Abstract
Sirtuins constitute a novel family of protein deacetylases and play critical roles in epigenetics, cell death, and metabolism. In spite of numerous experimental studies, the key and most complicated stage of its NAD+-dependent catalytic mechanism remains to be elusive. Herein by employing Born-Oppenheimer ab initio QM/MM molecular dynamics simulations, a state-of-the-art computational approach to study enzyme reactions, we have characterized the complete deacetylation mechanism for a sirtuin enzyme, determined its multistep free-energy reaction profile, and elucidated essential catalytic roles of the conserved dynamic cofactor binding loop. These new detailed mechanistic insights would facilitate the design of novel mechanism-based sirtuin modulators.
Keywords: ab initio QM/MM molecular dynamics simulation, enzyme catalysis, free energy and umbrella sampling, protein deacetylation, reaction mechanisms
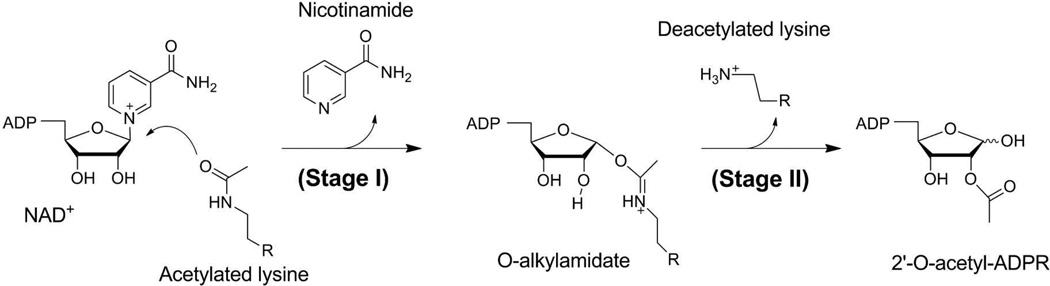
Sirtuins, also known as Sir2 (Silent information regulator 2), catalyse the NAD+-dependent protein post-translational modification, and have emerged as critical regulators of many cellular pathways.1–5 The abnormal sirtuin activity has been implicated in various diseases, including diabetes, obesity, neurodegenerative disorders and cancer.6–8 A thorough understanding of sirtuin chemistry is not only of fundamental importance, but also of high medicinal importance, since there is enormous current interest to develop new mechanism-based sirtuin modulators.9–13 The most common reaction catalyzed by sirtuin enzymes is the NAD+-dependent protein deacetylation.14 Its overall catalytic process has been suggested to proceed in two consecutive stages, as shown in Scheme 1.15, 16 The initial stage is relatively simple, which involves the cleavage of the nicotinamide and the nucleophilic attack for the formation of a positively charged O-alkylamidate intermediate.15, 17 Our previous ab initio QM/MM MD simulations of Sir2Tm, one of the best structurally characterized sirtuin homologues,18–21 determined that it employs a highly dissociative and concerted displacement mechanism.22 This computational characterization was subsequently confirmed by experimental kinetic isotope studies23 and another ab initio QM/MM study.24
Scheme 1.
The overall deacetylation process catalyzed by sirtuins.
The second stage of sirtuin catalysis, which includes the rate-determining step, is very complicated and has largely remained ambiguous.4, 9, 15, 25–30 Herein by employing Born–Oppenheimer ab initio QM/MM molecular dynamics with umbrella sampling, a state-of-the-art approach to simulate enzyme reactions, we have characterized the second stage of the deacetylation reaction catalyzed by Sir2Tm for the first time. The initial structure was modeled based on the X-ray crystal structure of Sir2Tm–S–alkylamidate intermediate complex (PDB ID: 3D81),20 which is the closest to the naturally occurring O-alkylamidate intermediate (INT1’) among all structures resolved.31 The QM sub–system was described at the B3LYP/6-31G(d) level and the QM/MM boundary was treated by the pseudobond approach with the improved parameters.32,33 All our QM/MM simulations have been carried out with modified Q-Chem34 and Tinker35 programs. This computational protocol has been successful in studying various enzyme systems and their catalytic mechanisms,22, 36–44 including the initial stage of Sir2Tm catalyzed deacetylation reactions.22 Computational details see supporting info.
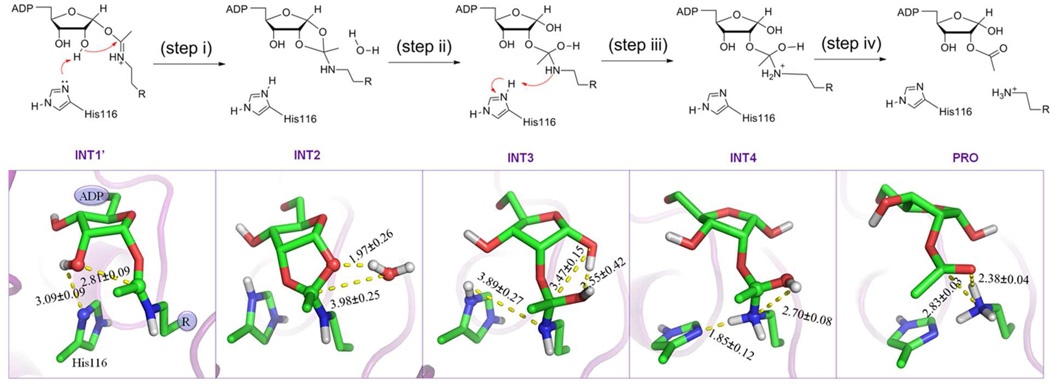
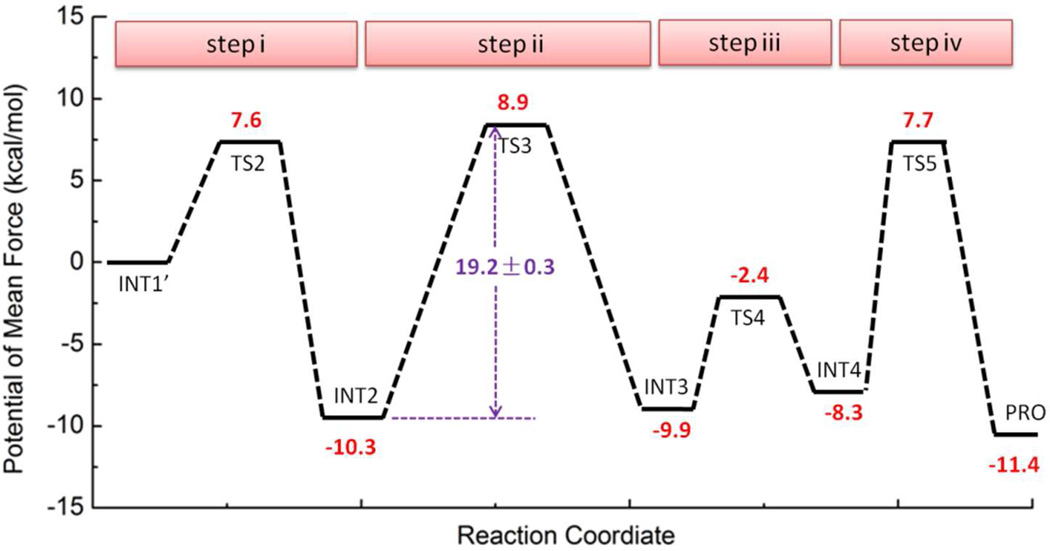
As illustrated in Figure 1, our computationally characterized second stage of the deacetylation reaction catalyzed by Sir2Tm proceeds in four consecutive steps: (i) formation of the bicyclic intermediate (INT2), in which His116 acts as a general base to facilitate the intra–molecular nucleophilic attack of the 2’ hydroxyl onto the positively charged iminium carbon; (ii) collapse of the bicyclic intermediate in the presence of water; (iii) proton transfer from the positively charged His116 to the imino group (−NH) of the tetrahedral intermediate; (iv) breakdown of the tetrahedral intermediate, in which a delocalized carbocation is formed from the cleavage of the indicated C–N bond before the proton-transfer. The determined complete free energy reaction profile is shown in Figure 2 and S1–4. For step iv, a 2-D free energy surface (Figure S4) has been determined since we found that the employment of 1-D reaction coordination is not sufficient to characterize this reaction step. We can see that among four steps, collapse of the bicyclic intermediate in the presence of water (step ii) is the rate-determining step. The calculated overall activation free energy barrier with B3LYP/6-31G(d) QM/MM simulations is 19.2 kcal mol−1, in good agreement with the experimental value of 18.6 kcal mol−1 estimated from the kcat value of 0.170 ± 0.006 s−1 for Sir2Tm with the transition state theory.45
Figure 1.
Reaction mechanism (top) and critical structures (bottom) determined for the second stage of the deacetylation reaction catalyzed by Sir2Tm. INT1': alkylamidate intermediate after nicotinamide release. INT2: bicyclic intermediate. INT3: tetrahedral intermediate. INT4: the protonated tetrahedral intermediate. PRO: 2'-O-AADPR and the deactylated product.
Figure 2.
Free energy profile for the second stage of the Sir2Tm deacetylation reaction, which is determined by B3LYP/6-31G(d) QM/MM molecular dynamics simulations and umbrella sampling. The statistical error is estimated by averaging the free energy difference between 10–20 ps and 20–30 ps.
In combination with our previously characterized reaction scheme for the initial stage with the very similar computational protocol,22 which has a calculated free energy barrier of 15.7 kcal/mol, we have determined the complete mechanism for the deacetylation reaction catalyzed by Sir2Tm and confirmed that the rate-determining step in the deacetylation reaction occurs after the acetyl group transfer. Besides that the determined free energy barriers are in excellent agreement with experimental kinetic studies,28 the simultaneous formation of the deacetylated product and 2’-O-AADPR is consistent with the experimental results that two products are released at the same time,46 and the direct participation of His116 in the second stage of deacetylation reaction is also supported by available mutation studies.29, 47 This mechanism can also easily account for the 18O incorporation studies,27, 29 in which 18O of the water is shown to be incorporated to be the carbonyl oxygen of the acetyl ester. Meanwhile, we have used ab initio QM/MM calculations to examine previously suggested alternative reaction schemes 4, 9, 26–29 as well as other potential mechanisms and found that they are energetically unfeasible (See supporting information part IV: Examination of other reaction schemes).
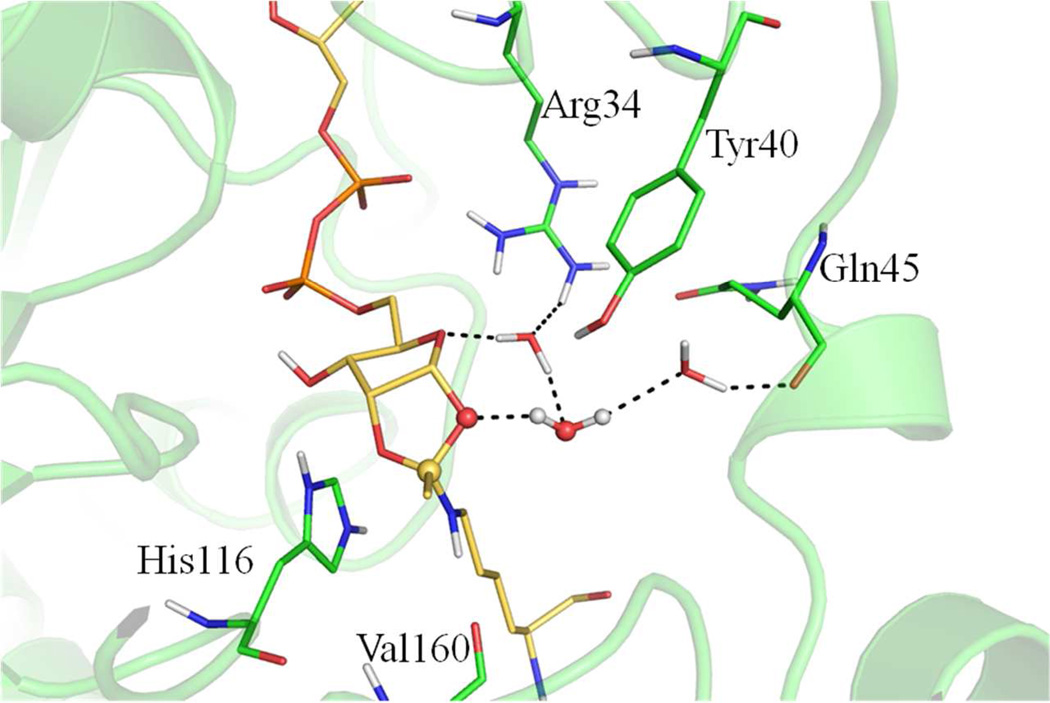
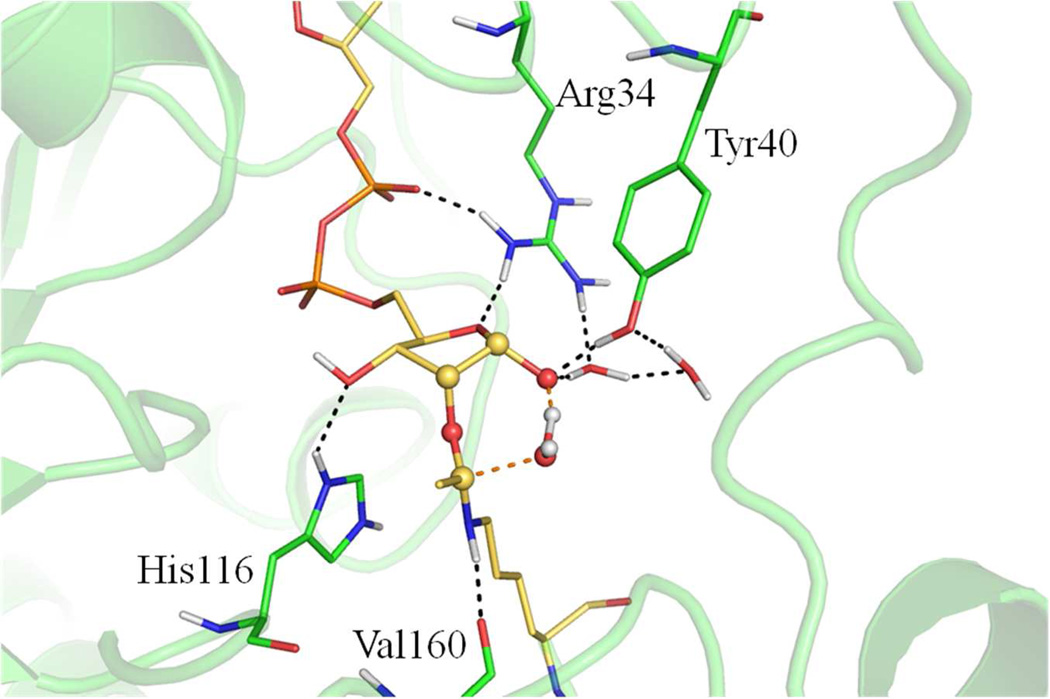
The collapse of the bicyclic intermediate (step ii) has long been the most mysterious part in the sirtuin deacetylation process, and our results highlighted it as the rate-limiting step. One very interesting finding is that during the classical MD simulation of the bicyclic intermediate, one water molecule comes into the active site through a water channel close to the dynamic cofactor binding loop and is positioned at a geometrically favored place to form a stable hydrogen bond with 1’O atom of the intermediate (Figure 3). This water molecule forms hydrogen bond with other adjacent solvent water molecules that interact with several residues in the cofactor binding loop, including Arg34 and Gln45, through a hydrogen bond network. Our ab initio QM/MM simulations indicate that the collapse of the bicyclic intermediate in the presence of the water employs a stepwise mechanism (Figure S2): First, the C-O1’ bond breaks to form a metastable intermediate at which both carbocation and oxyanion are stabilized by their interactions with the water; Second, the nucleophilic attack of water onto the carbocation is concerted with the proton transfer from the water to the O1’ atom, yielding the tetrahedral intermediate (INT3). At the transition state of the rate-determining step (TS3), we observed a 7-membered ring structure (see Figure 4), which is stabilized by forming hydrogen bonds with two conserved residues of the cofactor binding loop, Tyr40 and Arg34.
Figure 3.
Hydrogen bond network with the critical solvent water at the bicyclic intermediate (INT2) state. The C-O1’ bond which is breaking in step ii is highlighted with ball and stick representation.
Figure 4.
Hydrogen bond network at the transition state of the rate-limiting step (TS3). The 7-membered ring is highlighted with ball and stick representation.
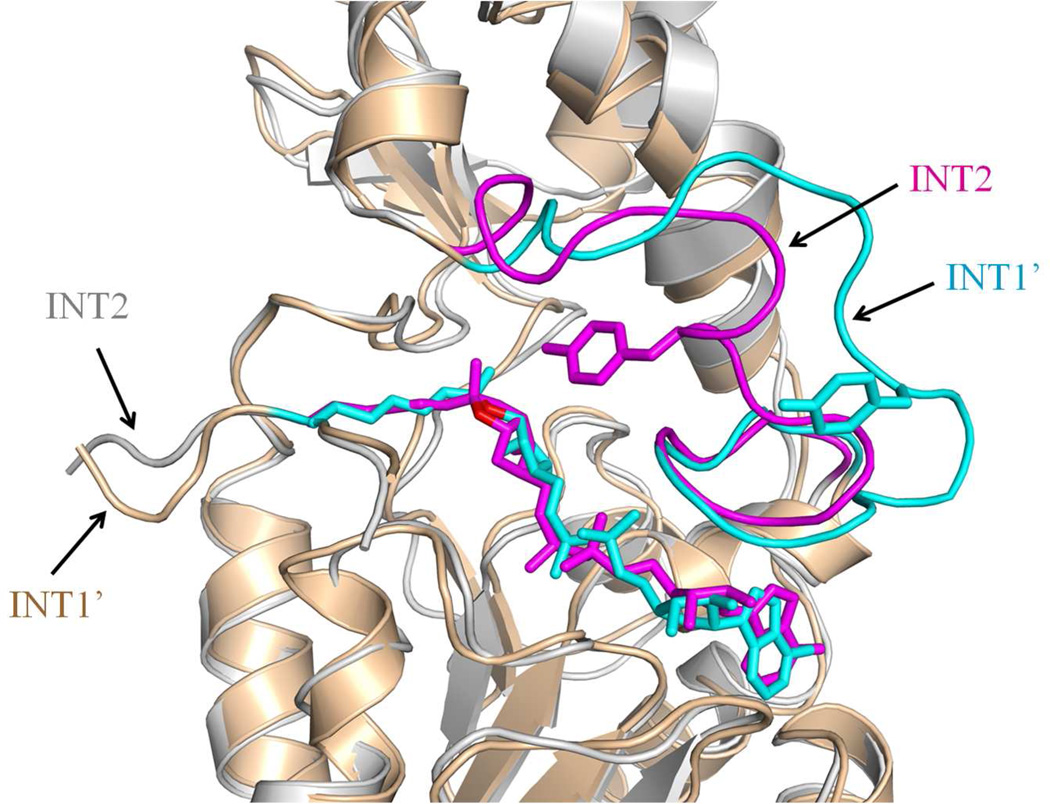
It should be noted that water molecules need to be shielded away from the O-alkylamidate intermediate state (INT1') to prevent the hydrolysis reaction which would yield an ADP ribose hydrolysis product. Based on the crystal structure study, it has been suggested that a cluster of conserved phenylalanines (Phe33, Phe48, and Phe162) play such an important shielding role.20 In our MD simulations of the INT1', we indeed found that orientation of these key residues remains stable and water molecules are prevented from being close to the peptidyl imidate, which significantly decrease the probability of hydrolysis (Figure S5 and S6). Then one intriguing question is how the water molecule can be well positioned to participate in the breakdown of the bicyclic intermediate (INT2). By a careful comparison between structural snapshots from simulations of INT1' and INT2, we have found that the most significant structural change is the conserved dynamic cofactor binding loop (residues 29 to 49), as shown in Figure 5. The loop movement shifts the positions of Phe33 and Phe48 to some extent that weakens their shielding function, while brings the conserved residue Tyr40 much closer to the bicyclic intermediate. Meanwhile, the formation of the bicyclic ring and the slight movement of Phe162 make some vacant space for water to come in (Figure S7). Due to the cooperation of all above factors, the water radial distribution at the INT2 state is completely different from that of INT1' (Figure S6), indicating that water is easily accessible to the bicyclic intermediate (INT2) while is shielded away from the O-alkylamidate intermediate state to prevent the hydrolysis.
Figure 5.
Superimposition of the active sites of INT1' (cyan) and INT2 (magenta). The representative snapshot of each state is taken at 20 ns of MD simulations, respectively. The intermediate and Tyr40 at both states are shown in sticks. The flexible loop region of Sir2Tm and O1’ atom of each intermediate are highlighted.
It is notable that this cofactor binding loop region (α2-α3 loop in Sir2Tm) has the most sequence homology in the sirtuin protein family, forms part of the cofactor NAD+ binding site, and is known to be highly flexible based on crystal structural studies.18–21, 48–52 Its importance on sirtuin deacetylation activity has in general been assumed to mainly come from its binding to the cofactor NAD+. Here our simulation results suggest that the observed high flexibility of this conserved cofactor binding loop plays an essential role in controlling the access of water molecules to reaction intermediates during the deacetylation. Furthermore, a more closed conformation of this loop at the INT2 state brings a highly conserved residue Tyr40 much closer to the bicyclic ring (Figure 5, S8). During the collapse of bicyclic intermediate step, Tyr40 moves even closer to the bicyclic ring, and forms a stable hydrogen bond with 1'O atom (Figure 4, S2); the OH(Tyr40)-O1' distance changes from 4.01±0.21Å at INT2 to 2.71±0.12Å at TS3. Thus our ab initio QM/MM MD simulations suggest that this Tyr40 residue plays an important role to facilitate the rate-liming step of the deacetylation process. In addition, we also noticed that another highly conserved loop residue Arg34 also forms a new hydrogen bond to catalyze the collapse of bicyclic intermediate (Figure 4, S2).
In summary, our state-of-the-art simulations have characterized the NAD+-dependent deacetylation catalyzed by a sirtuin enzyme for the first time, which proceeds with water access and participation at the bicyclic intermediate. The mechanism is consistent with current available experimental data and provides deeper insight into inner workings of sirtuin functioning. Meanwhile, our results elucidated the critical catalytic role of the conserved dynamic cofactor binding loop, in particular for Tyr40 and Arg34. In comparison with previous Born-Oppenheimer ab initio QM/MM molecular dynamics simulations of enzyme reactions, this multi-step reaction appears to be among the most complicated, and one reaction step (step iv) requires the determination of a two-dimensional free energy surface. Thus the present work further extends the applicability of this state-of-the-art computational approach.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH (R01-GM079223). We thank NYU-ITS and XSEDE for providing computational resources.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Figures S1–8; Complete citation for reference 34, 50, 51; Computational details; Examination of other reaction schemes. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Finkel T, Deng CX, Mostoslavsky R. Recent Progress in the Biology and Physiology of Sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haigis MC, Sinclair DA. Mammalian Sirtuins: Biological Insights and Disease Relevance. Annu. Rev. Pathol. Mech. Dis. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R. Are Sirtuins Viable Targets for Improving Healthspan and Lifespan? Nat. Rev. Drug Discov. 2012;11:443–461. doi: 10.1038/nrd3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The Biochemistry of Sirtuins. Annu. Rev. Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 5.Cen Y, Youn DY, Sauve AA. Advances in Characterization of Human Sirtuin Isoforms: Chemistries, Targets and Therapeutic Applications. Curr. Med. Chem. 2011;18:1919–1935. doi: 10.2174/092986711795590084. [DOI] [PubMed] [Google Scholar]
- 6.Uciechowska U, Sippl W, Jung M. NAD(+) -dependent Histone Deacetylases (Sirtuins) as Novel Therapeutic Targets. Med. Res. Rev. 2010;30:861–889. doi: 10.1002/med.20178. [DOI] [PubMed] [Google Scholar]
- 7.Houtkooper RH, Williams RW, Auwerx J. Metabolic Networks of Longevity. Cell. 2010;142:9–14. doi: 10.1016/j.cell.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakrabarty SP, Balaram H, Chandrasekaran S. Sirtuins: Multifaceted Drug Targets. Curr. Mol. Med. 2011;11:709–718. doi: 10.2174/156652411798062412. [DOI] [PubMed] [Google Scholar]
- 9.Hirsch BM, Zheng W. Sirtuin Mechanism and Inhibition: Explored with N(ε)-acetyllysine Analogs. Mol. BioSyst. 2011;7:16–28. doi: 10.1039/c0mb00033g. [DOI] [PubMed] [Google Scholar]
- 10.Dittenhafer-Reed KE, Feldman JL, Denu JM. Catalysis and Mechanistic Insights into Sirtuin Activation. Chembiochem. 2011;12:281–289. doi: 10.1002/cbic.201000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinke R, Carlino L, Kannan S, Jung M, Sippl W. Computer- and Structure-Based Lead Design for Epigenetic Targets. Bioorg. Med. Chem. 2011;19:3605–3615. doi: 10.1016/j.bmc.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 12.Asaba T, Suzuki T, Ueda R, Tsumoto H, Nakagawa H, Miyata N. Inhibition of Human Sirtuins by in Situ Generation of an Acetylated Lysine-ADP-Ribose Conjugate. J. Am. Chem. Soc. 2009;131:6989–6996. doi: 10.1021/ja807083y. [DOI] [PubMed] [Google Scholar]
- 13.Cen Y. Sirtuins Inhibitors: The Approach to Affinity and Selectivity. Biochim. Biophys. Acta. 2010;1804:1635–1644. doi: 10.1016/j.bbapap.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Hirschey MD. Old Enzymes, New Tricks: Sirtuins Are NAD(+)-dependent Deacylases. Cell Metab. 2011;14:718–719. doi: 10.1016/j.cmet.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauve AA. Sirtuin Chemical Mechanisms. Biochim. Biophys. Acta. 2010;1804:1591–1603. doi: 10.1016/j.bbapap.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith BC, Hallows WC, Denu JM. Mechanisms and Molecular Probes of Sirtuins. Chem. Biol. 2008;15:1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith BC, Denu JM. Sir2 Deacetylases Exhibit Nucleophilic Participation of Acetyl Lysine in NAD+ Cleavage. J. Am. Chem. Soc. 2007;129:5802–5803. doi: 10.1021/ja070162w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoff KG, Avalos JL, Sens K, Wolberger C. Insights into the Sirtuin Mechanism From Ternary Complexes Containing NAD(+) and Acetylated Peptide. Structure. 2006;14:1231–1240. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Cosgrove MS, Bever K, Avalos JL, Muhammad S, Zhang X, Wolberger C. The Structural Basis of Sirtuin Substrate Affinity. Biochemistry. 2006;45:7511–7521. doi: 10.1021/bi0526332. [DOI] [PubMed] [Google Scholar]
- 20.Hawse WF, Hoff KG, Fatkins DG, Daines A, Zubkova OV, Schramm VL, Zheng W, Wolberger C. Structural Insights into Intermediate Steps in the Sir2 Deacetylation Reaction. Structure. 2008;16:1368–1377. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bheda P, Wang JT, Escalante-Semerena JC, Wolberger C. Structure of Sir2Tm Bound to a Propionylated Peptide. Protein Sci. 2011;20:131–139. doi: 10.1002/pro.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu P, Wang S, Zhang Y. Highly Dissociative and Concerted Mechanism for the Nicotinamide Cleavage Reaction in Sir2Tm Enzyme Suggested By Ab Initio QM/MM Molecular Dynamics Simulations. J. Am. Chem. Soc. 2008;130:16721–16728. doi: 10.1021/ja807269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cen Y, Sauve A. A Transition State of ADP-ribosylation of Acetyllysine Catalyzed by Archaeoglobus Fulgidus Sir2 Determined by Kinetic Isotope Effects and Computational Approaches. J. Am. Chem. Soc. 2010;132:12286–12298. doi: 10.1021/ja910342d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Z, Shi T, Ouyang S, Li H, Yu K, Zhu W, Luo C, Jiang H. Investigation of the Catalytic Mechanism of Sir2 Enzyme with QM/MM Approach: SN1 vs SN2? J. Phys. Chem. B. 2010;114:11927–11933. doi: 10.1021/jp1054183. [DOI] [PubMed] [Google Scholar]
- 25.Avalos JL, Boeke JD, Wolberger C. Structural Basis for the Mechanism and Regulation of Sir2 Enzymes. Mol. Cell. 2004;13:639–648. doi: 10.1016/s1097-2765(04)00082-6. [DOI] [PubMed] [Google Scholar]
- 26.Khan AN, Lewis PN. Use of Substrate Analogs and Mutagenesis to Study Substrate Binding and Catalysis in the Sir2 Family of NAD-dependent Protein Deacetylases. J. Biol. Chem. 2006;281:11702–11711. doi: 10.1074/jbc.M511482200. [DOI] [PubMed] [Google Scholar]
- 27.Sauve AA, Celic I, Avalos J, Deng HT, Boeke JD, Schramm VL. Chemistry of Gene Silencing: The Mechanism of NAD(+)-dependent Deacetylation Reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 28.Sauve AA, Schramm VL. Sir2 Regulation by Nicotinamide Results from Switching Between Base Exchange and Deacetylation Chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 29.Smith BC, Denu JM. Sir2 Protein Deacetylases: Evidence for Chemical Intermediates and Functions of a Conserved Histidine. Biochemistry. 2006;45:272–282. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Zhang H, He B, Du J, Lin H, Cerione RA, Hao Q. The Bicyclic Intermediate Structure Provides Insights into the Desuccinylation Mechanism of Human Sirtuin 5 (SIRT5) J. Biol. Chem. 2012;287:28307–28314. doi: 10.1074/jbc.M112.384511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan H, Marmorstein R. Structural Basis for Sirtuin Activity and Inhibition. J. Biol. Chem. 2012;287:42428–42435. doi: 10.1074/jbc.R112.372300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Lee TS, Yang W. A Pseudobond Approach to Combining Quantum Mechanical and Molecular Mechanical Methods. J. Chem. Phys. 1999;110:46–54. [Google Scholar]
- 33.Zhang Y. Improved Pseudobonds for Combined Ab Initio Quantum Mechanical/Molecular Mechanical Methods. J. Chem. Phys. 2005;122:024114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 34.Shao Y, Fusti-Molnar L, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert ATB, Slipchenko LV, Levchenko SV, O'Neill DP, et al. Advances in Methods and Algorithms in a Modern Quantum Chemistry Program Package. Phys. Chem. Chem. Phys. 2006;8:3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 35.Ponder JW. Software Tools for Molecular Design, version 4.2. 2004. TINKER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu P, Wang S, Zhang Y. How Do SET-domain Protein Lysine Methyltransferases Achieve the Methylation State Specificity? Revisited by Ab Initio QM/MM Molecular Dynamics Simulations. J. Am. Chem. Soc. 2008;130:3806–3813. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ke Z, Wang S, Xie D, Zhang Y. Born-Oppenheimer Ab Initio QM/MM Molecular Dynamics Simulations of the Hydrolysis Reaction Catalyzed by Protein Arginine Deiminase 4. J. Phys.Chem. B. 2009;113:16705–16710. doi: 10.1021/jp9080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ke Z, Zhou Y, Hu P, Wang S, Xie D, Zhang Y. Active Site Cysteine Is Protonated in the PAD4 Michaelis Complex: Evidence from Born-Oppenheimer Ab Initio QM/MM Molecular Dynamics Simulations. J. Phys.Chem. B. 2009;113:12750–12758. doi: 10.1021/jp903173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Hu P, Zhang Y. Ab Initio Quantum Mechanical/Molecular Mechanical Moleculardynamics Simulation of Enzyme Catalysis: The Case of Histone Lysine Methyltransferase SET7/9. J. Phys.Chem. B. 2007;111 doi: 10.1021/jp067147i. 3758-37564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu R, Wang S, Zhou N, Cao Z, Zhang Y. A Proton-Shuttle Reaction Mechanism for Histone Deacetylase 8 and the Catalytic Role of Metal Ions. J. Am. Chem. Soc. 2010;132:9471–9479. doi: 10.1021/ja103932d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Wang S, Zhang Y. Catalytic Reaction Mechanism of Acetylcholinesterase Determined by Born-Oppenheimer Ab Initio QM/MM Molecular Dynamics Simulations. J. Phys. Chem. B. 2010;114:8817–8825. doi: 10.1021/jp104258d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ke Z, Smith GK, Zhang Y, Guo H. Molecular Mechanism for Eliminylation, a Newly Discovered Post-Translational Modification. J. Am. Chem. Soc. 2011;133:11103–11305. doi: 10.1021/ja204378q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Zhang Y. Serine Protease Acylation Proceeds with a Subtle Re-Orientation of the Histidine Ring at the Tetrahedral Intermediate. Chem. Commun. 2011;47:1577–1579. doi: 10.1039/c0cc04112b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lior-Hoffmann L, Wang L, Wang S, Geacintov NE, Broyde S, Zhang Y. Preferred WMSA Catalytic Mechanism of the Nucleotidyl Transfer Reaction in Human DNA Polymerase Kappa Elucidates Error-Free Bypass of a Bulky DNA Lesion. Nucleic. Acids Res. 2012;40:9193–9205. doi: 10.1093/nar/gks653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Avalos JL, Bever KM, Wolberger C. Mechanism of Sirtuin Inhibition by Nicotinamide: Altering the NAD(+) Cosubstrate Specificity of a Sir2 Enzyme. Mol. Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 46.Borra MT, Langer MR, Slama JT, Denu JM. Substrate Specificity and Kinetic Mechanism of the Sir2 Family of NAD(+)-dependent Histone/Protein Deacetylases. Biochemistry. 2004;43:9877–9887. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- 47.Min J, Landry J, Sternglanz R, Xu RM. Crystal Structure of a SIR2 Homolog-NAD Complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 48.Sanders BD, Jackson B, Marmorstein R. Structural Basis for Sirtuin Function: What We Know and What We Don't. Biochim. Biophys. Acta. 2010;1804:1604–1616. doi: 10.1016/j.bbapap.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finnin MS, Donigian JR, Pavletich NP. Structure of the Histone Deacetylase SIRT2. Nat. Struct. Bio. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 50.Schuetz A, Min J, Antoshenko T, Wang CL, Allali-Hassani A, Dong A, Loppnau P, Vedadi M, Bochkarev A, Sternglanz R, et al. Structural Basis of Inhibition of the Human NAD+-dependent Deacetylase SIRT5 by Suramin. Structure. 2007;15:377–389. doi: 10.1016/j.str.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 51.Jin L, Wei WT, Jiang YB, Peng H, Cai JH, Mao C, Dai H, Choy W, Bemis JE, Jirousek MR, et al. Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes. J. Biol. Chem. 2009;284:24394–24405. doi: 10.1074/jbc.M109.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao KH, Harshaw R, Chai XM, Marmorstein R. Structural Basis for Nicotinamide Cleavage and ADP-ribose Transfer by NAD(+)-dependent Sir2 Histone/Protein Deacetylases. Proc. Natl. Acad. Sci. USA. 2004;101:8563–8568. doi: 10.1073/pnas.0401057101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.