Abstract
The opportunistic pathogen Pseudomonas aeruginosa may cause both acute and chronic-persistent infections in predisposed individuals. Acute infections require the presence of a functional type III secretion system (T3SS), while chronic P.aeruginosa infections are characterized by the formation of drug-resistant biofilms. The T3SS and biofilm formation are reciprocally regulated by the signaling kinases LadS, RetS, and GacS. RetS down-regulates biofilm formation and up-regulates expression of the T3SS through a unique mechanism. RetS forms a heterodimeric complex with GacS and thus prevents GacS autophosphorylation and downstream signaling. The signals that regulate RetS are not known but RetS possesses a distinctive periplasmic sensor domain that is believed to serve as receptor for the regulatory ligand. We have determined the crystal structure of the RetS sensory domain at 2.0 Å resolution. The structure closely resembles those of carbohydrate binding modules of other proteins, suggesting that the elusive ligands are likely carbohydrate moieties. In addition to the conserved beta-sandwich structure the sensory domain features two alpha helices which create a unique surface topology. Protein-protein cross-linking and fluorescence energy transfer experiments also revealed that the sensory domain dimerizes with a dissociation constant of Kd=580±50 nM, a result with interesting implications for our understanding of the underlying signaling mechanism.
Keywords: RetS, type III secretion, biofilm formation, sensor kinase, sensory domain, carbohydrate binding, signal transduction, two-component system, periplasmic domain
Introduction
The Gram-negative bacterium Pseudomonas aeruginosa poses a major challenge to the medical community. Antibiotic-resistant strains of this pathogen may cause chronic-persistent as well as acute infections in transplant patients and other immunocompromised individuals 1-5. Most prominently, chronic P.aeruginosa-associated lung infections are the leading cause of mortality among people with cystic fibrosis 6.
Beyond its ability to colonize human tissues P.aeruginosa can persist in a variety of other milieus including plants and soil. This remarkable versatility may be attributed to a diverse array of virulence mechanisms, which enable the bacterium to adapt to vastly different environmental challenges. Expression and activation of these virulence mechanisms are carefully controlled by complex regulatory networks to ensure an optimal adaptive response. The type III secretion system (T3SS), for instance, is a hallmark of acute infections but is not active in chronic infections 7,8, which are instead characterized by the formation of antibiotic-resistant biofilms 9.
The two-component signaling system (TCS) is the primary means of bacteria to translate complex environmental cues into adaptive gene expression patterns. A canonical TCS is composed of a histidine kinase (HK) and a cognate receiver response regulator (RR). Signaling in response to a stimulus involves HK autophosphorylation and subsequent transfer of the phosphate group to a cognate RR. Frequently, the receiver proteins are transcription factors where the RR domains are coupled with DNA binding domains. Phosphorylation and dephosphorylation control gene expression by modulating the affinity of these transcription factors for their DNA binding sites.
P. aeruginosa harbors a particularly broad array of over 60 TCSs to modulate its gene expression10, including the expression of genes related to the T3SS and biofilm formation. Remarkably, T3SS and biofilm formation are regulated in a coordinated but reciprocal fashion by the signaling kinases RetS, LadS, and GacS 11-13. RetS and LadS are hybrid sensor kinases, combining both HK and RR domains in a single polypepetide, while GacS is a canonical signaling kinase requiring the response regulator GacA for downstream signaling. RetS is pivotal for the transcription of genes associated with cytotoxicity and acute infections, including the T3SS. On the other hand, a retS mutant displayed a hyperadhesive phenotype and showed elevated levels of transcription for the psl and pel operons which are associated with the synthesis of biofilm oligosaccharides, suggesting that RetS down-regulates biofilm formation in the wild-type strain 10,11. RetS accomplishes its task by blocking the synthesis of RsmZ, a small regulatory RNA. RsmZ had previously been shown to bind to and sequester the translational repressor RsmA. Free RsmA blocks biofilm formation and favors the expression of the T3SS 14,15. The kinases LadS and GacS, on the other hand, directly counteract RetS, as they stimulate the expression of the psl and pel operons by up-regulating the expression of the RsmA-antagonist RsmZ 16,17. Underlying this reciprocal regulation of the T3SSS and biofilm formation is an entirely novel regulatory mechanism that was uncovered in a recent study by Goodman et al. and involves direct contacts between RetS and GacS 18. Signaling kinases are usually homodimeric and autophosphorylation occurs almost always in trans. According to the proposed model an unknown environmental signal causes the RetS homodimer to dissociate to form a heterodimeric RetS-GacS complex. The asymmetric geometry of the heterodimer is thought to block RsmZ biosynthesis by preventing GacS trans-autophosphorylation. Remarkably, neither the kinase activity nor the RR domains of RetS are required for its unusual interactions with GacS 18.
Although the signal that causes RetS-GacS hetero-dimer formation is unknown, the periplasmic RetS sensory domain is believed to serve as the receptor for the elusive ligand. The sensory domain encompasses about 160 amino acids and belongs to the large but poorly characterized 7TM-DISM2 domain family, which has been hypothesized to constitute a new class of carbohydrate binding proteins 19. However, experimental support for this model is lacking. The here reported crystal structure of the RetS sensory domain not only offers insights into the nature of the molecular signal but it represents the first reported structure for any member of this domain family. Dimerization of the sensory domain and the stability of the dimer were also examined because the currently held model assumes a mechanism where ligand binding shifts the equilibrium from RetS and GacS homo-dimers to a RetS-GacS complex.
Experimental Procedures
Cloning, expression, and purification of the RetS sensory domain
The boundaries of the RetS sensory domain, containing amino acids 27 to 185, are defined by two transmembrane helices. The gene fragment coding for the entire periplasmic domain (retS27-185) was PCR-amplified from P. aeruginosa genomic DNA (ATCC 17933D). During PCR a tobacco etch virus (TEV) protease recognition site and the appropriate recombination sites (attB1 and attB2) were added to retS27-185. Subsequently, the amplicon was recombined into pDONR201 (Invitrogen) to produce the plasmid pDONR201-retS27-185. After verifying the nucleotide sequence via DNA sequencing, retS27-185 was recombined into the destination vector pDEST-HisMBP 20 to create the expression vector pDEST-HMBP-retS27-185. This vector is designed to produce RetS27-185 fused to the carboxy-terminal end of amino-terminally hexahistidine-tagged E. coli maltose binding protein (MBP).
Vector pDONR201-retS41-185, containing a shortened segment of the sensory domain, was generated via PCR with the appropriate primer and using pDONR201-retS27-185 as a template. Following sequence verification retS41-185 was recombined into pDEST-HisMBP to create the expression vector pDEST-HMBP-retS41-185. The RetS41-185-S45C variant used for the dimerization studies was generated via site-directed mutagenesis with pDONR201-retS41-185 serving as template. Subsequent recombination yielded the pDESTHMBP-retS41-185-S45C vector.
The protein expression and purification protocols for RetS27-185, RetS41-185 (RetSperi), and RetS41-185-S45C (RetSperi-S45C) were identical. Single colonies of E. coli BL21(DE3) CodonPlus RIL cells (Stratagene, La Jolla, CA) containing the expression plasmid were used to inoculate 100 ml of Luria broth supplemented with glucose at 2 g/L, 100 μg/ml ampicillin, and 30 μg/ml chloramphenicol. The cell culture was grown with shaking (225 rpm) to saturation overnight at 37 °C and then diluted 66-fold into six liters of fresh medium. When the cell density reached mid-log phase (OD600=0.5), the temperature was reduced to 30 °C and isopropyl-β-D-thiogalacto-pyranoside (IPTG) was added to a final concentration of 1 mM. After four hours cells were harvested by centrifugation at 5,000 × g for 15 minutes.
All of the following steps were carried out at 4 °C. Cells were re-suspended using a buffer containing 50 mM Tris-HCl, 150 mM NaCl, and 25 mM imidazole, pH 7.4 (buffer A) and 5 μl EDTA-free protease inhibitor cocktail (Sigma P8849) per milliliter of buffer (10 mL of buffer per gram of cell mass). Cells were lyzed through sonication and insoluble debris removed by centrifuging the cell extract at 40,000 × g for 30 minutes. The supernatant was filtered through a 0.45 μm polyethersulfone membrane and loaded onto to a 30 mL Ni-NTA chromatography column (Qiagen, Valencia, CA) pre-equilibrated in buffer A. The column was washed to baseline with buffer A and eluted with a linear imidazole gradient to 250 mM over 10 column volumes. Peak elution fractions were combined and His-TEV(S219V)-Arg (1 mg/100 mg of total protein) was added to effect the cleavage of His-MBP. The TEV protease-digest reaction mixture was dialyzed overnight against buffer A. After dialysis, the protein solution was filtered and applied to a 40-mL Ni-NTA Superflow column (Qiagen) pre-equilibrated with buffer A. Flow-through fractions containing the sensory domain were pooled and dialyzed overnight into buffer of 50 mM MES 50 mM NaCl, pH 6.0 (buffer B). The sample was then applied to a 5 mL Heparin column (GE Healthcare,) pre-equilibrated with buffer B and was eluted with a linear salt gradient to 1 M NaCl. Peak fractions were concentrated and loaded onto a HiPrep 26/60 Sephacryl S-200 HR column (GE Healthcare), pre-equilibrated in a buffer of 25 mM Tris-HCl, 150 mM NaCl, pH 7.4 (buffer C). The protein was judged to be >95% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Preparation of selenomethionine-labeled RetSperi
Selenomethionine-substituted RetSperi (SeMet-RetSperi) was produced in the same non-auxotrophic strain of E.coli as used for routine protein production. The incorporation of externally-added selenomethionine into the recombinant protein was accomplished by suppressing methionine biosynthesis 21. Initially, a 1L overnight culture of the expression strain was grown in LB medium also containing 100 μg/ml ampicillin and 30 μg/ml chloramphenicol. The cells were washed twice in 100 mL of M9 selenomethionine growth media (Medicilon Inc, Chicago, Illinois). The cells were re-suspended in 100 ml of M9 selenomethionine growth media and used to inoculate four 1L cultures containing M9 selenomethionine growth media and antibiotics. These cultures were grown with agitation at 37°C until the OD600nm reading reached 0.2. At this point IPTG was added to a final concentration of 1 mM and the temperature lowered to 30°C. Cells were harvested the following morning. The purification of selenomethione-containing protein followed the same protocols as that for the native protein with the exception that 2mM DTT was added to all but final buffer, which contained 1mM tris (2-Carboxyethyl) phosphine (TCEP) instead.
Limited proteolysis of the RetS sensory domain
A 1 mg/ml stock solution of thermolysin (Roche Molecular Biochemicals) in thermolysin buffer (10 mM Tris-HCl, 0.2 M NaCl, and 2 mM CaCl2, pH 8.0) was used for the limited proteolysis experiments. The RetS27-185 stock solution consisted of the protein at 1 mg/mL in buffer C. The five individual reactions were composed of 25 μL of RetS27-185 stock solution, 25 μL of 2X thermolysin buffer, and 0.5 μL of serial 1:4 dilutions of the thermolysin stock solution. The reactions were allowed to proceed for 1 hour at 37 °C before the protease was inactivated by the addition of 0.5 μL of 0.5 M EDTA. Reaction products were initially visualized by SDS-PAGE. The precise molecular weights of select fragments were determined using LC-electrospray ionization mass-spectrometry. The peptide fragments that corresponded to the observed molecular masses were determined using the FindPep program 22.
Crystallization of RetSperi and SeMet-RetSperi
High-throughput crystallization screening was conducted in the sitting-drop format by combining a solution of 17 mg/mL RetSperi in buffer C with commercially available crystallization matrices at volume ratios of 3:1, 1:1, and 1:3, where the protein solution was maintained at a constant volume of 0.3 μL throughout. Preliminary crystals were obtained from condition D4 of the IndexHT screen (Hampton Research). Hit optimization was carried out using the hanging drop vapor diffusion method at 18 °C. In the optimized conditions crystals for both the native protein and SeMet-RetSperi were obtained from drops containing a 1:1 mixture of 17 mg/mL proteins in their respective storage buffers and a crystallization solution composed of 0.1M citric acid, 22.5% w/v polyethylene glycol 3350 and 10% v/v glycerol.
Data collection, structure solution, and refinement
Crystals of RetSperi were loop-mounted and flash-frozen in liquid nitrogen. Data sets were collected at beamline X-29A of the National Synchrotron Light Source using an ADSC Q315 CCD detector. Data were processed at the beamline using the HKL2000 program suite 23. Details of data collection and processing are provided in Table I. The SeMet-RetSperi structure was determined using the single-wavelength anomalous dispersion (SAD) method. The location of heavy atoms, initial phase calculations, phase improvement through density modification, initial maps, and automated model building steps were all completed in the PHENIX program suite 24. PHENIX built nearly complete models for both molecules in the asymmetric unit. Iterative cycles of manual model adjustment using COOT 25 followed by refinement in PHENIX rapidly converged to produce the final structures. Model quality was assessed with PROCHECK 26 and the atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) 27 with accession code 3JYB.
Table I.
Data collection and refinement statistics for the SeMet-RetSperi crystal
| Data collection statistics | |
| Wavelength (Å) | 0.97900 |
| Space Group | P212121 |
| Unit cell parameters (Å) | a=51.164; b=67.193; c = 86.214 |
| Molecules/asymmetric unit | 2 |
| Resolution (Å) (last shell 2.11 - 2.04) | 30.00 - 2.04 |
| Total reflections | 255,128 |
| Unique reflections | 19,459 |
| Completeness (%) | 99.9 (99.2)a |
| I/σ | 26.3 (3.40) |
| Rmerge (%)b | 9.0 (39.5) |
| Refinement | |
| Resolution (highest resolution shell) (Å) | 29.62- 2.04 (2.1-2.04) |
| No. reflections | 17870 (1265) |
| Rworkc/Rfreed (%) | 19.9 ( 20.1)/ 23.5 (27.5) |
| Number of total atoms | 2403 |
| Number of protein atoms | 2298 |
| Number of water atoms | 105 |
| Overall mean B factor value ( Å2) | 18.747 |
| r.m.s deviations-bond lengths (Å) | 0.007 |
| r.m.s deviations-bond angles (°) | 1.119 |
The values in parentheses relate to the highest resolution shell.
Rmerge = Σ |I-<I>|/Σ I where I is the observed intensity and <I> is the average intensity obtained from multiple observations of symmetry-related reflections after the rejection of significant outliers.
R = Σ | |Fo| - |Fc| | / Σ |Fo|
Rfree defined by Brunger. 37
Protein-protein cross-linking
RetSperi was dialyzed into a buffer of 20 mM NaH2PO4 and 30 mM NaCl, pH7.4. A 2 mM stock solution of Bis(Sulfosuccinimidyl) suberate (BS3) was prepared in water. The 50 μl reaction mixtures contained 10 μM RetSperi and 500 μM BS3. Cross-linking reactions were quenched by adding 20 μl of each reaction to 10 μl 150 mM Tris-HCl, pH7.4. Cross-linking reactions containing MBP instead of RetSperi were used as negative controls. Cross-linked samples were analyzed via SDS-PAGE.
Quantitative oligomerization assay
The RetSperi-S45C variant was fluorescently labeled with Alexa Fluor 488 and Alexa Fluor 555 (Invitrogen) according to the manufacturer's instructions. Following overnight labeling the modified proteins were separated from the unincorporated dye molecules through buffer exchange into a buffer of 20 mM Tris-HCl and 150 mM NaCl, pH 7.4. The degree of labeling of both the Alexa Fluor 555-labeled form of the RetSperi variant (RetSperi-555) and the Alexa Fluor 488-labeled form of the RetSperi variant (RetSperi-488) were near 100% as assessed by UV-spectroscopy in conjunction with the estimated molar extinction coefficients of RetSperi and the respective fluorophores.
The fluorescence resonance energy transfer (FRET) measurements involved titrating a solution of 3 nM RetSperi-488 with RetSperi-555. Triplicate set-ups of 80 μl reactions containing 3 nM RetSperi-488 and 0 to 100 μM RetSperi-555 were transferred to a 96-well half area black polystyrene assay plate (Corning). Background fluorescence produced by RetSperi-555 was accounted for by measuring and subtracting the signal of solutions that did not contain RetSperi-488 but identical concentrations of RetSperi-555.
A TECAN infinite M200 fluorescence intensity scanner (Tecan, inc.) was used for the experiment. The excitation wavelength was set to 430 nm, while the emission spectrum was recorded in between in the wavelength range between 510 and 646 nm using a 4 nm step-size. The integration time was set to 20 μs and all scans were carried out at a room temperature of 24 °C. Dimer formation was monitored by recording the decrease in the peak fluorescence of RetSperi-488 at λ=522 nm. Data were fit to equation (I) using Matlab (The MathWorks).
| (I) |
R5T and R4T are the total concentrations of RetSperi-555 and RetSperi-488, respectively, F is the fluorescence measured at λ=522 nm, Fmax is the fluorescence measured at λ=522 nm in absence of RetSperi-555, Fmin the residual fluorescence of RetSperi-488 due to incomplete quenching even when all RetSperi-488 is bound to RetSperi-555, and Kd is the dissociation constant of the RetSperi dimer. Derivation of equation (I) is outlined in supplementary file S1.
Results
The RetS sensory domain assumes a beta-sandwich fold reminiscent of carbohydrate binding proteins
In order to ensure that the complete sensory domain was contained in the cloned construct, the entire periplasmic region of RetS encompassing residues 27 to 185 was initially cloned and over-expressed. Subsequently, the domain boundaries were more precisely mapped through limited proteolysis of RetS27-185 in order to facilitate crystallization. Mass-spectrometric analysis of the obtained fragments identified a relatively stable fragment comprising RetS residues 41 to 185 (RetSperi). RetSperi, over-expressed from a newly engineered plasmid, readily crystallized. SeMet-RetSperi crystals, while very small (~ 0.05*0.02*0.01mm3), produced x-ray diffraction to a resolution of 2.0 Å.
Forming an asymmetric dimer there are two independent RetSperi molecules in asymmetric unit of the crystal. The structural differences between the backbone atoms of the two molecules are small as reflected in a root-mean-square-deviation of 0.4 Å. The electron density throughout the structure was excellent. However, the amino-terminal regions ranging from Ala-41 to Asn-47 are disordered and produced no discernable electron density in either molecule. All non-glycine residues of the final model reside either in the most favorable or in the allowed regions of the Ramachandran plot; and the overall geometry was comparable to other structures solved at the same resolution.
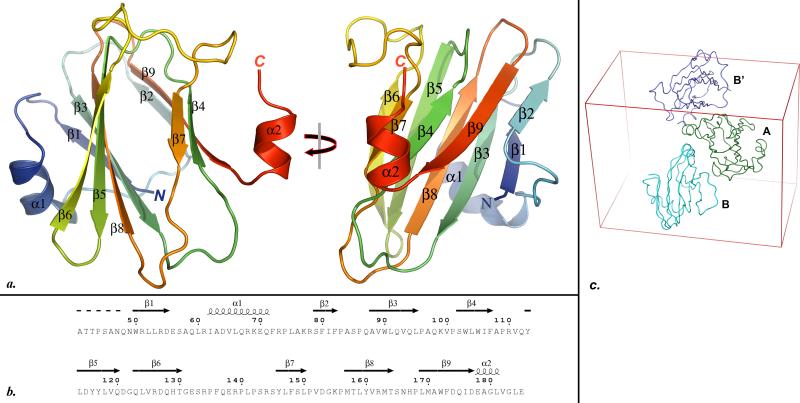
A cartoon drawing of the final RetSperi structure model is depicted in figure Fig. 1a, while the correlation between protein sequence and tertiary structure is visualized in figure Fig. 1b. The sensory domain of RetS adopts β-sandwich or jelly-roll fold formed by two opposing antiparallel β-sheets. The two sheets have a β1-β3-β8-β5-β6 and a β2-β9-β4-β7 topology, respectively. The β-sandwich structure is augmented by two alpha-helices, α-1 follows strand β-1 and the short α-2 helix is formed by the carboxy-terminal residues of RetSperi. The observed fold is characteristic of carbohydrate binding modules (CBM). Consistent with this notion, a DALI-guided 28 search of the PDB identified CBMs and enzymes involved in carbohydrate degradation as the closest structural homologs of RetSperi. While the structural conservation extends to both beta sheets, helix α-1 appears to be unique to RetSperi. The position of this helix directly above the β1-β3-β8-β5-β6 sheet is interesting because in most of the related CBMs the equivalent β-sheets form the carbohydrate binding sites. We examined the temperature factors as well as intermolecular and intramolecular contacts of helix α-1 to evaluate whether or not this helix might be conformationally flexible and its conspicuous position perhaps the result of packing contacts. However, we found no intermolecular contacts involving α-1 within a 5 Å radius. On the other hand, α-1 forms a total of 18 intramolecular contacts with neighboring residues if a 4 Å distance cutoff is applied, burying a surface area of 525 Å2. Furthermore, the positions of the two α-1 helices are identical for both molecules in the asymmetric unit and the average temperature factor of ~20 Å2 for the main-chain atoms also suggest a stable conformation. Therefore the unique topology of this region suggests that the 7TM-DISM2 domains constitute a novel class of carbohydrate binding proteins.
Fig. 1.
a. Two orthogonal views of the PYMOL 41-generated cartoon representation of the RetSperi structure. All secondary structure elements are labeled. b. Alignment of the secondary structure elements identified from the crystal structure with the primary structure RetSperi. This figure was generated with ESPript 42. c. The asymmetric unit of the crystal consists of molecules A and B. The contacts between A and B closely resemble the packing contacts between A and the symmetry-related B’ molecule, suggesting that the dimer in the asymmetric unit arose from crystal packing contacts.
Qualitative and quantitative evidence for RetSperi dimerization
The reversible oligomerization of RetS is believed to play a pivotal role in the regulation of RetS function. Consequently, we sought to establish whether or not RetSperi dimerization contributes to the stability of the RetS homodimer. Intuitively, we would have expected to observe a symmetric dimer in the crystal, but the two molecules that form the asymmetric unit display no two-fold symmetry. While unusual this asymmetric dimer could nevertheless represent a biologically relevant dimer. However, when we compared the intermolecular contacts within the asymmetric unit to packing contacts between symmetry-related asymmetric units in the crystal, we found them to be very similar. These packing arrangements are visualized in figure Fig. 1c, where molecules A and B represent the original asymmetric unit and B’ a molecule from a symmetry-related asymmetric unit. The buried surface area of the A•B and A•B’ interfaces are very similar at 610 and 629 Å2, respectively. There are twenty-one interacting residue pairs at A•B interface and twenty-three such pairs at the A•B’ interface when a 4 Å distance cutoff is applied (this cutoff was chosen because it represents the default value in programs such as Ligplot29 that are designed to analyze intermolecular contacts). Twenty of these interactions are found at both interfaces. Distances for the four contacts that did not match ranged between 3.7 and 4 Å, suggesting that they make only small contributions to the stability of the respective intermolecular interactions. These similarities suggest that the dimer observed in the asymmetric unit arose due to crystal packing contact and does not represent the biological unit of the protein. Otherwise, if the interactions in the asymmetric unit were indeed representative of the solution state then RetSperi should be forming higher order oligomers or even polymerize in solution. This was not observed in the cross-linking experiments described below.
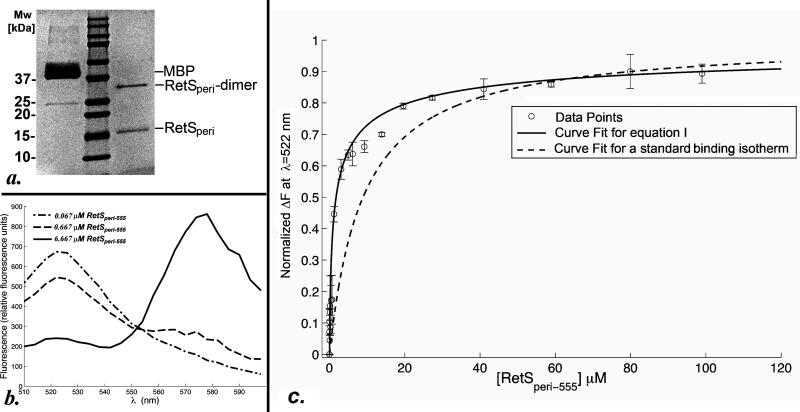
Since the non-physiological pH of 3.0 of the crystallization conditions could have caused a biological RetSperi oligomer to dissociate, protein-protein cross-linking using chemical BS3 was employed to probe for dimer formation at neutral pH. Monomeric E.coli MBP served as negative control. After cross-linking MBP still migrated as a monomer on an SDS-polyacrylamide gel, while a substantial portion of RetSperi had shifted to a higher band corresponding to the molecular weight of a RetSperi dimer (Figure Fig. 2a). Higher order oligomers were not observed.
Fig. 2.
a. SDS-PAGE analysis of the BS3-mediated cross-linking experiment with RetSperi. Shown in the lanes are from left to right: MBP, the molecular weight makers, and RetSperi. b. Sample plots of fluorescence spectra obtained during the FRET-based dimerization assay at three different concentrations of RetSperi-555. The spectra are generated by subtracting the background fluorescence produced by the same concentrations of RetSperi-555 in absence of RetSperi-488 from the raw data. c. Shown is the resulting curve when equation (I) is fit to the FRET data where . The error bars signify the standard deviation of the ΔF calculated from the readings obtained from triplicate set-ups. For comparative purposes the data were also fit to the conventional binding isotherm (supplementary file S1. part c.), which does not consider the increasing amounts RetSperi-555 dimer. This fit is shown as a dashed line.
Although the cross-linking result demonstrates RetSperi dimerization, a substantial fraction of RetSperi failed to cross-link even when large excess of cross-linking agent and extended reaction times were used. This finding is consistent with the presence of monomer-dimer equilibrium rather than a purely dimeric state. A FRET-based assay was developed to validate this hypothesis and quantify the strength of the RetSperi-RetSperi interactions. A low concentration of Alexa-Fluor-488-labeled RetSperi-S45C (RetSperi-488) was titrated with an Alexa-Fluor-555-labeled form of the same RetSperi variant (RetSperi-555). Dimerization was monitored by observing the quenching of the Alexa Fluor 488 fluorescence at λ=522 nm. The concentration of the RetSperi-488 was maintained more than a hundred-fold below the dissociation constant (Kd) for RetSperi dimer formation because here virtually all of the protein should be monomeric. This optimal concentration of RetSperi-488 was determined in an iterative process where titration experiments were carried out at concentrations of 50, 10, and finally 3 nM of RetSperi-488 and fit to equation (I). The two higher concentrations of the protein resulted in a poor fit and a Kd value that suggested that a substantial proportion of RetSperi-488 was dimeric at the outset of the experiment. A second important consideration relates to the fact that, as the concentration of RetSperi-555 is increased during the titration experiment, a large fraction of this protein is already dimeric and therefore not available for binding to RetSperi-488. In order to properly model RetSperi dimerization it was therefore necessary to express the concentration of the RetSperi-555 monomer as a function of the total concentration of RetSperi-555 and the Kd. Usually, equations for binding isotherms are arranged to express the amount of complex formed in terms of the variable concentration of one of the binding partners. However, when this was done for the modified isotherm the resulting cubic equation was solvable but the obtained solution was awkwardly long (supplemental file S.1 part b.). Therefore, the data were fit to the rearranged and thus simpler equation (I). This equation is valid under the assumption that the fluorescent labels do not influence the monomer-dimer equilibrium, which is based on the observation that the S45C mutation used to mediate thiol-based labeling is located in a structurally disordered region that does not appear to constitute a pivotal part of the sensory domain. Background-corrected example graphs are presented in figure Fig. 2b. The resulting FRET data are plotted in figure Fig. 2c; and fitting equation (I) to these data yielded a dissociation constant of Kd=580± 50 nM for RetSperi dimerization. If RetSperi-555 dimerization is not taken into consideration and the data are simply fit to a conventional isotherm, the fit is poor and the obtained Kd incorrect (dashed line in figure Fig. 2c).
Discussion
The putative ligand binding site of RetSperi
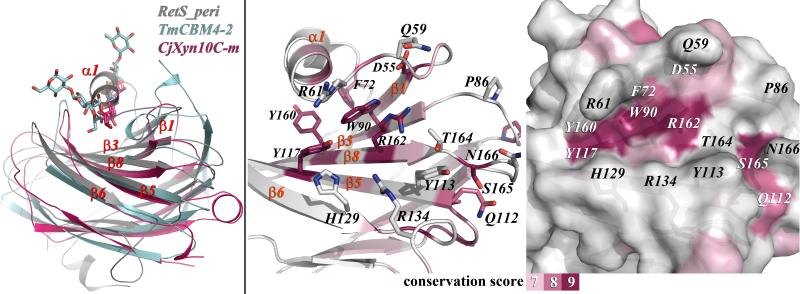
The crystal structure suggests that RetS is regulated by a carbohydrate-based moiety. However, since the natural ligand of RetS is currently not known, the common strategy for mapping the ligand binding site involving a mutational analysis coupled with in vitro binding studies could not be employed. Nevertheless, insights could be obtained by comparing RetSperi to the altogether eight structurally homologous proteins, where the positions of the ligand binding sites are known. Protein-carbohydrate complexes have been reported for the related family 4 CBM of Lam16A from Thermotoga maritima (TmCBM4-2, PDB code 1gui) 30, the family 6 CBM SdAga16B-CBM6-2 of the β-Agarase Aga16B from Saccharophagus degradans (PDB code 2CDO) 31, and the family 15 CBM of the Cellvibrio japonicas Xylanase 10C, (CjXyn10C-m, PDB code 1US2) 32. In addition, the binding sites of the following proteins have also been mapped and were therefore included in the analysis: the carbohydrate-recognition domain of the human glycoprotein sorting receptor p58/ERGIC-53 (PDB code 1R1Z) 33, the carbohydrate recognition domain of the cargo receptor Emp46p from Saccharomyces cerevisiae (PDB code 2A6W) 34, the carbohydrate binding module of xylanase 10A from the thermophilic bacterium Rhodothermus marinus (RmXyn10A-CBM4-2, PDB code 1K42) 35, the catalytic domain of the endoglucanase CelB from Streptomyces lividans, (SlCelB-2, PDB code 1NLR) 36, and family 32 carbohydrate-binding protein YeCBM32 from Yersinia enterolitica (PDB code 2JDA) 37. Although all eight proteins share the same fold, their ligand binding sites cluster to not one but two regions when superimposed. In YeCBM32 and SdAga16B-CBM6-2 the ligand binding sites are located between the two beta sheets, involving residues equivalent to the carboxy-terminal end of β-2, the amino-terminal end of β-9, and residues from the two connected loop regions in RetSperi. This pocket is not present in RetSperi suggesting that its binding cleft is located elsewhere.
In the other six homologous structures the critical residues map to structurally conserved section of the proteins that corresponds to the β1-β3-β8-β5-β6 sheet in RetSperi. In the TmCBM4-2 and CjXyn10C-m complexes, the entire beta-sheet forms an extended cleft, where their respective oligosaccharide ligands are bound, while the ligand binding site in the other four proteins are smaller, covering only part of the beta-sheet. In RetSperi helix α-1 alters the topology of this the corresponding surface area. In fact, α-1 effectively occupies the same space as the bound ligands in TmCBM4-2 and CjXyn10C-m (Figure Fig. 3a.). However, together with residues from the beta sheet and several surrounding loops α-1 does create three large grooves that appear well-suited for ligand binding. The pocket that most closely coincides with the prevalent binding pockets observed in the related proteins is formed by the loop connecting β-1 and α-1, the amino terminal regions of strands β3 and β5, the carboxy-terminal ends of strands β1, β8, and β6 and the loop regions between β-2 and β-3 as well as those between β-8 and β-9 (Figures Fig. 3b and 3c).
Fig. 3.
a. Superposition of RetSperi, TmCBM4-2, and CjXyn10C-m. TmCBM4-2 and CjXyn10C-m have both been crystallized in complex with xylopentaose. The positions of the ligands in the latter complexes coincide with helix α-1 in RetSperi. b. The backbone structure of RetSperi has been colored according to the level of sequence conservation among proteins with related 7TM-DISM2 domains. Also, shown are side chains and labels of the seven broadly conserved amino acids that are concentrated in this section of the protein and of those amino acids that form the putative binding site. c. The solvent-accessible surface area of RetSperi, viewed from the same perspective and also colored–coded according to the level of sequence conservation, reveals a large cavity that is proposed to constitute the ligand binding site.
The hypothesis that this site indeed constitutes the ligand binding pocket may be evaluated from two additional angles. The solvent-accessible surface area of RetSperi was color-coded according to the degree of sequence conservation using the ConSurf 38. Those regions with the three highest conservation scores were colored brightly, while the remainder of the surface was kept white (Figures Fig. 3b and 3c.). Remarkably, seven of the ten amino acids that received the highest conservation scores map to the β1-β3-β8-β5-β6 beta sheet. Four of these seven residues, Trp-90, Tyr-117, Arg-162 and Ser-164 are part of the pocket that was identified from the structural comparison with other binding sites. Trp-90 is one of only two amino acids that are strictly conserved in all thirty-two of the homologous protein sequences that were used for this analysis.
Lastly, the predicted binding site may also be assessed by analyzing the types of amino acids that form this pocket. Tryptophan, arginine, asparagine, glutamic acid, tyrosine, and histidine display the highest propensities for participating in carbohydrate binding 39. The highly conserved Trp-90, Tyr-117, and Arg-162 fall into this category. Other residues in the binding pocket also feature prominently in carbohydrate binding sites: Gln-59, Arg-61, Gln-112, Arg-134, and Asn-166, and the aromatic residues Tyr-113 and His-129. Pro-86 and Thr-164 constitute the only unusual residues among the amino acids that are form the putative ligand binding site (Figures Fig. 3b. and 3c.). In summary, three independent indicators -structural conservation, sequence conservation, and composition of the putative binding pocket- all point toward the same location of the ligand binding site and suggest that the elusive ligand is a carbohydrate moiety. Ultimate confirmation of these findings, however, will require the identification of the natural ligand of RetS.
Implications of RetSperi dimerization for the RetS-GacS signaling mechanism
RetS is an unusual sensor kinase because at least one of its signaling modes does not require its kinase activity but relies on direct interactions with GacS kinase to prevent phosphate transfer in the GacS/GacA TCS 18. The kinase domains of both enzymes appear to be sufficient for this interaction. The model assumes ligand binding to the periplasmic sensory domain disrupts the RetS dimer, but how is this signal communicated across the inner cell membrane? The hypothesis that the RetS sensory domain mediates the up-regulation of T3SS-related gene expression also appears to be at odds with a previous study, where deletion of the sensory domain did not cause a decrease but a slight increase in virulence associated with the T3SS 13. Our finding that RetSperi alone is capable of dimer formation offers a possible explanation for this apparent contradiction as it suggests that the sensory domain is required for repression as well as activation of the T3SS. Ligand-free RetSperi likely dimerizes and prevents RetS-GacS interactions by stabilizing the RetS homodimer. The unknown ligand could act by disrupting the interface between the two sensory domains, and thus favor RetS-GacS hetero-dimer formation. Consistent with the previously published results this model would explain why a deletion of the sensory domain did not result in a repression of the T3SS, since such a deletion, just as ligand binding, would favor a RetS-GacS complex.
Conclusion
The crystal structure of the RetS sensory domain, the first representative structure for the 7TM-DISM2 domain family, revealed a fold closely related to carbohydrate binding modules, suggesting that RetS function is regulated by a carbohydrate-based ligand. While the crystal structure does not unambiguously reveal the ligand binding site, structural homology, sequence conservation, and amino acid composition suggest that the ligand likely binds to a pocket formed by one of the two beta sheets and helix α-1. RetSperi dimerizes with a submicromolar dissociation constant, suggesting that dimerization of the RetS sensory domain plays a role in both stabilizing the RetS homo-dimer and, upon ligand binding, in shifting the equilibrium towards RetS-GacS hetero-dimer formation.
Supplementary Material
Acknowledgments
Funding for this work was provided by Jeffress Memorial Trust Grant number J-910 and the Nationial Scientist Development Grant 09SDG2260401 from the American Heart Association both awarded to Florian Schubot. Funding for data collected at beamline-29 NSLS is provided by DOE/DER and NIH/NCRR. The authors also wish to thank Dr. Keith Ray for his assistance with the mass spectrometry work.
Abbreviations used
- BS3
Bis(Sulfosuccinimidyl) suberate
- CBM
carbohydrate binding module
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- FRET
Fluorescence resonance energy transfer
- IPTG
isopropyl-β-D-thiogalacto-pyranoside
- MBP
Maltose Binding Protein
- PDB
Protein Data Bank
- rms
root mean square
- SAD
single-wavelength anomalous dispersion
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCS
two-component signaling system
- TCEP
tris(2-Carboxyethyl) phosphine
- TEV
tobacco etch virus
- T3SS
type III secretion system
References
- 1.Richards MJ, Edwards JR, Culver DH, Gaynes RP. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Critical Care Medicine. 1999;27(5):887–892. doi: 10.1097/00003246-199905000-00020. [DOI] [PubMed] [Google Scholar]
- 2.Crouch Brewer S, Wunderink RG, Jones CB, Leeper KV., Jr. Ventilator-associated pneumonia due to Pseudomonas aeruginosa. Chest. 1996;109(4):1019–1029. doi: 10.1378/chest.109.4.1019. [DOI] [PubMed] [Google Scholar]
- 3.Labrec EH, Schneider H, Magnani TJ, Formal SB. Epithelial cell penetration as an essential step in the pathogenesis of bacillary dysentery. Journal of Bacteriology. 1964;88:1503–1518. doi: 10.1128/jb.88.5.1503-1518.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garau J, Gomez L. Pseudomonas aeruginosa pneumonia. Current Opinion in Infectious Diseases. 2003;16(2):135–143. doi: 10.1097/00001432-200304000-00010. [DOI] [PubMed] [Google Scholar]
- 5.Cheng KH, Leung SL, Hoekman HW, Beekhuis WH, Mulder PG, Geerards AJ, Kijlstra A. Incidence of contact-lens-associated microbial keratitis and its related morbidity. Lancet. 1999;354(9174):181–185. doi: 10.1016/S0140-6736(98)09385-4. [DOI] [PubMed] [Google Scholar]
- 6.Mulcahy H, O'Callaghan J, O'Grady EP, Macia MD, Borrell N, Gomez C, Casey PG, Hill C, Adams C, Gahan CG, Oliver A, O'Gara F. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infection and Immunity. 2008;76(2):632–638. doi: 10.1128/IAI.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaver CM, Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infection and Immunity. 2004;72(12):6969–6977. doi: 10.1128/IAI.72.12.6969-6977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology (Reading, England) 2001;147(Pt):2659–2669. doi: 10.1099/00221287-147-10-2659. [DOI] [PubMed] [Google Scholar]
- 9.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407(6805):762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 10.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7(5):745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 11.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. PNAS. 2006;103(1):171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heeb S, Blumer C, Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. Journal of Bacteriology. 2002;184(4):1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laskowski MA, Kazmierczak BI. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infection and Immunity. 2006;74(8):4462–4473. doi: 10.1128/IAI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liaw SJ, Lai HC, Ho SW, Luh KT, Wang WB. Role of RsmA in the regulation of swarming motility and virulence factor expression in Proteus mirabilis. Journal of Medical Microbiology. 2003;52(Pt):19–28. doi: 10.1099/jmm.0.05024-0. [DOI] [PubMed] [Google Scholar]
- 15.Ang S, Horng YT, Shu JC, Soo PC, Liu JH, Yi WC, Lai HC, Luh KT, Ho SW, Swift S. The role of RsmA in the regulation of swarming motility in Serratia marcescens. Journal of Biomedical Science. 2001;8(2):160–169. doi: 10.1007/BF02256408. [DOI] [PubMed] [Google Scholar]
- 16.Kay E, Humair B, Denervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. Journal of Bacteriology. 2006;188(16):6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. PNAS. 2006;103(1):171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23(2):249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anantharaman V, Aravind L. Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics. 2003;4(1):34. doi: 10.1186/1471-2164-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nallamsetty S, Austin BP, Penrose KJ, Waugh DS. Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 2005;14(12):2964–2971. doi: 10.1110/ps.051718605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doublie S. Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol Biol. 2007;363:91–108. doi: 10.1007/978-1-59745-209-0_5. [DOI] [PubMed] [Google Scholar]
- 22.Gattiker A, Bienvenut WV, Bairoch A, Gasteiger E. FindPept, a tool to identify unmatched masses in peptide mass fingerprinting protein identification. Proteomics. 2002;2(10):1435–1444. doi: 10.1002/1615-9861(200210)2:10<1435::AID-PROT1435>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski ZM. W. HKL2000. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the steriochemical quality of protein structures. J Appl Cryst. 1993;26:282–291. [Google Scholar]
- 27.Sussman JL, Lin D, Jiang J, Manning NO, Prilusky J, Ritter O, Abola EE. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 6 Pt 1):1078–1084. doi: 10.1107/s0907444998009378. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Curr Protoc Bioinformatics. 2006 doi: 10.1002/0471250953.bi0505s14. Chapter 5:Unit 5 5. [DOI] [PubMed] [Google Scholar]
- 29.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 30.Boraston AB, Nurizzo D, Notenboom V, Ducros V, Rose DR, Kilburn DG, Davies GJ. Differential oligosaccharide recognition by evolutionarily-related beta-1,4 and beta-1,3 glucan-binding modules. J Mol Biol. 2002;319(5):1143–1156. doi: 10.1016/S0022-2836(02)00374-1. [DOI] [PubMed] [Google Scholar]
- 31.Henshaw J, Horne-Bitschy A, van Bueren AL, Money VA, Bolam DN, Czjzek M, Ekborg NA, Weiner RM, Hutcheson SW, Davies GJ, Boraston AB, Gilbert HJ. Family 6 carbohydrate binding modules in beta-agarases display exquisite selectivity for the non-reducing termini of agarose chains. J Biol Chem. 2006;281(25):17099–17107. doi: 10.1074/jbc.M600702200. [DOI] [PubMed] [Google Scholar]
- 32.Pell G, Szabo L, Charnock SJ, Xie H, Gloster TM, Davies GJ, Gilbert HJ. Structural and biochemical analysis of Cellvibrio japonicus xylanase 10C: how variation in substrate-binding cleft influences the catalytic profile of family GH-10 xylanases. J Biol Chem. 2004;279(12):11777–11788. doi: 10.1074/jbc.M311947200. [DOI] [PubMed] [Google Scholar]
- 33.Velloso LM, Svensson K, Pettersson RF, Lindqvist Y. The crystal structure of the carbohydrate-recognition domain of the glycoprotein sorting receptor p58/ERGIC-53 reveals an unpredicted metal-binding site and conformational changes associated with calcium ion binding. J Mol Biol. 2003;334(5):845–851. doi: 10.1016/j.jmb.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 34.Satoh T, Sato K, Kanoh A, Yamashita K, Yamada Y, Igarashi N, Kato R, Nakano A, Wakatsuki S. Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p. J Biol Chem. 2006;281(15):10410–10419. doi: 10.1074/jbc.M512258200. [DOI] [PubMed] [Google Scholar]
- 35.Simpson PJ, Jamieson SJ, Abou-Hachem M, Karlsson EN, Gilbert HJ, Holst O, Williamson MP. The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase. Biochemistry. 2002;41(18):5712–5719. doi: 10.1021/bi012093i. [DOI] [PubMed] [Google Scholar]
- 36.Sulzenbacher G, Shareck F, Morosoli R, Dupont C, Davies GJ. The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 A resolution. Biochemistry. 1997;36(51):16032–16039. doi: 10.1021/bi972407v. [DOI] [PubMed] [Google Scholar]
- 37.Abbott DW, Hrynuik S, Boraston AB. Identification and characterization of a novel periplasmic polygalacturonic acid binding protein from Yersinia enterolitica. J Mol Biol. 2007;367(4):1023–1033. doi: 10.1016/j.jmb.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 38.Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009;37(Database issue):D323–327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malik A, Ahmad S. Sequence and structural features of carbohydrate binding in proteins and assessment of predictability using a neural network. BMC Struct Biol. 2007;7:1. doi: 10.1186/1472-6807-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355(6359):472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 41.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; San Carlos, CA, USA: 2001. http://wwwpymolorg. [Google Scholar]
- 42.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31(13):3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.