicted to encode a periplasmic TRAP component. The defect in this strain has been shown to prevent sialic acid transport, thereby providing direct evidence for the proposed function of TRAP transporters in sialic acid uptake (E. R. Vimr, S. M. Steenbergen, R. Caughlan, J. Garfinkle, and T. E. Fuller, unpublished data). To the extent that sialic acid uptake is required for the virulence of diverse pathogenic microorganisms, new therapeutic agents aimed at blocking transport offer a new class of nonantibiotic drugs that could have utility for treating a wide range of human or animal infectious diseases.
FIG. 4.
Bacterial solute transporters. Solute (solid circles) channels (solid rectangles) for uptake through the cytoplasmic membrane (CM) from the extracellular environment (Out) to the cytoplasm (In) are indicated by the large rectangles representing integral membrane transport proteins. (A) Facilitated diffusion. (B) ABC transporter, where intermediate-size open circles indicate the periplasmic binding component and the hatched circles indicate the ATPase. (C) Symporter of the major facilitator superfamily of membrane transporters, where the small circles indicate protons or metal ions coupled to solute uptake. (D) TRAP transporter, indicating features in common with ABC and secondary transporters and the addition of a second (hatched rectangle) membrane protein of unknown function that is necessary for solute uptake.
Unlike the potential secondary or TRAP transporters identified in Fig. 3, the putative sialic acid permease of Streptococcus pyogenes (spy0236 and spy0237) is predicted to be an ABC system, while spy0234 is predicted to be a lipoprotein transporter (Fig. 3G). Thus, it is evident that there can be a great deal of diversity among potential sialic acid transporters of even a single genus such as Streptococcus.
Relationships between NanAEK Proteins
Once internalized by secondary, TRAP, or ABC transporters, sialic acid may be cleaved to produce ManNAc and pyruvate in a lyase reaction catalyzed by NanA. Pyruvate enters intermediary metabolism as either phosphoenolpyruvate (Embden-Meyerhof-Parnas pathway) or acetyl coenzyme A (tricarboxylic or citric acid cycle), whereas ManNAc is converted to fructose-6-P by the concerted action of NanEK and NagAB, thereby completing the dissimilation of sialic acid for entrance of the hexosamine carbon backbone into glycolysis. As shown by the dendrogram in Fig. 5, the NanE (blue panel) and NanK (green panel) homologues follow the expected phylogenetic relationships, indicating that the gram-positive gene products cluster separately from their gram-negative counterparts. The progenitors of the 2-epimerases (NanE), which convert ManNAc-6-P to GlcNAc-6-P, and ManNAc-specific kinases (NanK) likely predate the split between gram-positive and gram-negative bacteria. In contrast, the distribution of NanA and its functional homologues does not follow the expected pattern of relationships seen for NanE and NanK (Fig. 5, orange panel). Thus, E. coli NanA does not cluster uniquely with gram-negative aldolases and appears not to share any standard or expected phylogenetic affinities with other aldolases. Not included in this analysis is the sialate aldolase from the unicellular eukaryote Trichomonas vaginalis, which is more similar at the primary structural level to the aldolases of H. influenzae and P. multocida than it is to E. coli NanA, suggesting horizontal transfer from a prokaryote donor to unicellular eukaryote recipient (43). Unlike bacterial sialate aldolases, the aldolase from T. vaginalis is a secreted enzyme (87), suggesting that T. vaginalis does not directly scavenge sialic acid but uses either or both of the breakdown products (ManNAc and pyruvate) for energy metabolism. Thus, it is possible that T. vaginalis lacks orthologues of either NanE or NanK if pyruvate is the preferred metabolite. The occurrence of NanA clearly related to certain bacterial aldolases appears to suggest that sialic acid metabolism by T. vaginalis may be important to its parasitic lifestyle. The horizontal transfer of nanA from a prokaryote to unicellular eukaryote is quite independent of the prokaryotic-to-vertebrate (or vice versa) transfer of nanA discussed above.
FIG. 5.
Phylogeny of selected sialic acid aldolase (NanA), epimerase (NanE), and kinase (NanK) polypeptides involved in microbial sialic acid catabolism. Colored blocks indicate phylogenetic relationships of the respective gene products as defined in the legend to Fig. 3. Primary structures were multiply aligned by using ClustalW and analyzed with the Phylogenetic Analyses and Reconstruct Phylogeny tools in MacVector version 7.2. The tree was constructed by using the neighbor-joining algorithm and absolute number of differences. Bootstrap values for 1,000 replicates are given as percentages at major nodes. Polypeptides are identified by their gene designation as they appear in Fig. 3.
Sialic Acid Uptake by NanT
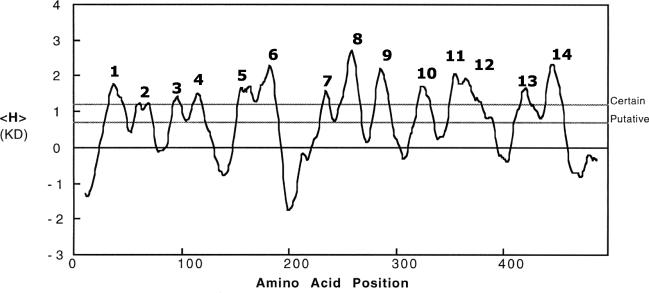
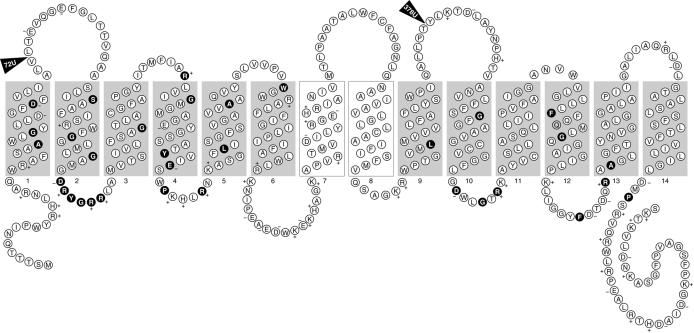
Mutants with defects in the nanT structural gene have only residual sialic acid uptake capacity, as shown by an inability to grow on sialic acid as the sole carbon source and a low rate of sialic acid transport (156, 157). These results suggest that NanT is the only sialic acid transporter in E. coli. The complete DNA sequence of nanT indicated that it encodes a transporter with 14 instead of the more usual 12 membrane-spanning domains found in most members of the major facilitator superfamily (82). As shown in Fig. 6, most of these 14 membrane-spanning domains are predicted with certainty by hydropathy analysis, although spanner 2 barely qualifies as an obvious spanning domain. Another model of NanT is based on conserved residues or motifs in the major facilitator superfamily (10, 56). Figure 7 shows the predicted secondary structure of NanT, where shaded regions indicate the 12 membrane-spanning domains and interconnecting loops containing the conserved residues shown in black. The two centrally located domains numbered 7 and 8 (unshaded) are not observed in other secondary transporters and thus lack conserved residues (82). Interestingly the amino acids comprising spanner 7 are predicted to form an amphipathic helix that may be important to the specificity of NanT for sialic acids. Direct evidence supporting the structure shown in Fig. 7 came from a survey of TnphoA insertions, two of which are shown in Fig. 7, that confirm the structure shown for the putative spanner 2 and spanners 9 and 10 (J. Hickman, S. Steenbergen, and E. Vimr, unpublished data). Saier (115) suggested that in addition to NanT, the sialate:H+ symporter subfamily 2.1.12 should include the S. cerevisiae lactate transporter encoded by JEN1 (26). We find no indication that the JEN1 product contains the unique NanT central domain, suggesting that if this domain is important for sialic acid uptake, NanT may recognize more than just the negative charge of its substrate(s).
FIG. 6.
Hydropathy analysis of NanT. E. coli NanT was analyzed for hydropathy (H) by using the Kyte-Doolittle (KD) index. Predicted membrane-spanning domains are indicated beginning from the N terminus.
FIG. 7.
Secondary structural model of NanT. The secondary structure of NanT was modeled on the basis of conserved residues in the major facilitator superfamily as described in the text. Amino acid residues (circles) are given as their single-letter designations, and where applicable the charge is also indicated. Black circles indicate conserved residues. Shaded rectangles indicate the 12 membrane-spanning domains in common with those of most superfamily members. The central, unshaded rectangles indicate those unique to NanT (82). Triangles indicate the site of TnphoA insertions with units of alkaline phosphatase activity.
The apparent affinity of E. coli NanT for sialic acid uptake, measured as Km for a nanA mutant, was estimated at 0.3 mM, a value similar to the Km of uptake by LacY for lactose transport (156). A similar analysis for a nanA+ stain produced a Km that was about 10-fold lower (107), suggesting that the active aldolase, by degrading internalized sialic acid, may have “pulled” uptake to account for the discrepancy in apparent uptake values in the two sets of studies. However, Rodriguez-Aparicio et al. (107) also concluded that sialic acid transport in E. coli involved a periplasmic component because of the apparent osmotic shock sensitivity of the system. The analysis of NanT shown in Fig. 6 and 7 clearly places it in the major facilitator superfamily, suggesting a nonspecific effect of the shock treatment on uptake. In contrast, the periplasmic components of TRAP transporters are expected to increase apparent affinity of uptake by locally increasing the solute concentration or through specific interactions with the membrane TRAP components (Fig. 4D). Certainly the uptake of sialic acid by H. influenzae, which is predicted to transport sialic acid by a TRAP system (Fig. 3B), is an efficient process as measured by chemical depletion assays (149). We suggest that the bacteria which use TRAP transporters for sialic acid uptake normally exist in an environment where sialic acid concentrations may be low or that there may be intense competition for free sialic acids by the microbiota sharing a common niche, such as the nasopharynx. However, it may be that the type (secondary or TRAP) of transporter is immaterial to fitness as long as specificity for sialic acid is maintained. Clearly much more work is required to understand the mechanisms of sialic acid uptake and the functional distinctions in terms of sialometabolism between secondary, TRAP, or ABC sialic acid transporters.
Role of Sialidases in Sialic Acid Catabolism
Sialidases (neuraminidases) are glycosylhydrolases that cleave the glycoketosidic linkages of Sia-O-acceptor substrates by an exohydrolytic reaction involving retention of configuration through a double-displacement mechanism (151). While sialidases are obviously encoded by the nan systems of V. cholerae (Fig. 3D) and S. pneumoniae (Fig. 3F), all strains of some organisms such as E. coli and H. influenzae appear to lack these enzymes, while other organisms such as P. multocida encode sialidases that are unlinked to nan (88). It is also possible that sialidases are infrequent traits within particular genera and depend on the choice of strain for identification, as in the case of the nanH gene in S. enterica (63).
Although the concentration of total sialic acid in humans serum is quite high (ca. 2 mM), almost all of it (99.9%) is bound to proteins or lipid acceptors under normal physiological conditions, making it unavailable to microbial metabolism (128). However, the low free sialic acid concentration in serum may simply reflect the rapid clearance of low-molecular-weight substances by the glomerular filtration barrier of the vertebrate kidney (51). Indeed, the concentration of free sialic acid in certain vertebrate tissues is dramatically greater than that of the bound nonulosonates, suggesting that a given animal may consist of subenvironments containing variable amounts of free sialic acids (6). In the colon of a vertebrate animal existing on a meat-rich diet, the high concentration of sialidase-positive commensals presumably allows sialidase-negative E. coli to successfully compete for free sialic acid (156). In contrast, an organism like V. cholerae colonizing a more rarefied intestinal environment presumably finds it advantageous to excrete its own sialidase for scavenging host sialoglycoconjugates from the small intestinal lumen (151). To compensate for loss of environmental sialic acid by diffusion, V. cholerae may need a high-affinity TRAP transporter to scavenge sialic acids released by its excreted sialidase (Fig. 3D). Similarly, H. influenzae (sialidase negative) and P. multocida (sialidase positive) are obligate commensals of the nasopharynx, an environment where free sialic acid concentrations may be low. A high-affinity TRAP transporter under these growth conditions may be beneficial to persistence. Other potential functions of microbial sialidases unrelated to nutrition, such as modulation of host innate immunity, have been discussed previously (148), as has the potential use of sialidases for interspecies competition between sialidase-positive and sialidase-negative bacteria occupying the same niche (124).
Although, as discussed above, the systemic concentration of free sialic acid is low, localized increases caused by inflammatory cues could create foci of highly concentrated free sialic acids independently of microbial sialidase action. This suggestion follows from observations that inflammatory neutrophils undergo an interleukin-8-inducible recruitment of intracellular sialidase(s) to the cell surface, where release of bound sialic acids from surface molecules and the surfaces of cells in the surrounding environment has the potential to raise local sialic acid concentrations (39). Thus, inflammation triggered by microbial products such as endotoxin (lipopolysaccharide [LPS]) may trigger events resulting in increased free sialic acids during an infection. Effective scavenging of these “endogenous” free sialates may involve the high-affinity uptake provided by TRAP or ABC transporters.
The sialidase gene, nanH, of S. enterica was the first chromosomal gene in this organism to be ascribed an origin by cross-kingdom horizontal gene transfer (63). Although the complete physiological function of nanH in the salmonellae is unknown, the evidence indicated that it was part of an active bacteriophage or phage remnant with homology to the lambdoid group (108). Subsequent studies showed that nanH is part of an intact prophage, Fels-1, that also encodes a novel superoxide dismutase (SodCIII), neither of which seems to be a component of the phage itself or involved in phage maturation (48). However, expression of nanH in S. enterica is high (ca. 2% of the total soluble protein), indicating that it is probably not involved in the prophage life cycle per se, because high expression continues during the lysogenic phase of the host-phage relationship (64). If nanH has no direct function in Fels-1 propagation, its persistent expression must confer some selective advantage to nanH-positive salmonellae. Possible functions include scavenging sialic acids for nutrition during colonization, systemic infection, or, possibly, the intracellular phase of infection and modulation of host cell surfaces after enzyme release following prophage induction. Indeed, enzyme release could be tied to the ordinary phage life cycle, whereby spontaneous prophage induction destroys a portion of the bacterial population but provides a benefit to the remaining population as a whole, perhaps by modifying effector responses of host inflammatory cells or molecules. Note that whereas nanH has been found only in LT2 isolates of S. enterica serovar Typhimurium, it is common (>50%) among S. enterica serovar Arizonae isolates (63).
S. enterica NanH was the first bacterial sialidase to have its structure solved in three dimensions (36), thereby supporting the sialidase superfamily hypothesis identifying the conserved active sites and overall architecture of the sialidase catalytic domain in other bacterial, viral, protozoan, and mammalian sialidases (63, 151). The enzyme is easy to purify in large amounts in its native form and is marketed by several commercial interests. The ease of genetic manipulation offered by S. enterica together with the availability of pure NanH and a three-dimensional structure should make it straightforward to determine its function in the host-bacterium interaction. Although the in vivo function of bacterial sialidase has been a topic of considerable speculation (54, 88, 139, 140), no definitive studies exist. From an evolutionary perspective, it is fascinating to speculate that phages are general mediators of horizontal gene transfer, a process that may be proceeding to this day at levels that are currently unknown but could be high (108). Thus, at the tertiary structural level S. enterica NanH may be more closely related to mammalian sialidases than it is to its bacterial counterparts, which would be consistent with cross-kingdom gene transfer (63). This putative relationship between a bacterial sialidase and a mammalian sialidase is not indicated by simple phylogenetic reconstruction (7), but the primary structures of most sialidases are poorly conserved, thus potentially blurring the true relationships among this group of enzymes.
SIALIC ACID SIGNALING
The prevalence of sialic acid in animals but scarcity in most other environments suggests that it would be an ideal substance for signaling that a microbe has entered an animal host or, if the microbe is an obligate commensal, then for sensing changes in the immediate surroundings in response to the localized free sialic acid concentration. There are at least two ways that a microorganism could sense the free sialic acid concentration: (i) binding of sialic acid extracellularly to a sensor-kinase of a regulatory system with two or more components or (ii) intracellular transport of sialic acid and modulation of a regulator by direct binding or binding of a metabolic breakdown product. The available evidence indicates that the second mechanism is used by at least E. coli to sense the environmental sialic acid concentration and respond accordingly through a global regulator encoded by nanR.
Regulation of nan Expression
The genetic organization of E. coli nan (Fig. 3A) is reminiscent of that of the lac system for lactose utilization, in which the regulator LacI is encoded by a poorly transcribed structural gene located immediately upstream of the lacZYA operon. Given this genetic organization and the initial evidence that the nan operon is inducible by sialic acid (156, 157), it was not surprising that nanR expressed in trans to a nanR null mutant repressed nanATEK-yhcH transcription (98). NanR thus behaves as a classical negative transcriptional regulator by blocking the binding of RNA polymerase or preventing activation of nan transcription from the nanA promoter (70). The operator sequence recognized by NanR includes three tandem repeats of the hexanucleotide sequence GGTATA (5′-GGTATAacaGGTATAaaGGTATA-3′) so that NanR (presumably as the homodimer) binds with a stoichiometry of from one to three molecules per molecule of DNA (70). Binding of more than one NanR molecule per operator is supported by DNase I footprint analysis, indicating that NanR protects a region of >30 bp corresponding to the three GGTATA repeats and overlapping the nanA −10 region and +1 transcriptional start site. No general consensus −35 region was identified, consistent with the need for an upstream catabolite activator protein (CAP) and positive regulation exerted from this site, as is typical for a class I promoter (70). NanR appears to be a member of the GntR-FadR superfamily of winged helix-turn-helix regulators, which currently includes over 250 members that repress or activate target genes involved in diverse metabolic processes (102). The superfamily was named after its first member, GntR, a repressor of gluconate catabolism in Bacillus subtilis (60). NanR is a member of the so-called FadR subfamily, which includes 40% of all members of the superfamily. The remaining regulators were placed into three other subfamilies on the basis of C-terminal similarities (102). FadR regulates fatty acid metabolism and is the only member of the superfamily whose structure has been solved in three dimensions cocrystallized with its operator (165). The exact mechanism of NanR binding is currently under investigation and should shed light on its evolutionary and functional relatedness to the superfamily.
Evidence that NanR responds to sialic acid binding comes from observations that sialic acid induces the nan operon in an aldolase-deficient mutant which is unable to initiate sialic acid breakdown, thereby preventing accumulation of the potential inducers ManNAc, pyruvate, and ManNAc-6-P, and endogenous induction by sialic acid produced biosynthetically in an E. coli K1 nanA mutant (156, 157). In addition, among a variety of sugars tested as potential inducers, only sialic acid was effective in vivo (70). In vitro evidence was equivocal but suggested that the thermodynamically disfavored α-isomer of sialic acid may bind NanR to cause the conformational change associated with derepression (70). Preliminary results suggest that sialic acid influences the equilibrium between NanR homodimers (active) and monomers (inactive) instead of simply perturbing the structure of the intact homodimer.
In addition to the function of NanR in regulating nan expression, the intergenic region between nanR and nanA includes a functional CAP site as noted above, a potential nitrogen-regulated site for binding the nitrogen assimilation control (NAC) protein, and two GATC Dam methylation sites, one overlapping the CAP site and another located immediately 5′ to the first tandem GGTATA repeat. Expression of nac is controlled by the alternative sigma factor NtrC, and in turn NAC either represses or activates genes involved in acquisition of nitrogen sources other than the preferred source, ammonia. NAC thus links the regulation of genes under control of the housekeeping sigma factor with NtrC. Operators activated by NAC have the consensus sequence ATA-N6-TNGTAT, while those repressed have a slightly different sequence, ATAA-N8-GAT (65). The potential nan NAC site, ATAAGCTTTCTGTAT, thus most closely resembles an activating operator, ATA-N6-CTGTAT, with one mismatch (underlined). Activation of nan transcription by NAC would make sense physiologically because sialic acid can serve as a sole nitrogen source in E. coli (82), suggesting that the NAC site in nan may be functional. However, microarray transcriptional analysis of NtrC-regulated genes did not provide evidence of nan induction or repression by nitrogen limitation (167).
The presence of overlapping GATC methylation sites is observed in a variety of different CAP sites, indicating that hemimethylation controls gene expression, as shown by microarray analysis of an E. coli dam mutant (92). In that study, nan expression was decreased in the dam mutant background, as expected from the need for positive control exerted by CAP binding (70). The presence of another GATC site located immediately upstream of the first tandem GGTATA repeat of the nan operator could affect NanR binding either positively or negatively. Expression of the nan operon thus is negatively regulated by NanR in response to sialic acid and positively regulated by CAP with hemimethylation functioning as an antiactivator. Additionally, NAC may still regulate nan under an unknown set of conditions, and potential hemimethylation of the GATC site proximal to the nan operator could affect the affinity of NanR binding.
NanR as a Global Regulator
To determine whether NanR regulates E. coli genes other than nan, we carried out microarray analysis under two sets of overlapping but not necessarily identical (in terms of outcome) conditions: (i) gene expression of an isogenic nanR null mutant compared to its wild-type parent and (ii) expression of wild-type genes from a culture grown on glycerol compared to the messages from a wild-type culture grown on sialic acid as sole carbon source (S. M. Steenbergen et al., unpublished data). Although we expected that there would be differences (either up or down) in expression of some genes in the two treatment groups, a subset of genes that included nan was affected in concert under the two sets of experimental conditions. Thus, in addition to nan, two apparent operons, yjhBC and yjhATS, were induced in the nanR mutant or by growth of the wild-type strain on sialic acid, indicating the probable direct involvement of NanR in regulating expression of these genes. Visual inspection of the putative transcriptional control regions upstream of the two yjh operons revealed potential operators that were nearly identical to that of the nan system, and relative mobility gel shift analysis with purified NanR confirmed specific binding to these sites (Steenbergen et al., unpublished data). The functions of the yjh gene products regulated by NanR are unlikely to be directly involved with sialic acid metabolism, because mutants with defects in the nan operon appear to have no residual growth on sialic acid. Alternatively, nan may be specific for Neu5Ac, while the gene products of the yjh operons might recognize other derivatives, such as Neu5Ac2en or any of a number of other sialic acids that are found in at least several percent of the total sialic acid concentration in human blood (24). Another possibility is that sialic acids signal that the bacteria are in a particular subenvironment where other sugars or metabolites unique to that environment also would be present. Coregulation by NanR for utilization of these other metabolites would make sense physiologically by allowing the according responses to be mediated through a single regulatory pathway involving release of gene expression from NanR repression. Computer-assisted analysis of the putative yjh gene products does not indicate the identity of these potential metabolites, although yjhB could encode a permease with homology to NanT and yjhA could encode a putative oligosugar-specific porin. The substrates of the Yjh polypeptides are thus likely to be carbohydrates or oligosaccharides.
Sialic acid also could act as an environmental signal through its immediate breakdown products, pyruvate and ManNAc. Thus, pyruvate by binding to the GntR-FadR family member PdhR would derepress aceEF (pyruvate dehydrogenase) expression and that of any other genes regulated by PdhR or other pyruvate response elements (100). Indirect regulation by sialic acid breakdown products would be NanR independent and likely to be observed only in cultures containing an active nan system exposed to an exogenous supply of sialic acid. Bacterial responses to sialic acid are therefore expected to be complex and to involve direct regulatory effects mediated by NanR and indirect effects exerted by PdhR and other regulators that may recognize ManNAc, ManNAc-6-P, or further-downstream products of sialic acid dissimilation such as the nagBA products (98). Results of microarray expression profiling indicate that expression of four dozen E. coli genes responds (either up or down) at least fourfold when cells are grown with sialic acid compared to glycerol. The use of cDNA derived from a glycerol-grown culture for these comparisons should exclude CAP as a major source of the observed differences in relative gene expression. Although we expect most of these changes in gene expression to be exerted through sialic acid breakdown products, it is possible that regulators other than NanR bind sialic acid. Even by limiting consideration to E. coli, the physiological responses to environmental sialic acid are proving to be highly complex, involving changes in expression of multiple genes either through the direct action of NanR or through indirect effects of sialic acid-derived metabolism on other regulatory proteins. Given the diversity of nan systems shown in Fig. 3, each organism is likely to maximize its responses to host sialic acid as a mechanism for colonization and persistence in vivo. Our initial results and the results of others (see below) are likely to be of general significance for understanding a range of host-pathogen-commensal interactions involving microbial responses to host-derived sialic acid. As an ubiquitous and frequently terminal sugar unit on most mammalian cell surfaces, sialic acid is one of the first and most prevalent molecules that a microbe would encounter when entering virtually any host subenvironment. The signaling functions of sialic acids in the host-microbe interaction are thus likely to have profound implications for microbial colonization, persistence, and, in certain cases, disease progression.
Communication between the host and resident microbiota mediated by monosaccharides is not unprecedented (62), suggesting a rich lexicon of chemical signals derived from the metabolism of host glycoconjugates. Furthermore, as discussed below, the host need not be a deaf bystander to the communication. Some bacteria scavenge host sialic acids and represent them in various oligosaccharide structural contexts at the cell surface, where host sialic acid-binding proteins can interpret these signals. This type of recognition by the host could ultimately prove inimical to the bacterium (67), but it is equally possible that bacteria can create a type of molecular cognitive dissonance in the host's ability to respond accordingly to the microbe (148).
Sialic Acid as a Regulator of Type 1 Fimbrial Phase Variation
The gene fimB, encoding a site-specific recombinase, and yjhATS are divergently transcribed from a 1.24-kb intergenic region that includes a putative operator with similarity (20 of 23 nucleotides) to the NanR-binding site located 5′ of the nanA structural gene (25). FimB regulates both on-to-off and off-to-on switching of type 1 fimbriae (fimAICDFGH) in E. coli and is the only recombinase capable of switching a cell from the afimbriate to the fimbriate state. Intergenic regions in E. coli K-12 tend to be short (averaging 118 bp [19]), and when they occur as longer stretches they are likely to function in genetic regulatory phenomena (109). The 1.24-kb grey hole (a region containing no discernible features) between E. coli fimB and the yjhATS operon has been noted as an unusually long noncoding region in what is otherwise a gene-dense organism.
Deletion of a region located >500 bp upstream from the fimB promoter removes the NanR-like binding site. This region appears to function as an antirepressor of fimB expression as measured by reporter fusion assays and effects of mutations in the region on fimbrial phase variation (45). Type 1 fimbriation is a known virulence factor in uropathogenesis (9, 34), a potential factor in Crohn's disease (23), and both necessary and sufficient for E. coli invasion of epithelial cells (83). El-Labany et al. (45) showed that sialic acid added exogenously to the growth medium reduced fimB expression, perhaps through an effect on hemimethylation of the cis-acting site mediated through one or more DNA-binding proteins. The effect of sialic acid in suppressing off-to-on phase variation of type 1 fimbriation in E. coli occurred at a concentration of sialic acid that is normally present in urine (45), suggesting a physiological role of sialic acid in controlling fimbrial phase variation during infection. Our results indicate that the effect of sialic acid on fimbrial phase variation could be mediated at least in part by NanR (70; Steenbergen et al., unpublished data). However, the control of type 1 fimbrial phase variation appears to involve more than just sialic acid, suggesting a potentially complex mechanism requiring other cis- and trans-acting factors (45).
Although we saw no evidence of changes in fimB expression (or of fimAICDFGH expression) in our microarray analyses (Steenbergen et al., unpublished data), the absolute effect of sialic acid on fimB expression was relatively low (45) and may not have been detected by our methods. In contrast, the effect of sialic acid or nanR deletion on expression of the divergently transcribed yjhATS operon was dramatic, confirming the importance of NanR to its expression. NanR thus appears to act as a classical repressor of nan, functioning in an antirepressor mechanism regulating fimB expression from a distance and as a repressor of yjhATS (and yjhBC) also by acting at a distance. The precise mechanism of NanR binding to its nanA operator and how it regulates promoters hundreds of base pairs from its binding site await investigation. Likely possibilities with precedent in other systems include DNA bending or looping to bring proteins into contact with other regulatory elements, supercoiling effects at downstream sites, or even oligomerization of bound effector molecules through cooperative binding that may mimic certain nucleoprotein structures.
We stress that NanR is found only in E. coli and very closely related bacteria such as S. enterica and Shigella spp. (70). Despite this limited distribution of NanR, potential regulatory molecules abound in the nan systems of less closely related gram-negative and gram-positive bacteria (Fig. 3), suggesting that there may be diverse mechanisms for sensing the host sialic acid concentration and responding accordingly. Many important regulatory phenomena involving sialic acids clearly remain to be elucidated in these organisms. The results of these studies should be of manifold importance to pathogenesis and for understanding the host-microbe interaction.
DECORATING THE CELL SURFACE WITH SIALIC ACID
Microorganisms that modify (decorate) their surfaces with sialic acids often masquerade as “self” to avoid, subvert, or inhibit host innate immunity (148). The selective advantage of cell surface sialic acid to microbial survival or persistence occurring during the elaborate host-microbe “minuet” is evidently so great that at least four distinct strategies of cell surface decoration have evolved. The biological functions of cell surface sialylation in the host-pathogen or commensal interaction have been reviewed previously (148), and that review should be consulted for topics not covered here.
De Novo Synthesis
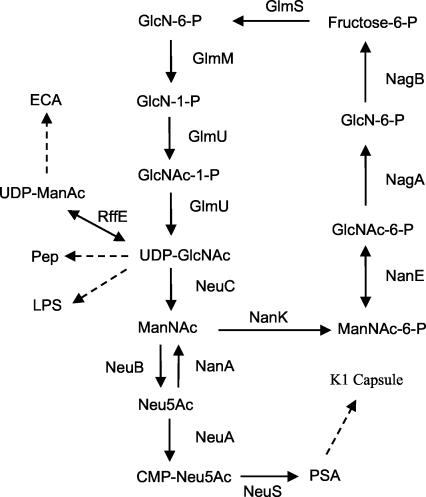
Bacterial sialic acid (as PSA) was discovered in the 1950s as the repeating subunit of the capsular polysaccharide of E. coli K1 (12). Much of our current understanding of capsule biosynthesis and synthesis of sialic acid itself has since come from analyses of this serotype or of group B meningococci, which synthesize an identical PSA capsule (150). Figure 8 shows that de novo sialic acid synthesis is thought to begin with the conversion of the common cell wall precursor UDP-GlcNAc to ManNAc by NeuC. The elimination reaction catalyzed by NeuC is thought to involve the formation of a 2-acetamidoglucal intermediate followed by the irreversible epimerization of this intermediate to ManNAc, as happens in mammalian systems (28). However, the NeuC orthologue, SiaA (SynX), in group B meningococci is postulated to be a GlcNAc-6-P epimerase that produces ManNAc-6-P, which would then be dephosphorylated by an unknown phosphatase (96). In either case, free ManNAc is produced as the obligate substrate for the condensing (ManNAc plus phosphoenolpyruvate) enzyme, NeuB (144, 152, 153).
FIG. 8.
Proposed pathways of sialic acid synthesis and degradation in E. coli. Metabolites of sialic acid degradation or synthesis are defined in the text. Note that cosubstrates and by-products are not indicated but can be found in reference 103. Abbreviations not defined in the text are as follows: ECA, enterobacterial common antigen; GlmS, GlcN-6-P synthase (l-glutamine:d-fructose-6-P amidotransferase); GlmU, GlcNAc-1-P uridyltransferase; RffE, UDP-GlcNAc epimerase. Dashed lines indicate that multiple reactions catalyze the indicated syntheses. The figure was adapted from reference 104.
Rodriguez-Aparicio et al. (106) and Ferrero et al. (47) have suggested that NanA synthesizes sialic acid in E. coli K1 as well as Mannheimia (formerly Pasteurella) haemolytica (11) by the condensation of ManNAc with pyruvate. Although NanA has long been known to carry out the condensation reaction under in vitro conditions (33), the suggestion that NanA is the sole enzyme of sialic acid biosynthesis in E. coli K1 is inconsistent with observations that NeuB is necessary for sialic acid biosynthesis in vivo (152, 153) and with the obvious function of the aldolase in sialic acid catabolism (156, 157). Once synthesized, free sialic acid is activated by CMP-sialic acid synthetase (NeuA), producing the obligate donor of sialic acid for all known prokaryotic and eukaryotic sialyltransferases. In E. coli K1, the biosynthetic genes neuDBACES (N-acetylneuraminic acid) appear to be transcribed constitutively, although an antitermination mechanism mediated by an unlinked gene encoding RfaH may help to coordinate capsule synthesis with the synthesis of LPS (161). The exact function of NeuD is unknown, but it is required for sialic acid synthesis (41), perhaps by stabilizing NeuB by direct heterooligomerization (40). There is no obvious orthologue of NeuD in group B meningococci; thus, de novo sialic acid biosynthesis in this organism appears to require only the orthologues of NeuB and NeuC. In mammals, the UDP-GlcNAc epimerase is homologous with NeuC (103), and the mammalian orthologue of NeuB uses ManNAc-6-P instead of free ManNAc for condensation with phosphoenolpyruvate. The sialic acid-9-P produced by mammalian NeuB must be dephosphorylated prior to activation by NeuA (111). Although the identity of this phosphatase is unknown, the C-terminal domain of mammalian NeuA is homologous with a phosphatase (YrbI) that has been shown to dephosphorylate KDO-8-P as a necessary prior step to CMP-KDO synthesis (164). Synthesis and activation of sialic acid and KDO thus appear to be mechanistically similar processes, as suggested by the phylogenetic relationships between some of the biosynthetic enzymes (7).
The synthesis and activation of Leg5Ac7Am and Pse5,7Ac appear to depend on orthologues of NeuABC. L. pneumophila neuA and neuB were shown to complement synthetase- and synthase-defective E. coli K1 mutants, respectively (80), while three separate neuB orthologues from C. jejuni were shown to complement the K1 synthase mutant (71). These complementation results are unusual because the hexosamine precursors of legionaminic and pseudaminic acids would appear to be structurally distinct from ManNAc, suggesting that orthologues of NeuC synthesize the hexosamine precursors, which can then serve as substrates for the NeuA and NeuB homologues. The differences between mammalian and bacterial de novo sialic acid synthesis noted above may reflect the varied uses of sialic acids in microbes for nutrition, for environmental signaling, and as sources of amino sugars for cell wall biosynthesis (98). It will now be interesting, and presumably straightforward, to determine the mechanism(s) of de novo sialic acid synthesis in unicellular fungi, where some studies have correlated the presence of sialic acid with a given strain's relative pathogenicity (1, 105).
Expression of the PSA capsule by E. coli K1 also depends on an intact neuE gene (154). Although the exact function of NeuE is unknown, the polypeptide was shown to include a C-terminal putative polyprenol-binding domain that would anchor NeuE to the inner membrane (133), where one of its functions could be to initiate PSA biosynthesis. However, transmission electron microscopy of thin sections from a neuE null mutant (Fig. 9) indicated intracellular accumulation of PSA, seen as lacunae of the type detected previously in capsule mutants with defects in PSA export (30). This translocation-defective phenotype of a neuE mutant would seem to exclude NeuE as a sialyltransferase, suggesting instead that it may function by coupling PSA synthesis to export. McGowen et al. also found no evidence for the proposed sialyltransferase function of NeuE (85). Alternatively, if the small amount of residual sialyltransferase activity in a neuS (polysialyltransferase) null mutant is carried out by NeuE, then it might function to initiate PSA biosynthesis (perhaps by transferring sialic acid to a lipid acceptor) in a translocation-competent form (133). Therefore, in the absence of NeuE, PSA may be synthesized but fail to engage the export apparatus, accumulating intracellularly as shown in Fig. 9. Zhou and Troy (166) have recently carried out an extensive in vitro study of the polyprenol-binding domain of NeuE, showing that this domain has the potential to function as a lipid-binding site. However, the relevance of these in vitro results to PSA biosynthesis is uncertain, in large part because the function or even participation of polyprenol in the in vivo polymerization process is unclear (132). The requirement in PSA biosynthesis for an initiator of some sort derives from the inability of NeuS to synthesize PSA when presented with exogenous CMP-Sia (133). In contrast, NeuS will elongate (albeit with weak processivity) oligosialic acid acceptors containing three or more sialic acid residues (131) and will efficiently elongate certain ganglioside (sialylated mammalian glycolipid) acceptors in vitro (27, 85). However, the fact remains that exogenous acceptors must already contain sialic acid, which does not help explain how at least the first sialic acid residue is added to the (unknown) endogenous acceptors in E. coli K1 PSA biosynthesis. Although some studies (reviewed by Troy [141]) have implicated polyisoprenol, phospholipid, or even protein as the endogenous acceptor, no definitive evidence yet addresses this important question.
FIG. 9.
Accumulation of intracellular PSA in an E. coli K1 neuE null mutant. In contrast to the case for the wild type (Fig. 2), the defect caused by loss of NeuE disrupts the export of PSA, which is found to accumulate in the cytoplasm, predominantly at one pole of the cells. This phenotype is observed in other E. coli K1 mutants with export defects.
Donor Scavenging
The second mechanism of cell surface sialylation was discovered in some strains of gonococci that cause the sexually transmitted disease gonorrhea in humans (94). Neisseria gonorrhoeae strains that use this method of sialylation lack NeuABC yet sialylate their surfaces with an extracellular sialyltransferase and CMP-Sia, which is present in small amounts as a normal constituent in human secretions. The gonococcal solution to cell surface sialylation thus involves a complete truncation of the precursor synthetic pathway, and this has so far been found only in N. gonorrhoeae. Whether N. gonorrhoeae once had the orthologues of neuABC and dispensed with them when it evolved an obligate sexually transmitted life style or whether it never had the precursor biosynthetic system and acquired just the sialyltransferase is not known. However, the gonococcal sialyltransferase is homologous with one of the Neisseria meningitidis sialyltransferases involved in lipooligosaccharide (LOS) sialylation (52) and has been definitively shown to be membrane bound and surface exposed, where it would have access to host CMP-Sia donor substrate (127).
trans-Sialidase
A third mechanism of cell surface sialylation was proposed first for the agent of American trypanosomiasis (Chagas' disease), Trypanosoma cruzi (99), and later for the African trypanosomes T. brucei gambiense and T. brucei rhodesiense (causing human sleeping sickness), T. brucei brucei, and T. congolense (causing Nagana, or African ruminant trypanosomiasis) (138) and the nasopharyngeal bacterium Corynebacterium diphtheriae (84). This mechanism of sialylation does not involve activated sialic acid or sialyltransferase, nor do any of these organisms, with the possible exception of C. diphtheriae, appear to synthesize or scavenge sialic acids. Instead, a surface sialidase(s) that normally transfers sialic acid from its acceptors to water (hydrolysis) may transfer it to a terminal galactosyl or N-acetylgalactosamine residue of an appropriate lactose or lactosamine acceptor, respectively. The trans-sialidase is capable of endogenous as well as host cell surface modification. Endogenous trans-sialidase acceptors are mucin-like molecules that when sialylated may protect bloodstream trypanosomes from innate (antibody-independent) immunity, whereas host cell surface remodeling of glycoproteins facilitates parasite adhesion and invasion (121, 122). T. cruzi trans-sialidase has also been shown to cause neuronal differentiation, stimulate interleukin-6 secretion from endothelial cells, and potentiate T-cell activation (29, 114, 138). In the African trypanosomes, trans-sialidase is expressed only in the insect form of the parasite and presumably protects it from gut proteases or complement components of an insect's blood meal (138). The function of the C. diphtheriae trans-sialidase either is unknown or has not been reported. The remodeling through sialylation of both the parasite and host cell surface glycoconjugates thus appears to be important to the virulence or persistence of vector-borne protozoan parasites, suggesting that trans-sialidase is an attractive target for potential new drug or vaccine development.
The crystal structure of trans-sialidase from T. cruzi and a comparison to the crystal structure of the Trypanosoma rangeli sialidase (which has only the hydrolase activity) indicate that the mechanism of trans-sialylation involves relatively few structural changes (amino acid substitutions) modifying the canonical sialidase active site (2). Thus, unlike most viral, bacterial, and (probably) mammalian sialidases, in which substrate binding does not trigger a dramatic conformational change, the T. cruzi sialidase active site undergoes structural changes triggered by substrate binding or hydrolysis that produces the binding site for the acceptor lactose or lactosamine moieties. Clearly it seems that trans-sialidase has the potential (through simple hydrolysis) to create its own novel acceptor-binding site (32). The mechanism of hydrolysis has recently been shown to involve a conserved tyrosine residue that functions in an unusual tyrosine-glutamate charge-relay system (163). This is the first instance where tyrosine has been unequivocally shown to function as a catalytic nucleophile and the first example of a glycosidase forming a glycoside instead of a glycosyl ester as a catalytic intermediate, suggesting that “some form of charge relay will probably prove to be a common feature among enzymes that process sialic and neuraminic acids” (163).
In addition to their conserved catalytic domains, a variety of bacterial and protozoan sialidases also include noncatalytic (often lectin-like) domains that could be involved in carbohydrate recognition and cell adhesion. For example, V. cholerae sialidase has two domains flanking the catalytic unit which share structural similarities with plant and animal lectins (35). These lectin-like domains could function in binding host cell glycoconjugates. Despite the a priori attractiveness of this idea, no evidence has been obtained demonstrating either ligand binding or that either of the lectin domains is capable of mediating cell adhesion. Other sialidase noncatalytic domains, such as those found in association with the P. multocida sialidases, are homologous to autotransporter polypeptides and could be important to enzyme secretion and surface localization (88). Thus, microbial sialidases may be excreted into the extracellular milieu, membrane bound, periplasmic, or cytoplasmic depending on the species in which they are expressed, consistent with their diverse functions in host-microbe interactions (151).
Precursor Scavenging
The most recent mechanism of cell surface sialylation, designated precursor scavenging (148), was elucidated through phenotypic analyses of H. influenzae nanA mutants (149). H. influenzae lacks NeuBC but encodes a NeuA orthologue as well as multiple sialyltransferases. As an obligate commensal and occasional pathogen capable of systemic or localized respiratory infections of the lungs and nasopharynx, it was hypothesized that H. influenzae scavenges free sialic acids from its host, presumably by using a TRAP transporter system (Fig. 3B), and then activates the internalized sialic acid with its NeuA orthologue and transfers sialic acid to LOS acceptors (148). LOSs are similar to LPS but lack the extended O-antigen chains (somatic antigens) of the latter type of gram-negative glycolipid.
Direct evidence that H. influenzae scavenges host sialic acid and uses it for cell surface decoration was recently obtained. Using the chinchilla model of otitis media, an inoculating dose of bacteria grown in the absence of sialic aid (unsialylated) was reisolated after animal passage and the LOS was characterized directly from recovered bacteria, unequivocally demonstrating LOS sialylation that must have resulted from scavenging, activation, and sialyltransfer in vivo (22). Mutants unable to sialylate due to defects in the CMP-Sia synthetase were profoundly attenuated for otitis media in the model, demonstrating the potential importance of cell surface sialylation to natural disease. However, chinchillas lack a bactericidal mechanism in the absence of exogenously added complement, suggesting that the function of sialylation as a disease factor in this model may have another explanation (22). One important alternative function may be to block phagocytosis through steric hindrance of charge repulsion conferred by the host-derived sialic acid expressed at the cell surface (136). Despite the ambiguous function of sialylation in the otitis media model, therapeutics aimed at blocking the TRAP transporter for sialic acid uptake may have utility in treating H. influenzae infections or other infections where the pathogen relies on precursor scavenging for the host-pathogen interaction. That the host-microbe interaction is dependent on precursor scavenging now seems certain for H. influenzae and may prove to be generally true for other microbes that scavenge host sialic acids (148).
REGULATION OF CELL SURFACE SIALYLATION
Although in principle the sialyltransferases or trans-sialidases of gonococci and protozoa, respectively, could be regulated by transcriptional mechanisms, the availability of CMP-Sia or sialylated glycoconjugate donors is probably the crucial feature determining the extent of cell surface decoration in these organisms. In contrast, meningococci and H. influenzae are known to phase vary between sialylated and unsialylated states (17, 59, 78, 134), whereas in E. coli K1 sialic acid is the inducer of its own catabolic system, creating the potential for a futile cycle in which the obligate PSA precursor is degraded by an inducible catabolic pathway (103). Similarly, H. influenzae is faced with the metabolic decision to degrade any scavenged sialic acid for nutrition or activate it for cell surface sialylation (149). Microorganisms have evolved various strategies for dealing with the need to regulate their cell surface sialylation.
Regulation of PSA Capsule Expression in E. coli K1
UDP-GlcNAc is the obligate common precursor for enterobacterial common antigen, LPS and LOS, peptidoglycan (Pep), and PSA biosynthesis (Fig. 8). Because PSA is not required for cell growth or survival, at least under most in vitro conditions, whereas LPS and Pep are, there would seem to be some mechanism to control the rate at which NeuC may tap the UDP-GlcNAc pool for PSA biosynthesis without threatening depletion for essential LPS or Pep biosynthesis. It is known that the LPS antiterminator RfaH also controls kpsMT-neuDBACES expression (130, 161), potentially to guard against overutilization of the UDP-GlcNAc pool for nonessential functions. As discussed above, the neu genes code for enzymes involved in sialic acid synthesis, activation, and polymerization, whereas kpsMT (for “kapsel” polysaccharide synthesis) encode the membrane and ATPase components of an ABC exporter for PSA extrusion (20). RfaH would thus appear to be a prime candidate for regulating cell wall polymer synthesis. However, the specific activities of neu-encoded enzymes appear to be constitutive throughout the E. coli K1 growth cycle, suggesting no limitation of UDP-GlcNAc production. In fact, the UDP-GlcNAc pool is among the highest among over 20 measurable metabolites in E. coli (81). Studies of GlmM (Fig. 8) suggest that this enzyme responds posttranslationally (autophosphorylation) to maintain the UDP-GlcNAc pool at a concentration in excess of that needed to ensure maximal cell growth and division (66, 86). Therefore, synthesis of PSA may not pose any threat to the cell's ability to maintain an adequate UDP-GlcNAc pool for most or all growth conditions.
E. coli K1 mutants with a null defect in neuA would be expected to accumulate free sialic acid (Fig. 8), but they do not because of nan induction and subsequent sialate breakdown catalyzed by NanA (103). This observation indicates that as a precursor of PSA capsule biosynthesis, sialic acid also functions as the inducer of its own catabolic system. How the cell avoids a futile cycle between synthesis and degradation of sialic acid was inferred to result from the tight coupling between sialic acid synthesis and activation such that free sialic acid normally never accumulates to a detectable level in the wild type (103, 104). In mammalian sialic acid biosynthesis, which involves a ManNAc-6-P intermediate, metabolic channeling is apparently accomplished by physically coupling two biosynthetic activities into one polypeptide (61). As indicated above, genetic evidence suggests physical interaction between NeuB and NeuD (41), suggesting that the entire process of sialic acid synthesis, activation, and polymerization may depend on a loosely organized “sialosome” to control PSA expression. Physical analysis of sialic acid flux has been aided by recent extension of analytical spectroscopic methods to the in situ measurement of metabolite pools (104).
The only environmental condition known to prevent PSA expression in E. coli K1 is growth at temperatures below about 30°C (31). This temperature effect is mediated through control of the kpsFEDUCS promoter by an incompletely described mechanism that may include integration host factor (30, 130). The kpsFEDUCS operon is thought to encode proteins (KpsED) that couple KpsMT to the outer membrane, a CMP-KDO synthetase (KpsU), and two enzymes (KpsCS) involved in lipidation of PSA (20, 90, 129, 150). Lipidation of the capsular PSA chains is thought to anchor them to the outer leaflet of the outer membrane through an acid-labile phosphodiester linkage to phosphatidic acid. Because a nonpolar mutation in kpsF reduced surface PSA by about 90%, KpsF was postulated to regulate capsule expression, although this effect did not seem to involve direct regulation of the kpsF promoter (30). Studies with group B meningococci subsequently indicated that KpsF is probably an isomerase involved in KDO precursor biosynthesis (143). However, KpsF also includes two so-called cystathionine-beta-synthase domains (13, 14). These domains are widespread in nature and are thought to function in a variety of regulatory phenomena by mediating protein-protein interactions. Because the role, if any, of KDO in PSA production is uncertain, KpsF may function primarily to modulate the rate of PSA synthesis by an as-yet-unknown mechanism. E. coli KpsF is membrane associated, where it could influence PSA expression by interacting with other components of the capsule synthesis and export apparatus (30).
Meningococcal PSA Phase Variation and LOS Sialylation
PSA appears to inhibit binding to and invasion of host cells but protects meningococci from innate immune mechanisms and is therefore necessary for systemic infection (158). Phase variation between capsular and acapsular states is postulated as a necessary step in meningococcal persistence and virulence (59). One mechanism controlling this phase variation is transposition of an insertion element into the genes for PSA expression (58). However, a more common mechanism appears to be slipped-strand mispairing in a poly(dC) tract of the capsule polymerase gene, siaD, causing premature translational arrest coupled with Rho-dependent termination (78). Evidence from the effects of the Rho inhibitor bicyclomycin on phase variation of a siaD reporter fusion expressed in a mismatch-defective E. coli host suggested coupling between transcriptional termination and the cell's mismatch repair system. This coupling between transcriptional termination and the mutator phenotype was suggested to account for the greater frequency of capsule switching from off to on than from on to off rate (78).
In addition to the PSA capsule, meningococci also sialylate their LOS, thereby imparting resistance to host innate immune mechanisms such as antibody-independent complement fixation (57, 69, 158). Given the diversity of mammalian siglecs and their specificity for distinct sialoglycoconjugates (38), it is likely that sialylated microorganisms interact with host siglec-expressing cells of the myeloid and lymphoid lineages (148). These interactions could modulate innate immunity by blocking phagocytosis or inhibiting the inflammatory response that is necessary for clearing extraintestinal bacteria. Paradoxically, siglecs were shown to increase uptake of LOS-sialylated meningococci by professional phagocytes (67). However, it was not clear whether the internalized bacteria were killed, leaving open the possibility that the siglec-mediated internalization facilitates bacterial spreading within the host. The wide variety of sialylated glycoconjugates expressed on different microbial cell surfaces suggests numerous opportunities for subverting host innate immunity through siglec-microbial sialoglycoconjugate interactions.
Regulation of Sialylated LOS Glycoforms in H. influenzae
LOSs are rough-type LPSs lacking the external O-antigen chains of smooth-type LPS. H. influenzae LOS seems to vary at random by using a common polymerase-stuttering mechanism involving strand slippage of tetranucleotide repeats in the 5′ ends of genes coding for glycosyltransferases or other proteins affecting the cell surface. This “Monte Carlo” strategy for achieving maximal variation presumably ensures the ability of H. influenzae to avoid extinction when faced with the problems of a small number of founder organisms and variation within and between its obligate human hosts (15, 134).
H. influenzae has three genes encoding LOS sialyltransferases designated Lic3A, LsgB, and SiaA (68). The functions of the sialylated glycoforms in the variant LOS synthesized are unknown but may involve differential adherence to epithelial cells (134). Loss of all sialylating capacity caused by a mutation in H. influenzae siaB (CMP-Sia synthetase) resulted in profound attenuation in the chinchilla model of otitis media (22). However, loss of sialylation shifts the overall population of LOS glycoforms, making it difficult to conclude that the attenuation of a siaB mutants is caused by the loss of sialylated cell surface molecules per se (68, 149). Thus, while the functions of the sialylated LOS glycoforms are unclear, sialylation is likely to profoundly affect the host-bacterium interaction of H. influenzae and other sialylated bacteria.
Synthesis of PSA Chemotypes
Chemotype refers to the type of glycoketosidic bond linking the repeating sialyl units of the PSA chains found in bacteria and mammals. Bacterial chemotypes may be either α2,8, as in E. coli K1, group B meningococci, M. haemolytica A2, or Moraxella nonliquefaciens; α2,9, as in group C meningococci; or repeating α2,8/2,9 linkages, as synthesized by E. coli K92 (44). In mammals, the α2,8 chemotype is an oncofetal antigen of the developing nervous system and in adult regions of the central nervous system where neuronal plasticity is maintained into adulthood (113). Similar polymers composed of repeating KDN residues have been detected in mammals, and there is no reason to believe that the α2,9 PSA chemotype does not exist in animals as well. The enzymes known as polysialyltransferases (polyST) are required for PSA chemotype biosynthesis in mammals and bacteria but show no obvious primary structural homologies, suggesting a remarkable functional convergence of glycosyltransferases that otherwise synthesize chemically identical polysaccharides (131).
An E. coli K1 mutant bearing a null defect in neuS contains all of the metabolic machinery for sialic acid synthesis and activation and the apparatus to modify (lipidate) and translocate PSA to the outer surface of the cell. If neuS is expressed in trans, PSA synthesis is restored and the mutant produces a normal extracellular PSA capsule. Using this system, Steenbergen and Vimr (131) showed that in trans expression of the neuS orthologues from E. coli K92 and groups B and C meningococci reconstituted the cognate capsules, demonstrating that polyST is the sole enzyme responsible for dictating chemotype in vivo. The construction and analysis of genetic hybrids of the E. coli K1 and K92 polyST structural genes indicated that the N-terminal domains of these enzymes function as critical determinants of linkage specificity, probably through their influence as part of the acceptor binding sites of these enzymes (131).
True Bacterial Protein Glycosylation
Most eukaryotes carry out some form of protein glycosylation by using a conserved N (asparagine)- or O (serine/threonine)-linked glycosylation system. Whereas O-linked modification of flagella with pseudaminic acid had been described (79), N-linked glycosylation was not thought to exist in eubacteria until it was reconstituted recently from Campylobacter spp. in E. coli (160). Because only the O-linked type of system uses sialic acid (Pse5,7Ac), the system with similarity to N-linked glycosylation will not be addressed here, but a discussion of it can be found in a recent review (135).
Currently flagellin is the only known protein target for modification with Pse5,7Ac in the Campylobacter spp. O-linked system. Total glycosylation of this protein is extensive (10% by weight), and the entire target and glycosylation system includes about 50 clustered genes (135). Orthologues of some of these genes are found in Helicobacter, Aeromonas, Clostridium, Caulobacter, and Methanococcus spp., suggesting that flagellin decoration may be a common modification. The genetic region responsible for pseudaminic biosynthesis contains genes encoding paralogues of sialic acid synthase (neuB2 and neuB3), synthetase (neuA2 and neuA3), and epimerase (neuC2), suggesting synthesis of at least two different structural derivatives. Phase variation of pseudaminic acids mediated by a slipped-strand mechanism will probably be shown to regulate the pathway, the function of which may be similar to that of fim described above. Thus, by altering glycosylation of an immunodominant antigen such as flagellin, the bacterial population may be able to avoid or at lest delay clearance by the host by altering its surface antigenicity. Whether sialylation of proteins other than flagellin occurs in other bacterial species awaits further investigation.
CONCLUSIONS AND FUTURE DIRECTIONS
The vanguard of research in sialobiology has always been in eukaryotic (mostly mammalian) systems. As few as 5 years ago the number of laboratories even tangentially involved in research on bacterial sialic acid metabolism was less than 10. The epithet that sialic acid “is just another sugar” frequently greeted attempts to establish research on microbial sialometabolism as a separate discipline. This review has documented at several levels, i.e., evolutionarily, structurally, genetically, biochemically, and physiologically, that this epithet is unsupported by the available evidence. To devotees of sialobiology, the reason for sialic acid's special position at the interface between host and microbe was obvious from the beginning (110). As terminal sugars of an animal's cellular glycocalyx, sialic acid is one of the first substances a microbe “sees” when entering a host. As a regulator of innate defense mechanisms (120), host sialic acid is an environmental substance to be copied (molecular mimicry), eaten (nutrition), or interpreted (cell signaling) so that different microbes may avoid clearance and, for some, probable extinction.
This review has attempted to make clear that genomics (DNA sequencing of entire microbial genomes) and rapid global screening methods such as gene expression analysis have deepened our appreciation of sialometabolism as a central feature of the host-microbe interaction. The challenge for the future will be to extend this knowledge, gained mostly from two decades of research on E. coli, to the wide range of other microbes (bacteria, fungi, and protozoans) that have incorporated various aspects of sialobiology into their individual life styles. Continued use of the standard tools of microbial genetics and physiology will remain essential for success, but increasingly the integration of sialometabolic research with the overall microbial response to the host environment will be necessary for further advances. Although the conceptual challenges to these integrated studies are great, the benefit will undoubtedly be a deepened understanding of the host-microbe interaction. Research on microbial sialometabolism also will continue to illuminate the many functions of sialic acids in complex metazoans. The combined results will have the potential to uniquely pinpoint and then target specific virulence attributes in many of the most significant pathogens still plaguing humans and their domesticated animals.
ADDENDUM IN PROOF
A relatively low-resolution (2.7 Å) crystal structure of the mouse NeuA (CMP-N-acetylneuraminic acid synthetase)-activating enzyme has been solved (S. Krapp, A. K. Munster-Kuhnel, J. T. Kaiser, R. Huber, J. Tiralongo, R. Gerardy-Schahn, and U. Jacob, J. Mol. Biol. 334:625-637, 2003). While the N-terminal catalytic domain of this enzyme is homologous with its E. coli NeuA counterpart, the mammalian enzyme’s C-terminal domain is suggested to be a phosphatase (mentioned above in “De novo synthesis”) that regulates synthetase activity through potential dephosphorylation of the N-terminal catalytic domain. However, Krapp et al. (2003) did not detect phosphate residues in the N-terminal domain and it seems equally valid to speculate that the putative phosphatase domain converts the Neu5Ac-9-P precursor to the obligate-free Neu5Ac substrate prior to the activation reaction (111). Because the pathway of sialic acid synthesis in E. coli does not include the phosphorylated Neu5Ac intermediate (Fig. 8), the bacterial synthetase would not require phosphatase.
H. influenzae lacks a de novo sialate biosynthetic pathway but decorates its surface using host-derived sialic acid (discussed under “Precursor scavenging”). Although this process is essential for virulence in the chinchilla model of otitis media (22), the exact in vivo function of cell surface sialylation is unknown. It was recently shown that an exogenous source of sialic acid is necessary for complete biofilm by H. influenzae in vitro and that a mutant defective in the CMP-sialate synthetase is attenuated for virulence in both the gerbil model of otitis media and the rat pulmonary challenge model (W. E. Swords, M. L. Moore, L. Godzicki, G. Bukofzer, M. J. Mitten, and J. VonCannon, Infect. Immun. 72:106-113, 2004). If host-derived sialic acid is essential for biofilm formation and the natural diseases caused by H. influenzae and other microbes that use TRAP transporters for scavenging environmental sialic acid (Fig. 3), inhibitors of sialate uptake have the potential to both treat acute infection and potentially block colonization as discussed under “Diversity of nan systems.”
Acknowledgments
We thank Jacqueline Plumbridge and Ian Blomfield for helpful discussions as well as all past and current members of the Laboratory of Sialobiology.
K.A.K. was supported in part by American Heart Association and Value-Added predoctoral fellowships. The support of research from the authors' laboratory by the National Institutes of Health and Governor's Venture Technology Individual Grants Program is gratefully acknowledged.
REFERENCES
- 1.Alviano, C. S., L. R. Travassos, and R. Schauer. 1999. Sialic acids in fungi: a minireview. Glycoconj. J. 16:545-554. [DOI] [PubMed] [Google Scholar]
- 2.Amaya, M. F., A. Buschiazzo, T. Nguyen, and P. M. Alzari. 2003. The high resolution structures of free and inhibitor-bound Trypanosoma rangeli sialidase and its comparison with T. cruzi trans-sialidase. J. Mol. Biol. 325:773-784. [DOI] [PubMed] [Google Scholar]
- 3.Ambrose, M. G., S. J. Freese, M. S. Reinhold, T. G. Warner, and W. F. Vann. 1992. 13C NMR investigation of the anomeric specificity of CMP-N-acetylneuraminic acid synthetase from Escherichia coli. Biochemistry 31:775-780. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, J. O., W. F. Doolittle, and C. L. Nesbo. 2001. Genomics. Are there bugs in our genome? Science 292:1848-1850. [DOI] [PubMed] [Google Scholar]
- 5.Angata, K., and M. Fukuda. 2003. Polysialyltransferases: major players in polysialic acid synthesis on the neural cell adhesion molecule. Biochimie 85:195-206. [DOI] [PubMed] [Google Scholar]
- 6.Angata, T., D. Nakata, T. Matsuda, K. Kitajima, and F. A. Troy II. 1999. Biosynthesis of KDN (2-keto-3-deoxy-d-glycero-d-galacto-nononic acid). Identification and characterization of a KDN-9-phosphate synthetase activity from trout testis. J. Biol. Chem. 274:22949-22956. [DOI] [PubMed] [Google Scholar]
- 7.Angata, T., and A. Varki. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102:439-469. [DOI] [PubMed] [Google Scholar]
- 8.Azuma, Y., A. Taniguchi, and K. Matsumoto. 2000. Decrease in cell surface sialic acid in etoposide-treated Jurkat cells and the role of cell surface sialidase. Glycoconj. J. 17:301-306. [DOI] [PubMed] [Google Scholar]
- 9.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079-1093. [DOI] [PubMed] [Google Scholar]
- 10.Baldwin, S. A. 1993. Mammalian passive glucose transporters: members of an ubiquitous family of active and passive transport proteins. Biochim. Biophys. Acta 1154:17-49. [DOI] [PubMed] [Google Scholar]
- 11.Barrallo, S., A. Reglero, B. Revilla-Nuin, H. Martinez-Blanco, L. B. Rodriguez-Aparicio, and M. A. Ferrero. 1999. Regulation of capsular polysialic acid biosynthesis by temperature in Pasteurella haemolytica A2. FEBS Lett. 445:325-328. [DOI] [PubMed] [Google Scholar]
- 12.Barry, G. T. 1957. The isolation of N-acetylneuraminic acid from colominic acid. Science 126:1230. [Google Scholar]
- 13.Bateman, A. 1999. The SIS domain: a phosphosugar-binding domain. Trends Biochem. Sci. 24:94-95. [DOI] [PubMed] [Google Scholar]
- 14.Bateman, A. 1997. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 22:12-13. [DOI] [PubMed] [Google Scholar]
- 15.Bayliss, C. D., D. Field, X. de Bolle, and E. R. Moxon. 2000. The generation of diversity by Haemophilus influenzae: response. Trends Microbiol. 8:435-436. [DOI] [PubMed] [Google Scholar]
- 16.Beau, J. M., R. Schauer, J. Haverkamp, J. P. Kamerling, L. Dorland, and J. F. Vliegenthart. 1984. Chemical behaviour of cytidine 5′-monophospho-N-acetyl-beta-d-neuraminic acid under neutral and alkaline conditions. Eur. J. Biochem. 140:203-208. [DOI] [PubMed] [Google Scholar]
- 17.Berrington, A. W., Y. C. Tan, Y. Srikhanta, B. Kuipers, P. van der Ley, I. R. Peak, and M. P. Jennings. 2002. Phase variation in meningococcal lipooligosaccharide biosynthesis genes. FEMS Immunol. Med. Microbiol. 34:267-275. [DOI] [PubMed] [Google Scholar]
- 18.Biswas, M., B. Singh, and A. Datta. 1979. Induction of N-acetylmannosamine catabolic pathway in yeast. Biochim. Biophys. Acta 585:535-542. [DOI] [PubMed] [Google Scholar]
- 19.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 20.Bliss, J. M., C. F. Garon, and R. P. Silver. 1996. Polysialic acid export in Escherichia coli K1: the role of KpsT, the ATP-binding component of an ABC transporter, in chain translocation. Glycobiology 6:445-452. [DOI] [PubMed] [Google Scholar]
- 21.Blix, G. 1936. Uber die kohlenhydratgruppen des submaxillarismucins. Z. Physiol. Chem. 240:43-54. [Google Scholar]
- 22.Bouchet, V., D. W. Hood, J. Li, J. R. Brisson, G. A. Randle, A. Martin, Z. Li, R. Goldstein, E. K. Schweda, S. I. Pelton, J. C. Richards, and E. R. Moxon. 2003. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. USA 100:8898-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boudeau, J., N. Barnich, and A. Darfeuille-Michaud. 2001. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39:1272-1284. [DOI] [PubMed] [Google Scholar]
- 24.Bulai, T., D. Bratosin, A. Pons, J. Montreuil, and J. P. Zanetta. 2003. Diversity of the human erythrocyte membrane sialic acids in relation with blood groups. FEBS Lett. 534:185-189. [DOI] [PubMed] [Google Scholar]
- 25.Burland, V., G. Plunkett III, H. J. Sofia, D. L. Daniels, and F. R. Blattner. 1995. Analysis of the Escherichia coli genome. VI. DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 23:2105-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casal, M., S. Paiva, R. P. Andrade, C. Gancedo, and C. Leao. 1999. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J. Bacteriol. 181:2620-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho, J. W., and F. A. Troy II. 1994. Polysialic acid engineering: synthesis of polysialylated neoglycosphingolipids by using the polysialyltransferase from neuroinvasive Escherichia coli K1. Proc. Natl. Acad. Sci. USA 91:11427-11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou, W. K., S. Hinderlich, W. Reutter, and M. E. Tanner. 2003. Sialic acid biosynthesis: stereochemistry and mechanism of the reaction catalyzed by the mammalian UDP-N-acetylglucosamine 2-epimerase. J. Am. Chem. Soc. 125:2455-2461. [DOI] [PubMed] [Google Scholar]
- 29.Chuenkova, M. V., and M. A. Pereira. 2001. The T. cruzi trans-sialidase induces PC12 cell differentiation via MAPK/ERK pathway. Neuroreport 12:3715-3718. [DOI] [PubMed] [Google Scholar]
- 30.Cieslewicz, M., and E. Vimr. 1997. Reduced polysialic acid capsule expression in Escherichia coli K1 mutants with chromosomal defects in kpsF. Mol. Microbiol. 26:237-249. [DOI] [PubMed] [Google Scholar]
- 31.Cieslewicz, M., and E. Vimr. 1996. Thermoregulation of kpsF, the first region 1 gene in the kps locus for polysialic acid biosynthesis in Escherichia coli K1. J. Bacteriol. 178:3212-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colman, P. M., and B. J. Smith. 2002. The trypanosomal trans-sialidase: two catalytic functions associated with one catalytic site. Structure (Cambridge) 10:1466-1468. [DOI] [PubMed] [Google Scholar]
- 33.Comb, D. G., and S. Roseman. 1960. The sialic acids. I. The structure and enzymatic synthesis of N-acetylneuraminic acid. J. Biol. Chem. 235:2529-2537. [PubMed] [Google Scholar]
- 34.Connell, I., W. Agace, P. Klemm, M. Schembri, S. Marild, and C. Svanborg. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. USA 93:9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crennell, S., E. Garman, G. Laver, E. Vimr, and G. Taylor. 1994. Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain. Structure 2:535-544. [DOI] [PubMed] [Google Scholar]
- 36.Crennell, S. J., E. F. Garman, W. G. Laver, E. R. Vimr, and G. L. Taylor. 1993. Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc. Natl. Acad. Sci. USA 90:9852-9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crocker, P. R. 2002. Siglecs: sialic-acid-binding immunoglobulin-like lectins in cell-cell interactions and signalling. Curr. Opin. Struct. Biol. 12:609-615. [DOI] [PubMed] [Google Scholar]
- 38.Crocker, P. R., and A. Varki. 2001. Siglecs, sialic acids and innate immunity. Trends Immunol. 22:337-342. [DOI] [PubMed] [Google Scholar]
- 39.Cross, A. S., S. Sakarya, S. Rifat, T. K. Held, B. E. Drysdale, P. A. Grange, F. J. Cassels, L. X. Wang, N. Stamatos, A. Farese, D. Casey, J. Powell, A. K. Bhattacharjee, M. Kleinberg, and S. E. Goldblum. 2003. Recruitment of murine neutrophils in vivo through endogenous sialidase activity. J. Biol. Chem. 278:4112-4120. [DOI] [PubMed] [Google Scholar]
- 40.Daines, D. A., and R. P. Silver. 2000. Evidence for multimerization of Neu proteins involved in polysialic acid synthesis in Escherichia coli K1 using improved LexA-based vectors. J. Bacteriol. 182:5267-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daines, D. A., L. F. Wright, D. O. Chaffin, C. E. Rubens, and R. P. Silver. 2000. NeuD plays a role in the synthesis of sialic acid in Escherichia coli K1. FEMS Microbiol. Lett. 189:281-284. [DOI] [PubMed] [Google Scholar]
- 42.Dawkins, R., and J. R. Krebs. 1979. Arms races between and within species. Proc. R. Soc. London B 205:489-511. [DOI] [PubMed] [Google Scholar]
- 43.de Koning, A. P., F. S. Brinkman, S. J. Jones, and P. J. Keeling. 2000. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol. Biol. Evol. 17:1769-1773. [DOI] [PubMed] [Google Scholar]
- 44.Devi, S. J., R. Schneerson, W. Egan, W. F. Vann, J. B. Robbins, and J. Shiloach. 1991. Identity between polysaccharide antigens of Moraxella nonliquefaciens, group B Neisseria meningitidis, and Escherichia coli K1 (non-O acetylated). Infect. Immun. 59:732-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Labany, S., B. K. Sohanpal, M. Lahooti, R. Akerman, and I. C. Blomfield. 2003. Distant cis-active sequences and sialic acid control the expression of fimB in Escherichia coli K-12. Mol. Microbiol. 49:1109-1118. [DOI] [PubMed] [Google Scholar]
- 46.Faillard, H. 1989. The early history of sialic acids. Trends Biochem. Sci. 14:237-241. [DOI] [PubMed] [Google Scholar]
- 47.Ferrero, M. A., A. Reglero, M. Fernandez-Lopez, R. Ordas, and L. B. Rodriguez-Aparicio. 1996. N-acetyl-d-neuraminic acid lyase generates the sialic acid for colominic acid biosynthesis in Escherichia coli K1. Biochem. J. 317:157-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-271. [DOI] [PubMed] [Google Scholar]
- 49.Forward, J. A., M. C. Behrendt, N. R. Wyborn, R. Cross, and D. J. Kelly. 1997. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J. Bacteriol. 179:5482-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 29:25-38. [DOI] [PubMed] [Google Scholar]
- 51.Gelberg, H., L. Healy, H. Whiteley, L. A. Miller, and E. Vimr. 1996. In vivo enzymatic removal of alpha 2→6-linked sialic acid from the glomerular filtration barrier results in podocyte charge alteration and glomerular injury. Lab. Investig. 74:907-920. [PubMed] [Google Scholar]
- 52.Gilbert, M., D. C. Watson, A. M. Cunningham, M. P. Jennings, N. M. Young, and W. W. Wakarchuk. 1996. Cloning of the lipooligosaccharide alpha-2, 3-sialyltransferase from the bacterial pathogens Neisseria meningitidis and Neisseria gonorrhoeae. J. Biol. Chem. 271:28271-28276. [DOI] [PubMed] [Google Scholar]
- 53.Gil-Serrano, A. M., M. A. Rodriguez-Carvajal, P. Tejero-Mateo, J. L. Espartero, M. Menendez, J. Corzo, J. E. Ruiz-Sainz, and A. C. A. M. Buendi. 1999. Structural determination of a 5-acetamido-3, 5, 7, 9-tetradeoxy-7-(3-hydroxybutyramido)-l-glycero-l-manno-nonulosonic acid-containing homopolysaccharide isolated from Sinorhizobium fredii HH103. Biochem. J. 342:527-535. [PMC free article] [PubMed] [Google Scholar]
- 54.Godoy, V. G., M. M. Dallas, T. A. Russo, and M. H. Malamy. 1993. A role for Bacteroides fragilis neuraminidase in bacterial growth in two model systems. Infect. Immun. 61:4415-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gottschalk, A. 1960. The chemistry and biology of sialic acids and related substances. University Press, Cambridge, United Kingdom.
- 56.Griffith, J. K., M. E. Baker, D. A. Rouch, M. G. Page, R. A. Skurray, I. T. Paulsen, K. F. Chater, S. A. Baldwin, and P. J. Henderson. 1992. Membrane transport proteins: implications of sequence comparisons. Curr. Opin. Cell Biol. 4:684-695. [DOI] [PubMed] [Google Scholar]
- 57.Hammerschmidt, S., C. Birkholz, U. Zahringer, B. D. Robertson, J. van Putten, O. Ebeling, and M. Frosch. 1994. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis. Mol. Microbiol. 11:885-896. [DOI] [PubMed] [Google Scholar]
- 58.Hammerschmidt, S., R. Hilse, J. P. van Putten, R. Gerardy-Schahn, A. Unkmeir, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192-198. [PMC free article] [PubMed] [Google Scholar]
- 59.Hammerschmidt, S., A. Muller, H. Sillmann, M. Muhlenhoff, R. Borrow, A. Fox, J. van Putten, W. D. Zollinger, R. Gerardy-Schahn, and M. Frosch. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211-1220. [DOI] [PubMed] [Google Scholar]
- 60.Hayden, D. J., and J. R. Guest. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 79:291-296. [DOI] [PubMed] [Google Scholar]
- 61.Hinderlich, S., R. Stasche, R. Zeitler, and W. Reutter. 1997. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J. Biol. Chem. 272:24313-24318. [DOI] [PubMed] [Google Scholar]
- 62.Hooper, L. V., and J. I. Gordon. 2001. Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology 11:1R-10R. [DOI] [PubMed]
- 63.Hoyer, L. L., A. C. Hamilton, S. M. Steenbergen, and E. R. Vimr. 1992. Cloning, sequencing and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Mol. Microbiol. 6:873-884. [DOI] [PubMed] [Google Scholar]
- 64.Hoyer, L. L., P. Roggentin, R. Schauer, and E. R. Vimr. 1991. Purification and properties of cloned Salmonella typhimurium LT2 sialidase with virus-typical kinetic preference for sialyl alpha 2-3 linkages. J. Biochem. (Tokyo) 110:462-467. [DOI] [PubMed] [Google Scholar]
- 65.Janes, B. K., C. J. Rosario, and R. A. Bender. 2003. Isolation of a negative control mutant of the nitrogen assimilation control protein, NAC, in Klebsiella aerogenes. J. Bacteriol. 185:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jolly, L., F. Pompeo, J. van Heijenoort, F. Fassy, and D. Mengin-Lecreulx. 2000. Autophosphorylation of phosphoglucosamine mutase from Escherichia coli. J. Bacteriol. 182:1280-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones, C., M. Virji, and P. R. Crocker. 2003. Recognition of sialylated meningococcal lipopolysaccharide by siglecs expressed on myeloid cells leads to enhanced bacterial uptake. Mol. Microbiol. 49:1213-1225. [DOI] [PubMed] [Google Scholar]
- 68.Jones, P. A., N. M. Samuels, N. J. Phillips, R. S. Munson, Jr., J. A. Bozue, J. A. Arseneau, W. A. Nichols, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2002. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J. Biol. Chem. 277:14598-14611. [DOI] [PubMed] [Google Scholar]
- 69.Kahler, C. M., and D. S. Stephens. 1998. Genetic basis for biosynthesis, structure, and function of meningococcal lipooligosaccharide (endotoxin). Crit. Rev. Microbiol. 24:281-334. [DOI] [PubMed] [Google Scholar]
- 70.Kalivoda, K. A., S. M. Steenbergen, E. R. Vimr, and J. Plumbridge. 2003. Regulation of sialic acid catabolism by the DNA binding protein NanR in Escherichia coli. J. Bacteriol. 185:4806-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karlyshev, A. V., D. Linton, N. A. Gregson, A. J. Lastovica, and B. W. Wren. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35:529-541. [DOI] [PubMed] [Google Scholar]
- 72.Kelly, D. J., and G. H. Thomas. 2001. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25:405-424. [DOI] [PubMed] [Google Scholar]
- 73.Kelm, S., and R. Schauer. 1997. Sialic acids in molecular and cellular interactions. Int. Rev. Cytol. 175:137-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klenk, E. 1941. Neuraminsaure, das spaltproduct eines neuen gehirnlipoids. Z. Physiol. Chem. 268:50-58. [Google Scholar]
- 75.Knirel, Y. A., N. A. Kocharova, A. S. Shashkov, and N. K. Kochetkov. 1986. The structure of the Pseudomonas aeruginosa immunotype 6 O-antigen: isolation and identification of 5-acetamido-3, 5, 7, 9-tetradeoxy-7-formamido-l-glycero-l-manno-nonulosonic acid. Carbohydr. Res. 145:C1-C4. [DOI] [PubMed] [Google Scholar]
- 76.Kooistra, O., E. Luneberg, Y. A. Knirel, M. Frosch, and U. Zahringer. 2002. N-Methylation in polylegionaminic acid is associated with the phase-variable epitope of Legionella pneumophila serogroup 1 lipopolysaccharide. Identification of 5-(N, N-dimethylacetimidoyl)amino and 5-acetimidoyl(N-methyl)amino-7-acetamido-3, 5, 7, 9-tetradeoxynon-2-ulosonic acid in the O-chain polysaccharide. Eur. J. Biochem. 269:560-572. [DOI] [PubMed] [Google Scholar]
- 77.Kundig, W., and S. Roseman. 1971. Sugar transport. II. Characterization of constitutive membrane-bound enzymes II of the Escherichia coli phosphotransferase system. J. Biol. Chem. 246:1407-1418. [PubMed] [Google Scholar]
- 78.Lavitola, A., C. Bucci, P. Salvatore, G. Maresca, C. B. Bruni, and P. Alifano. 1999. Intracistronic transcription termination in polysialyltransferase gene (siaD) affects phase variation in Neisseria meningitidis. Mol. Microbiol. 33:119-127. [DOI] [PubMed] [Google Scholar]
- 79.Logan, S. M., J. F. Kelly, P. Thibault, C. P. Ewing, and P. Guerry. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46:587-597. [DOI] [PubMed] [Google Scholar]
- 80.Luneberg, E., N. Zetzmann, D. Alber, Y. A. Knirel, O. Kooistra, U. Zahringer, and M. Frosch. 2000. Cloning and functional characterization of a 30 kb gene locus required for lipopolysaccharide biosynthesis in Legionella pneumophila. Int. J. Med. Microbiol. 290:37-49. [DOI] [PubMed] [Google Scholar]
- 81.Maharjan, R. P., and T. Ferenci. 2003. Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal. Biochem. 313:145-154. [DOI] [PubMed] [Google Scholar]
- 82.Martinez, J., S. Steenbergen, and E. Vimr. 1995. Derived structure of the putative sialic acid transporter from Escherichia coli predicts a novel sugar permease domain. J. Bacteriol. 177:6005-6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martinez, J. J., M. A. Mulvey, J. D. Schilling, J. S. Pinkner, and S. J. Hultgren. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 19:2803-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mattos-Guaraldi, A. L., L. C. Formiga, and A. F. Andrade. 1998. Trans-sialidase activity for sialic acid incorporation on Corynebacterium diphtheriae. FEMS Microbiol. Lett. 168:167-172. [DOI] [PubMed] [Google Scholar]
- 85.McGowen, M. M., J. Vionnet, and W. F. Vann. 2001. Elongation of alternating alpha 2, 8/2, 9 polysialic acid by the Escherichia coli K92 polysialyltransferase. Glycobiology 11:613-620. [DOI] [PubMed] [Google Scholar]
- 86.Mengin-Lecreulx, D., and J. van Heijenoort. 1996. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J. Biol. Chem. 271:32-39. [DOI] [PubMed] [Google Scholar]
- 87.Meysick, K. C., K. Dimock, and G. E. Garber. 1996. Molecular characterization and expression of a N-acetylneuraminate lyase gene from Trichomonas vaginalis. Mol. Biochem. Parasitol. 76:289-292. [DOI] [PubMed] [Google Scholar]
- 88.Mizan, S., A. Henk, A. Stallings, M. Maier, and M. D. Lee. 2000. Cloning and characterization of sialidases with 2-6′ and 2-3′ sialyl lactose specificity from Pasteurella multocida. J. Bacteriol. 182:6874-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nees, S., and R. Schauer. 1974. Induction of neuraminidase from Clostridium perfringens and the correlation of the enzyme with acylneuraminate pyruvate-lyase. Behring. Inst. Mitt. 55:68-78. [Google Scholar]
- 90.Nsahlai, C. J., and R. P. Silver. 2003. Purification and characterization of KpsT, the ATP-binding component of the ABC-capsule exporter of Escherichia coli K1. FEMS Microbiol. Lett. 224:113-118. [DOI] [PubMed] [Google Scholar]
- 91.Ophir, T., and D. L. Gutnick. 1994. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl. Environ. Microbiol. 60:740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oshima, T., C. Wada, Y. Kawagoe, T. Ara, M. Maeda, Y. Masuda, S. Hiraga, and H. Mori. 2002. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 45:673-695. [DOI] [PubMed] [Google Scholar]
- 93.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parsons, N. J., P. V. Patel, E. L. Tan, J. R. Andrade, C. A. Nairn, M. Goldner, J. A. Cole, and H. Smith. 1988. Cytidine 5′-monophospho-N-acetyl neuraminic acid and a low molecular weight factor from human blood cells induce lipopolysaccharide alteration in gonococci when conferring resistance to killing by human serum. Microb. Pathog. 5:303-309. [DOI] [PubMed] [Google Scholar]
- 95.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 96.Petersen, M., W. Fessner, M. Frosch, and E. Luneberg. 2000. The siaA gene involved in capsule polysaccharide biosynthesis of Neisseria meningitidis B codes for N-acylglucosamine-6-phosphate 2-epimerase activity. FEMS Microbiol. Lett. 184:161-164. [DOI] [PubMed] [Google Scholar]
- 97.Petter, J. G., and E. R. Vimr. 1993. Complete nucleotide sequence of the bacteriophage K1F tail gene encoding endo-N-acylneuraminidase (endo-N) and comparison to an endo-N homolog in bacteriophage PK1E. J. Bacteriol. 175:4354-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Plumbridge, J., and E. Vimr. 1999. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 181:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Previato, J. O., A. F. Andrade, M. C. Pessolani, and L. Mendonca-Previato. 1985. Incorporation of sialic acid into Trypanosoma cruzi macromolecules. A proposal for a new metabolic route. Mol. Biochem. Parasitol. 16:85-96. [DOI] [PubMed] [Google Scholar]
- 100.Quail, M. A., and J. R. Guest. 1995. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol. Microbiol. 15:519-529. [DOI] [PubMed] [Google Scholar]
- 101.Rabus, R., D. L. Jack, D. J. Kelly, and M. H. Saier, Jr. 1999. TRAP transporters: an ancient family of extracytoplasmic solute-receptor-dependent secondary active transporters. Microbiology 145:3431-3445. [DOI] [PubMed] [Google Scholar]
- 102.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 103.Ringenberg, M., C. Lichtensteiger, and E. Vimr. 2001. Redirection of sialic acid metabolism in genetically engineered Escherichia coli. Glycobiology 11:533-539. [DOI] [PubMed] [Google Scholar]
- 104.Ringenberg, M. A., S. M. Steenbergen, and E. R. Vimr. 2003. The first committed step in the biosynthesis of sialic acid by Escherichia coli K1 does not involve a phosphorylated N-acetylmannosamine intermediate. Mol. Microbiol. 50:961-975. [DOI] [PubMed] [Google Scholar]
- 105.Rodrigues, M. L., A. S. Dobroff, J. N. Couceiro, C. S. Alviano, R. Schauer, and L. R. Travassos. 2002. Sialylglycoconjugates and sialyltransferase activity in the fungus Cryptococcus neoformans. Glycoconj. J. 19:165-173. [DOI] [PubMed] [Google Scholar]
- 106.Rodriguez-Aparicio, L. B., M. A. Ferrero, and A. Reglero. 1995. N-acetyl-d-neuraminic acid synthesis in Escherichia coli K1 occurs through condensation of N-acetyl-d-mannosamine and pyruvate. Biochem. J. 308:501-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rodriguez-Aparicio, L. B., A. Reglero, and J. M. Luengo. 1987. Uptake of N-acetylneuraminic acid by Escherichia coli K-235. Biochemical characterization of the transport system. Biochem. J. 246:287-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roggentin, P., R. Schauer, L. L. Hoyer, and E. R. Vimr. 1993. The sialidase superfamily and its spread by horizontal gene transfer. Mol. Microbiol. 9:915-921. [DOI] [PubMed] [Google Scholar]
- 109.Rogozin, I. B., K. S. Makarova, D. A. Natale, A. N. Spiridonov, R. L. Tatusov, Y. I. Wolf, J. Yin, and E. V. Koonin. 2002. Congruent evolution of different classes of non-coding DNA in prokaryotic genomes. Nucleic Acids Res. 30:4264-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Roseman, S. 2001. Reflections on glycobiology. J. Biol. Chem. 276:41527-41542. [DOI] [PubMed] [Google Scholar]
- 111.Roseman, S., G. W. Jourdian, D. Watson, and R. Rood. 1961. Enzymatic synthesis of sialic acid 9-phosphates. Proc. Natl. Acad. Sci. USA 47:958-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roth, J., A. Kempf, G. Reuter, R. Schauer, and W. J. Gehring. 1992. Occurrence of sialic acids in Drosophila melanogaster. Science 256:673-675. [DOI] [PubMed] [Google Scholar]
- 113.Rutishauser, U. 1998. Polysialic acid at the cell surface: biophysics in service of cell interactions and tissue plasticity. J. Cell Biochem. 70:304-312. [DOI] [PubMed] [Google Scholar]
- 114.Saavedra, E., M. Herrera, W. Gao, H. Uemura, and M. A. Pereira. 1999. The Trypanosoma cruzi trans-sialidase, through its COOH-terminal tandem repeat, upregulates interleukin 6 secretion in normal human intestinal microvascular endothelial cells and peripheral blood mononuclear cells. J. Exp. Med. 190:1825-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saier, M. H., Jr. 1999. A proposal for classification of transmembrane transport proteins in living organisms, p. 61-136. In L. J. Van Winkle (ed.), Biomembrane transport. Academic Press, San Diego, Calif.
- 116.Saito, M., and A. Rosenberg. 1984. Identification and characterization of N-acetyl-2, 3-didehydro-2-deoxyneuraminic acid as a metabolite in mammalian brain. Biochemistry 23:3784-3788. [DOI] [PubMed] [Google Scholar]
- 117.Salzberg, S. L., O. White, J. Peterson, and J. A. Eisen. 2001. Microbial genes in the human genome: lateral transfer or gene loss? Science 292:1903-1906. [DOI] [PubMed] [Google Scholar]
- 118.Schauer, R. 2000. Achievements and challenges of sialic acid research. Glycoconj. J. 17:485-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schauer, R. 1982. Chemistry, metabolism, and biological functions of sialic acids. Adv. Carbohydr. Chem. Biochem. 40:131-234. [DOI] [PubMed] [Google Scholar]
- 120.Schauer, R. 1985. Sialic acids and their role as biological masks. Trends Biochem. Sci. 10:357-360. [Google Scholar]
- 121.Schenkman, R. P., F. Vandekerckhove, and S. Schenkman. 1993. Mammalian cell sialic acid enhances invasion by Trypanosoma cruzi. Infect. Immun. 61:898-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schenkman, S., D. Eichinger, M. E. Pereira, and V. Nussenzweig. 1994. Structural and functional properties of Trypanosoma trans-sialidase. Annu. Rev. Microbiol. 48:499-523. [DOI] [PubMed] [Google Scholar]
- 123.Schwarzkopf, M., K. P. Knobeloch, E. Rohde, S. Hinderlich, N. Wiechens, L. Lucka, I. Horak, W. Reutter, and R. Horstkorte. 2002. Sialylation is essential for early development in mice. Proc. Natl. Acad. Sci. USA 99:5267-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shakhnovich, E. A., S. J. King, and J. N. Weiser. 2002. Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: a paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect. Immun. 70:7161-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sharon, N., and W. Weis. 1998. Carbohydrates and glycoconjugates from cellulose and polysialic acids to the control of intracellular protein trafficking: new insights into carbohydrate structure and function. Curr. Opin. Struct. Biol. 8:545-547. [Google Scholar]
- 126.Shashkov, A. S., L. N. Kosmachevskaya, G. M. Streshinskaya, L. I. Evtushenko, O. V. Bueva, V. A. Denisenko, I. B. Naumova, and E. Stackebrandt. 2002. A polymer with a backbone of 3-deoxy-d-glycero-d-galacto-non-2-ulopyranosonic acid, a teichuronic acid, and a beta-glucosylated ribitol teichoic acid in the cell wall of plant pathogenic Streptomyces sp. VKM Ac-2124. Eur. J. Biochem. 269:6020-6025. [DOI] [PubMed] [Google Scholar]
- 127.Shell, D. M., L. Chiles, R. C. Judd, S. Seal, and R. F. Rest. 2002. The Neisseria lipooligosaccharide-specific alpha-2,3-sialyltransferase is a surface-exposed outer membrane protein. Infect. Immun. 70:3744-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sillanaukee, P., M. Ponnio, and I. P. Jaaskelainen. 1999. Occurrence of sialic acids in healthy humans and different disorders. Eur. J. Clin. Invest. 29:413-425. [DOI] [PubMed] [Google Scholar]
- 129.Silver, R. P., K. Prior, C. Nsahlai, and L. F. Wright. 2001. ABC transporters and the export of capsular polysaccharides from gram-negative bacteria. Res. Microbiol. 152:357-364. [DOI] [PubMed] [Google Scholar]
- 130.Simpson, D. A., T. C. Hammarton, and I. S. Roberts. 1996. Transcriptional organization and regulation of expression of region 1 of the Escherichia coli K5 capsule gene cluster. J. Bacteriol. 178:6466-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Steenbergen, S. M., and E. R. Vimr. 2003. Functional relationships of the sialyltransferases involved in expression of the polysialic acid capsules of Escherichia coli K1 and K92 and Neisseria meningitidis groups B or C. J. Biol. Chem. 278:15349-15359. [DOI] [PubMed] [Google Scholar]
- 132.Steenbergen, S. M., and E. R. Vimr. 1990. Mechanism of polysialic acid chain elongation in Escherichia coli K1. Mol. Microbiol. 4:603-611. [DOI] [PubMed] [Google Scholar]
- 133.Steenbergen, S. M., T. J. Wrona, and E. R. Vimr. 1992. Functional analysis of the sialyltransferase complexes in Escherichia coli K1 and K92. J. Bacteriol. 174:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Swords, W. E., P. A. Jones, and M. A. Apicella. 2003. The lipo-oligosaccharides of Haemophilus influenzae: an interesting array of characters. J. Endotoxin Res. 9:131-144. [DOI] [PubMed] [Google Scholar]
- 135.Szymanski, C. M., S. M. Logan, D. Linton, and B. W. Wren. 2003. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol. 11:233-238. [DOI] [PubMed] [Google Scholar]
- 136.Telang, S., E. Vimr, J. R. Mahoney, I. Law, H. Lundqvist-Gustafsson, M. Qian, and J. W. Eaton. 2001. Strain-specific iron-dependent virulence in Escherichia coli. J. Infect. Dis. 184:159-165. [DOI] [PubMed] [Google Scholar]
- 137.Thibault, P., S. M. Logan, J. F. Kelly, J. R. Brisson, C. P. Ewing, T. J. Trust, and P. Guerry. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276:34862-34870. [DOI] [PubMed] [Google Scholar]
- 138.Tiralongo, E., S. Schrader, H. Lange, H. Lemke, J. Tiralongo, and R. Schauer. 2003. Two trans-sialidase forms with different sialic acid transfer and sialidase activities from Trypanosoma congolense. J. Biol. Chem. 278:23301-23310. [DOI] [PubMed] [Google Scholar]
- 139.Tong, H. H., L. E. Blue, M. A. James, and T. F. DeMaria. 2000. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect. Immun. 68:921-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tong, H. H., M. James, I. Grants, X. Liu, G. Shi, and T. F. DeMaria. 2001. Comparison of structural changes of cell surface carbohydrates in the eustachian tube epithelium of chinchillas infected with a Streptococcus pneumoniae neuraminidase-deficient mutant or its isogenic parent strain. Microb. Pathog. 31:309-317. [DOI] [PubMed] [Google Scholar]
- 141.Troy, F. A., II. 1992. Polysialylation: from bacteria to brains. Glycobiology 2:5-23. [DOI] [PubMed] [Google Scholar]
- 142.Tsvetkov, Y. E., A. S. Shashkov, Y. A. Knirel, and U. Zahringer. 2001. Synthesis and identification in bacterial lipopolysaccharides of 5, 7-diacetamido-3, 5, 7, 9-tetradeoxy-d-glycero-d-galacto- and -d-glycero-d-talo-non-2-ulosonic acids. Carbohydr. Res. 331:233-237. [DOI] [PubMed] [Google Scholar]
- 143.Tzeng, Y. L., A. Datta, C. Strole, V. S. Kolli, M. R. Birck, W. P. Taylor, R. W. Carlson, R. W. Woodard, and D. S. Stephens. 2002. KpsF is the arabinose-5-phosphate isomerase required for 3-deoxy-d-manno-octulosonic acid biosynthesis and for both lipooligosaccharide assembly and capsular polysaccharide expression in Neisseria meningitidis. J. Biol. Chem. 277:24103-24113. [DOI] [PubMed] [Google Scholar]
- 143a.U.S. Army Medical Research and Materiel Command. May 1997. U.S. patent 5,631,283.
- 144.Vann, W. F., J. J. Tavarez, J. Crowley, E. Vimr, and R. P. Silver. 1997. Purification and characterization of the Escherichia coli K1 neuB gene product N-acetylneuraminic acid synthetase. Glycobiology 7:697-701. [DOI] [PubMed] [Google Scholar]
- 145.VanValen, L. 1973. A new evolutionary law. Evol. Theor. 1:1-30. [Google Scholar]
- 146.Van Winkle, L. J. 1999. Biomembrane transport. Academic Press, San Diego, Calif.
- 147.Varki, A. 2001. Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am. J. Phys. Anthropol. Suppl. 33:54-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vimr, E., and C. Lichtensteiger. 2002. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 10:254-257. [DOI] [PubMed] [Google Scholar]
- 149.Vimr, E., C. Lichtensteiger, and S. Steenbergen. 2000. Sialic acid metabolism's dual function in Haemophilus influenzae. Mol. Microbiol. 36:1113-1123. [DOI] [PubMed] [Google Scholar]
- 150.Vimr, E., S. Steenbergen, and M. Cieslewicz. 1995. Biosynthesis of the polysialic acid capsule in Escherichia coli K1. J. Ind. Microbiol. 15:352-360. [DOI] [PubMed] [Google Scholar]
- 151.Vimr, E. R. 1994. Microbial sialidases: does bigger always mean better? Trends Microbiol. 2:271-277. [DOI] [PubMed] [Google Scholar]
- 152.Vimr, E. R. 1992. Selective synthesis and labeling of the polysialic acid capsule in Escherichia coli K1 strains with mutations in nanA and neuB. J. Bacteriol. 174:6191-6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vimr, E. R., W. Aaronson, and R. P. Silver. 1989. Genetic analysis of chromosomal mutations in the polysialic acid gene cluster of Escherichia coli K1. J. Bacteriol. 171:1106-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vimr, E. R., and S. M. Steenbergen. 1993. Mechanisms of polysialic acid assembly in Escherichia coli K1: a paradigm for microbes and mammals, p. 73-91. In J. Roth, U. Rutishauser, and F. A. Troy II (ed.), Polysialic acid: from microbes to man. Birkhauser Verlag, Basel, Switzerland.
- 155.Reference deleted.
- 156.Vimr, E. R., and F. A. Troy. 1985. Identification of an inducible catabolic system for sialic acids (Nan) in Escherichia coli. J. Bacteriol. 164:845-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vimr, E. R., and F. A. Troy. 1985. Regulation of sialic acid metabolism in Escherichia coli: role of N-acylneuraminate pyruvate-lyase. J. Bacteriol. 164:854-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vogel, U., and M. Frosch. 1999. Mechanisms of neisserial serum resistance. Mol. Microbiol. 32:1133-1139. [DOI] [PubMed] [Google Scholar]
- 159.von Itzstein, M., W. Y. Wu, G. B. Kok, M. S. Pegg, J. C. Dyason, B. Jin, T. Van Phan, M. L. Smythe, H. F. White, S. W. Oliver, et al. 1993. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363:418-423. [DOI] [PubMed] [Google Scholar]
- 160.Wacker, M., D. Linton, P. G. Hitchen, M. Nita-Lazar, S. M. Haslam, S. J. North, M. Panico, H. R. Morris, A. Dell, B. W. Wren, and M. Aebi. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298:1790-1793. [DOI] [PubMed] [Google Scholar]
- 161.Wang, L., S. Jensen, R. Hallman, and P. R. Reeves. 1998. Expression of the O antigen gene cluster is regulated by RfaH through the JUMPstart sequence. FEMS Microbiol. Lett. 165:201-206. [DOI] [PubMed] [Google Scholar]
- 162.Warren, L. 1963. The distribution of sialic acids in nature. Comp. Biochem. Physiol. 10:153-171. [DOI] [PubMed] [Google Scholar]
- 163.Watts, A. G., I. Damager, M. L. Amaya, A. Buschiazzo, P. Alzari, A. C. Frasch, and S. G. Withers. 2003. Trypanosoma cruzi trans-sialidase operates through a covalent sialyl-enzyme intermediate: tyrosine is the catalytic nucleophile. J. Am. Chem. Soc. 125:7532-7533. [DOI] [PubMed] [Google Scholar]
- 164.Wu, J., and R. W. Woodard. 2003. Escherichia coli YrbI is 3-deoxy-d-manno-octulosonate 8-phosphate phosphatase. J. Biol. Chem. 278:18117-18123. [DOI] [PubMed] [Google Scholar]
- 165.Xu, Y., R. J. Heath, Z. Li, C. O. Rock, and S. W. White. 2001. The FadR DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli. J. Biol. Chem. 276:17373-17379. [DOI] [PubMed] [Google Scholar]
- 166.Zhou, G. P., and F. A. Troy II. 2003. Characterization by NMR and molecular modeling of the binding of polyisoprenols and polyisoprenyl recognition sequence peptides: 3D structure of the complexes reveals sites of specific interactions. Glycobiology 13:51-71. [DOI] [PubMed] [Google Scholar]
- 167.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]