Abstract
The opportunistic pathogen Pseudomonas aeruginosa ranks among leading causes of nosocomial infections. The type III secretion system (T3SS) aids acute P. aeruginosa infections by injecting potent cytotoxins into host cells to suppress the host's innate immune response. Expression of all T3SS-related genes is strictly dependent upon the transcription factor ExsA. Consequently, ExsA and the biological processes that regulate ExsA function are of great biomedical interest. The presented work focuses on the ExsA-ExsC-ExsD-ExsE signaling cascade that ties host cell contact to the up-regulation of T3SS gene expression. Prior to T3SS induction, the anti-activator protein ExsD binds to ExsA and blocks ExsA-dependent transcription by interfering with ExsA dimerization and promoter interactions. Upon host cell contact, ExsD is sequestered by the T3SS chaperone ExsC resulting in the release of ExsA and an up-regulation of the T3SS. Previous studies have shown that the ExsD-ExsA interactions are not freely reversible. Because independently folded ExsD and ExsA were not found to interact, it has been hypothesized that folding intermediates of the two proteins form the complex. Here we demonstrate for the first time that ExsD alone is sufficient to inhibit ExsA-dependent transcription in vitro and that no other cellular factors are required. More significantly, we show that independently folded ExsD and ExsA are capable of interacting, but only at 37°C and not at 30°C. Guided by the crystal structure of ExsD, we designed a monomeric variant of the protein and demonstrate that ExsD trimerization prevents ExsD from inhibiting ExsA-dependent transcription at 30°C. We propose that this unique mechanism plays an important role in T3SS regulation.
Keywords: ExsA, ExsD, Pseudomonas aeruginosa, thermoregulation, type III secretion
Introduction
The opportunistic human pathogen Pseudomonas aeruginosa poses a significant medical threat due to its high levels of natural and acquired antibiotic resistance [1-7]. P. aeruginosa utilizes a broad array of virulence mechanisms to establish and sustain infections. The type III secretion system (T3SS) is a hallmark of acute infections and aids infection by translocating at least four distinct effector proteins into the eukaryotic host cell [8-10]. Inside the host, these effectors act to subvert the host-immune response by interfering with critical signal transduction pathways [11-18]. The needle complex that constitutes the secretion apparatus is assembled from multiple copies of 27 distinct proteins. Protein translocation through the T3SS is powered by the proton motive force, while a cytoplasmic ATPase (PscN in P. aeruginosa) is thought to mediate targeting and unfolding of the transported effectors at the base of the needle complex [19, 20]. Because expression, assembly, and operation of the T3SS are energy-intensive, T3SS-related gene expression is tightly regulated via a number of regulatory pathways and closely tied to host infection [21-23]. The ExsA-ExsC-ExsD-ExsE (ExsACDE) signaling cascade constitutes perhaps the most direct link between opening of the T3SS channel and activation of T3SS-gene expression [22]. The AraC-type transcriptional activator, ExsA, facilitates the recruitment of RNA polymerase to the transcription initiation site and is required for transcription from all 10 T3SS-related promoters including its own expression as well as genes of the other members of the signaling cascade: exsC, exsE, and exsD [24]. While unusual, the underlying regulatory mechanism appears to be relatively straightforward: Prior to the host cell contact-induced opening of the secretion channel, ExsC and ExsE form a tight 2:1 complex, while the antiactivator ExsD sequesters ExsA to prevent transcription activation [25]. Host cell contact triggers the opening of the T3SS channel. Now, ExsE is secreted thereby releasing ExsC, which in turn binds to ExsD to activate ExsA-mediated transcription [22, 23].
Recent work has focused on the question of how ExsD inhibits ExsA function. Thibault et al. determined that ExsD and ExsA form a 1:1 complex which fails to bind to ExsA-dependent promoters in vitro, suggesting that ExsD interferes with ExsA-promoter interactions [26]. Brutinel et al. subsequently discovered that ExsD also interferes with ExsA self-association using a monohybrid study [27]. While both phenomena have not been connected experimentally, the enhancement of DNA binding affinity through self-association is a widespread feature of DNA binding protein factors. In both studies, researchers reported an unusual feature of the signaling mechanism. ExsD could only bind to ExsA when both proteins were synthesized at the same time. Thibault et al. unsuccessfully added ExsD to see if it would interfere with ExsA-DNA interactions in EMSA studies [26]. When attempting to reconstitute the entire signaling cascade in vitro, Brutinel et al. observed that ExsC was indeed capable of dissociating the ExsD-ExsA complex [27]. Perhaps mirroring a scenario where the bacterial cell loses host cell contact, the addition of ExsE to the sample readily brought about the formation of an ExsC-ExsE complex. However, the released ExsD protein was not able to rebind to ExsA, suggesting that the signaling process is not freely reversible. To explain this phenomenon, it was proposed that concurrent expression of both ExsD and ExsA might be required, because folding intermediates of either ExsD, ExsA, or both proteins might actually associate to form this complex [27]. Under this scenario, dissociation of the ExsD-ExsA complex would allow for complete folding of the subunit(s) and create a formidable kinetic barrier preventing reassociation of the complex. In the present study, we demonstrate that, rather than the folding of either protein, it is ExsD self-trimerization that accounts for the observed irreversible dissociation of the ExsD-ExsA complex. We also demonstrate that this barrier may be overcome by shifting the temperature from 30°C to 37°C.
Results
ExsA-mediated in vitro transcription is not inhibited by wild-type ExsD at 30°C, but is strongly inhibited at 37°C
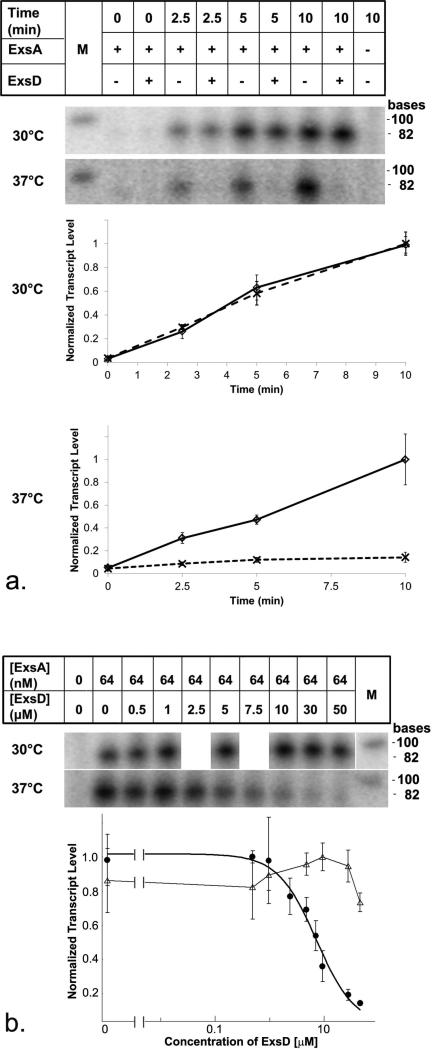
The above cited work demonstrated that ExsD interferes with ExsA dimerization and ExsA-promoter interactions [26]. In vitro transcription studies subsequently confirmed that ExsA is necessary and sufficient for activation of T3SS promoters [28]. Yet, what has not been explicitly shown is that ExsD alone can block ExsA-mediated transcription in vitro. We believe that this distinction is important, because the interaction of ExsA with RNA polymerase could significantly impact the interactions between ExsD and ExsA; for example, by stabilizing the ExsA dimer or binding of ExsA to the promoter region. We designed and optimized an efficient in vitro transcription assay using purified ExsA, ExsD, and P. aeruginosa RNA polymerase. In the process, we also developed a new expression and purification protocol for the transcriptional regulator ExsA, which produces a highly homogeneous sample suitable for structural studies. Unlike previous preparations, ExsA purified according to the described protocol does not require detergent and may be concentrated up to 50 μM. Figure S1 in the supplemental material contains the SDS-polyacrylamide gel lanes of all three purified samples. Our initial in vitro transcription experiments closely mirrored the protocol used in previously published experiments [28]. As anticipated, activation of the PexsD promoter requires the presence of ExsA, and it was established that transcript production proceeded in a linear fashion up to at least 20 minutes under the given experimental conditions (Supplementary Fig. S2). Also, confirming results of previous studies, even the addition of vast excess of ExsD (up to 50 μM) had no significant effect on the rate of transcription (Fig. 1a). We now applied this assay to examine previously unexplored parameters of ExsA-dependent transcription. Although temperature is likely important in the context of infection, to this point all studies of the ExsACDE cascade, including our initial assays, had been performed at or below 30°C. Because the body temperature of a human host is approximately 37°C, we conducted a second set of in vitro transcription assays at this temperature. At 37°C, the overall rate of ExsA-dependent transcription increased by about 13% compared to the 30°C assay experiment (Supplementary Fig. S3). When ExsD was included in the assay at the higher temperature, ExsD now strongly inhibited ExsA-dependent transcription, suggesting that the elevated temperature had alleviated the kinetic barrier that had previously prevented ExsD-ExsA interactions (Fig. 1a). Dose-response studies yielded a half maximal inhibitory concentration (IC50) value of 7.7 μM for ExsD under the given experimental conditions, which is indicative of a relatively weak inhibitor (Fig. 1b). To determine if the observed effect of ExsD is specific to ExsA-dependent transcription, we repeated the in vitro transcription reactions, this time using a template containing the constitutively expressed RNA-1 promoter [29]. ExsD had no effect on transcript levels from this promoter at 37°C (Supplementary Fig. S4), thus demonstrating that the observed inhibition is specific to ExsA-dependent promoters.
Fig. 1. Temperature-dependent regulation of ExsA by ExsD.
a. Autoradiograms and graphical representations for the in vitro transcription of an 86 nucleotide transcript from an ExsA-dependent PexsD promoter template. Assays were performed at 30°C and 37°C and in absence and presence of 50 μM ExsD. In both graphs, the ExsD-free data are represented by solid trend lines and empty diamonds, whereas the data obtained in the presence of ExsD are represented by dashed trend lines and crosses. b. Dose response data and fit generated by measuring in vitro transcription levels at the 10-minute time point at 37°C in the presence of increasing amounts of ExsD protein (filled circles). Data (empty triangles) and dashed trend line obtained when the same experiment was conducted at 30°C. The 30°C gel strip has been digitally manipulated to align the fewer data points with those generated in the experiments performed at 37°C. In order to obtain a reliable dose-response curve, two additional concentrations were included in the 37°C experiment.
A single mutation generating the ExsDM59R variant disrupts ExsD trimer formation
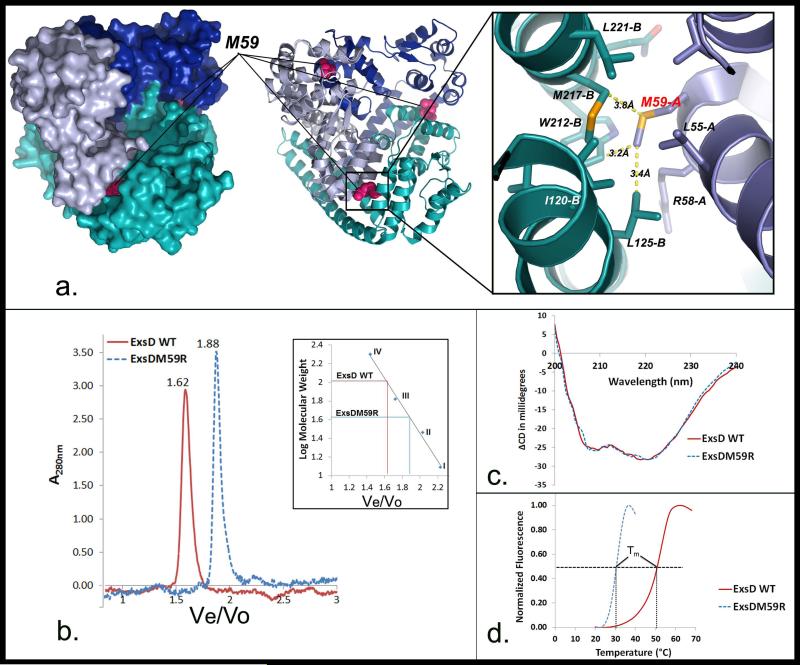
The studies described above demonstrate that independently folded ExsD and ExsA interact in vitro at 37°C. While it was still conceivable that the elevated temperature causes partial unfolding of either protein to permit binding as the original model posits, we sought to test an alternative hypothesis. A striking feature of ExsD is the apparent plasticity of its oligomeric state depending on the interacting partner. ExsD forms a 2:2 complex with ExsC and a 1:1 complex with ExsA, while analytical ultracentrifugation studies and the ExsD crystal structure suggest that ExsD self-associates to form a trimer in absence of the other two proteins [30, 31]. Our recent work suggests that the ExsC-ExsD complex actually consists of two ExsD monomers bound to an obligate ExsC dimer, rather than a dimer of dimers [32]. Therefore, ExsD appears to either form a homotrimer or enter 1:1 interactions with ExsC or ExsA, indicating that the mutually exclusive interactions of ExsD with ExsA and ExsC also compete with ExsD self-association. We hypothesized that dissociation of the ExsD trimer at 37°C prior to ExsA binding might account for the unusual kinetic phenomenon that prevents the association of the two proteins at 30°C. To test this hypothesis, we sought to engineer a monomeric variant of ExsD by disrupting the trimer through site-directed mutagenesis. We have previously reported the crystal structure of ExsD which contained three molecules in the asymmetric unit related by an almost perfect three-fold symmetry axis [30]. In the trimer, each ExsD molecule forms two distinct protein-protein interfaces, both covering approximately 1200 Å2 of surface area. Guided by the detailed structural maps of these interfaces, we focused our efforts on a hydrophobic patch formed by residues from helix α1 of one molecule and helices α6 and α9 from the other molecule (Fig. 2a). Because Met59 and Met217 are positioned at the heart of this interface, we decided to initially target these two residues. We reasoned that replacing one or both of these residues with amino acids possessing large charged side chains should not only disrupt the interface, but also increase the polarity of this part of the structure to prevent aggregation of the variant protein due to the exposure of a large hydrophobic surface. Both single point mutants were constructed, however, because the ExsDM59R variant already displayed the desired properties, the ExsDM217R variant was not characterized. Following purification of the variant protein, we conducted an analytical gel filtration study to estimate the approximate molecular weight of the ExsDM59R variant (Fig. 2b). Using a calibration curve obtained from analyzing a set of standard proteins, the retention time of the variant protein gave an apparent molecular weight of 42 kDa. While this is somewhat larger than the actual 32 kDa mass of an ExsD monomer, the discrepancy is readily explained by the distinctively non-globular shape of the molecule [30]. In contrast, wild-type ExsD (Fig. 2b) eluted significantly earlier from the gel filtration column and gave an estimated molecular weight of 102 kDa, consistent with a homotrimeric complex as previously reported [30].
Fig. 2. Characterization of the ExsDM59R variant.
a. Three views of the ExsD trimer (PDB code 3FD9). The mutated methionine 59 is highlighted at the three interfaces. The rightmost view provides a close-up of the intermolecular contacts of the mutated M59 residue. The letters behind the residue names denote the chain identifications of the different ExsD molecules in the trimer. b. Elution profiles for ExsDM59R and wild-type ExsD from an analytical gel filtration column. The inset shows the calibration curve for the column and the resulting apparent molecular weights for the two proteins. The four standards used to calibrate the column were I. cytochrome C (MW = 12.4 kDa), II. carbonic anhydrase (MW = 29 kDa), III. bovine serum albumin (MW = 66 kDa), and IV. β-amylase (MW = 200 kDa). c. Overlay of the circular dichroism spectra of ExsDM59R and wild-type ExsD. d. Differential scanning fluorimetry profiles for ExsDM59R and wild-type ExsD. The melting temperature (Tm) is defined as the temperature where 50% of the protein is unfolded, i.e., the inflection points of the curves. A Tm of 50.8°C was obtained for wild-type ExsD, while ExsDM59R had a Tm of 30.7°C.
To examine if the ExsDM59R variant had undergone a dramatic conformational change as a result of the mutation, we also compared circular dichroism spectra of wild-type ExsD and the variant. The spectra are virtually identical (Fig. 2c), thus indicating the observed difference in elution volumes from the gel filtration column is due to an altered oligomeric state of ExsDM59R rather than a conformational change.
Monomeric ExsDM59R efficiently inhibits ExsA-dependent transcription in vitro at 30°C
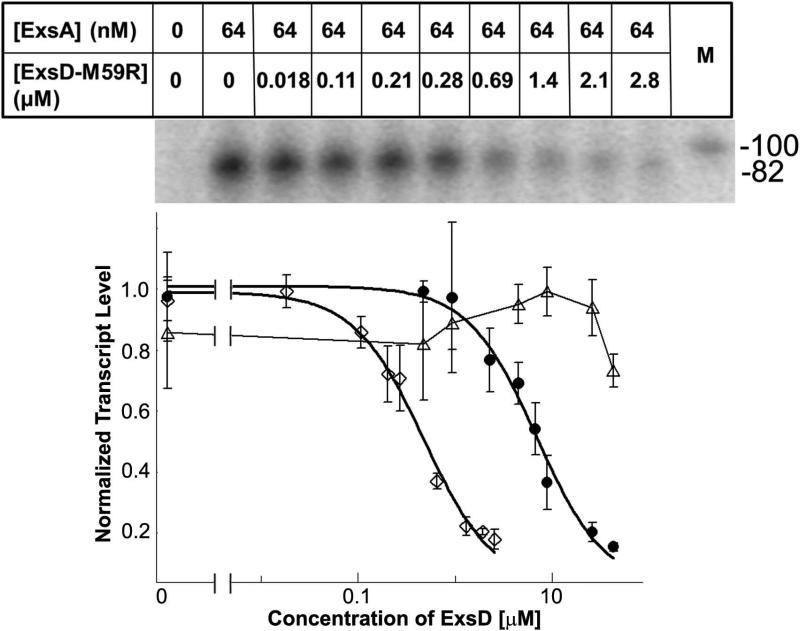
After confirming that the engineered ExsDM59R variant was indeed monomeric, we tested this protein in our in vitro transcription assay at 30°C (Fig. 3). In agreement with our hypothesis, this variant strongly inhibits ExsA-dependent transcription even at 30°C. A dose-response curve for ExsDM59R produced an IC50 value of approximately 0.5 μM, which is 15-fold lower than that obtained for wild-type ExsD at 37°C under otherwise identical conditions. This indicates that ExsDM59R is a stronger inhibitor than wild-type ExsD. In order to verify that ExsDM59R is specific for inhibiting ExsA-dependent transcription, an ExsA-independent RNA-1 promoter template was also tested at 30°C. As anticipated, ExsDM59R had no impact on the transcript levels when this promoter was used (Supplementary Fig. S4).
Fig. 3. Effect of ExsDM59R on ExsA-dependent transcription.
Dose response data and curve fit generated by measuring in vitro transcription levels in the presence of increasing amounts of ExsDM59R protein at the 10-minute time point and at 30°C (clear diamonds). To highlight the differences, data shown in figure 1b are reproduced. Clear triangles show the results for the titration experiment with wild-type ExsD at 30°C, while filled circles show data obtained at 37°C.
ExsDM59R disrupts ExsA dimerization, but does not interfere with ExsA promoter binding
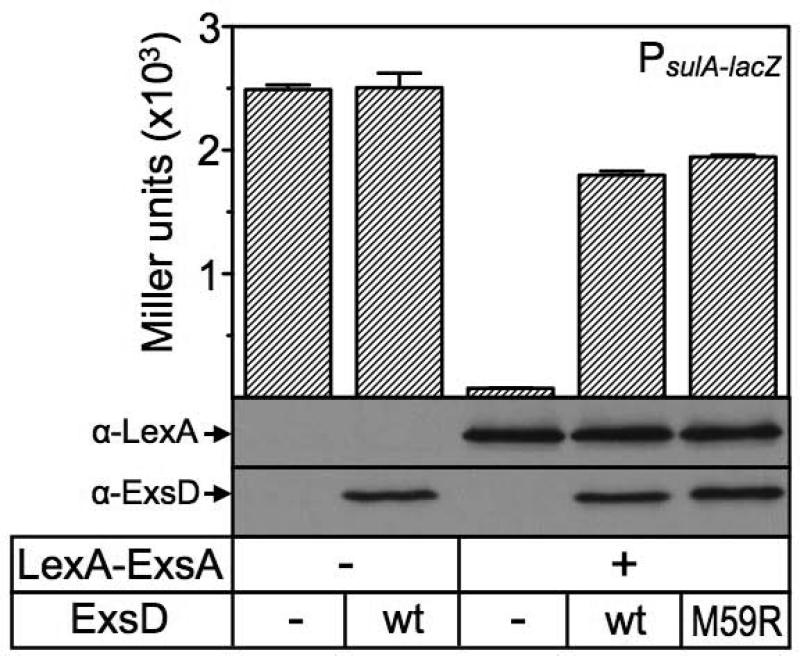
We replicated two experiments previously conducted with wild-type ExsD to directly examine the effect of ExsDM59R on ExsA dimerization and DNA binding. In these assays, ExsDM59R behaved in a manner that is indistinguishable from the wild-type protein. Using a bacterial mono-hybrid assay, we were able to demonstrate that ExsDM59R expression efficiently disrupts ExsA dimerization (Fig. 4). However, in subsequent EMSA studies, ExsDM59R did not measurably interfere with ExsA-DNA interactions (data not shown). Even pre-incubating ExsA and ExsDM59R at 37°C prior to running the EMSA gel did not affect the outcome of the assay. Based on the in vitro transcription results, we had anticipated that the ExsD variant would also interfere with ExsA-DNA binding interactions despite not being co-expressed with ExsA.
Fig. 4. ExsDM59R disrupts ExsA dimerization.
E. coli strain SU101 carrying a PsulA-lacZ transcriptional reporter was transformed with two different plasmids, as previously described [27]. The first plasmid was either a vector control (-, pSR658) or pSR658 expressing a LexA-ExsA fusion protein. The second plasmid was either a vector control (-, pJN105Δα) or pJN105Δα expressing either wild-type ExsD or the M59R variant. The resulting strains were cultured in LB medium containing the appropriate antibiotics and 50 μM IPTG to A600 = 1.0 and assayed for β-galactosidase activity. The reported values represent the average of three independent experiments.
Discussion
The ExsACDE signaling cascade displays a number of remarkable features, the most striking perhaps being that signal transduction does not involve the type of phosphate transfer events that mediate most of the signaling between extracellular milieu and bacterial gene expression. Instead, signal transduction in the ExsACDE pathway was shown to be based entirely on the competitive association and dissociation of various bimolecular complexes formed by the four involved proteins [23, 25, 33]. The newly discovered role of ExsD trimer formation in the regulation of T3SS gene expression adds an intriguing new wrinkle to the mechanism. Interestingly, as ITC studies have shown, ExsD self-association does not interfere with heterocomplex formation between ExsC and ExsD, suggesting that the affinity of the ExsC-ExsD complex is sufficiently high to overcome this obstacle [31]. The actual Kd for the ExsD-ExsA interactions is not known, however, consistent with its position at the bottom of the regulatory cascade, the complex is readily disrupted by ExsC [27]. Our data suggest that the affinity of the ExsD-ExsA complex falls perhaps in the hundreds of nanomolar range. Therefore, one would anticipate that the requirement for dissociation of the ExsD trimer prior to binding poses a more significant barrier for the ExsD-ExsA complex.
The thermoregulatory effect observed in the in vitro transcription assay appears to be produced by the self-association of ExsD into trimers. While the ExsDM59R variant has lost the ability to trimerize, we do not believe that the temperature increase from 30°C to 37°C causes a dramatic shift in the monomer-trimer equilibrium of wild-type ExsD. Rather, we propose that the temperature increase weakens this homotrimer sufficiently to permit the thermodynamically favored association of ExsD and ExsA. Two pieces of experimental data support the idea that ExsD is still primarily trimeric at 37°C. The IC50 value for the inhibition of ExsA-dependent transcription by wild-type ExsD is significantly larger than that observed for the ExsDM59R variant, suggesting that the wild-type protein still has a poorer affinity for ExsA. Second, our differential scanning fluorimetry experiments reveal that the ExsDM59R variant itself is significantly more temperature sensitive than the wild-type protein, presumably because the additional surface areas buried at the trimer interfaces stabilize the wild-type protein (Fig. 2d). Even though the melting temperature (Tm) of a protein depends on many factors, such as buffer conditions and the presence of ligands, we observed a striking drop in Tm for the variant, which would indicate that monomeric ExsD is not stable at 37°C unless it is associated with a different protein, such as ExsA or ExsC.
The observation that ExsDM59R did not affect ExsA-DNA interactions in the EMSA experiments is intriguing. In conjunction with the results of our in vitro transcription experiments, which indirectly demonstrate that ExsDM59R does interact with ExsA at 30°C, these findings appear to suggest that the presence of RNA polymerase is required for the association of ExsD and ExsA. Perhaps the binding of ExsA to RNA polymerase triggers a conformational change in the transcription factor that reveals the otherwise obscured ExsD binding site.
Even though ExsD self-association has been documented in two independent studies, its biological role is not clear [30, 31]. We carried out an initial comparison of wild-type P. aeruginosa and a mutant strain carrying an M59R mutation in the chromosomal exsD gene using a PexsD-lacZ transcriptional reporter [25]. In this study, we found no difference in reporter activity under non-inducing or inducing conditions at 30°C (data not shown). These results were perhaps not completely unexpected and highlight the fundamental question of when the cellular concentration of free ExsD will most likely reach significant levels. Because ExsD is not a secreted protein and expression is positively regulated by ExsA, the protein may accumulate inside the cell during a prolonged period of T3SS induction. However, the pool of free ExsD is diminished by its associations with ExsA and ExsC. Increases in the cellular levels of ExsE, on the other hand, should cause the concentration of free ExsD to rise. In the context of an infection, this could reflect a scenario where a bacterial cell releases from the host cell at a later stage, causing the closure the T3SS channel, and thereby, an intracellular accumulation of the ExsE-ExsC complex. While the temperature increase causes an overall rise in the expression levels of T3SS genes [34, 35], the ability of ExsD to re-associate with ExsA at 37°C may serve to dampen this effect at this stage, and thus, fine-tune virulence gene expression. The ExsDM59R variant provides a useful system for our molecular studies, however, due to its temperature sensitivity, it is not well suited for testing this model in vivo. Perhaps, a mutation that attenuates, but does not fully disrupt ExsD self-association, would be a better choice for these studies. In vivo, the temperature change from 30°C to 37°C is likely to affect the expression of multiple P. aeruginosa genes, and it might be instructive to examine the role of ExsD self-association in the larger context of these changes through computational modeling studies.
In summary, we have shown in vitro that independently expressed ExsD does not inhibit ExsA-dependent transcription at 30°C, but inhibits efficiently at 37°C. A monomeric ExsD variant strongly inhibits ExsA-dependent transcription at 30°C, suggesting that the temperature effect is caused by ExsD self-association. ExsD self-association appears to have no impact on basal T3SS expression levels at 30 or 37°C. Instead, we propose that trimerization limits the effective concentration of ExsD and stabilizes the protein. Re-association of the accumulated ExsD protein with ExsA may assist in fine-tuning T3SS gene expression at a later stage of an infection.
Materials and Methods
Recombinant protein expression and purification
ExsA and ExsD were overexpressed in E. coli from a vector constructed by Gateway recombinational cloning (Invitrogen, Carlsbad, CA, USA). A tobacco etch virus (TEV) protease recognition site and the appropriate att recombination sites (attB1 and attB2) were added to the exsA and exsD genes during PCR, and the amplicons were subsequently recombined into pDONR201 (Invitrogen). The nucleotide sequences of the ORFs were verified, then recombined into the destination vector pDEST-HisMBP [36] to create the expression vectors pFS-HMBPExsD and pFS-HMBPExsA. These vectors were designed to produce either ExsA or ExsD as a fusion to the C-terminus of an N-terminally His6-tagged E. coli maltose-binding protein (MBP).
Single colonies of E. coli BL21(DE3) CodonPlus RIL cells (Stratagene, La Jolla, CA, USA) containing either expression plasmid were used to inoculate 125 mL of Luria broth (LB) supplemented with 2 g/L dextrose, 100 μg/mL ampicillin, and 30 μg/mL chloramphenicol. The cultures were grown with shaking (225 rpm) to saturation overnight at 37°C and then diluted 66-fold into 6 L of fresh medium. ExsA cultures were grown to an OD600 of 1.0, ExsD cultures were grown to an OD600 of 0.5, and ExsDM59R cultures were grown to an OD600 of 0.8. All three cultures were induced with IPTG at a final concentration of 1 mM. The induction temperature for the ExsA cultures was 18°C, and they were shaken for six hours. ExsD cultures were induced at 28°C for four hours, and ExsDM59R cultures were induced at 17°C overnight. Cells were harvested by centrifugation at 5,000 × g for 15 minutes. The cell pastes were resuspended in 200 mL of 500 mM NaCl, 25 mM imidazole, 50 mM Tris-HCl (pH 7.4), 2 mM DTT (buffer A), along with three tablets of Complete, EDTA-free Protease Inhibitor Cocktail (Roche Applied Science, Indianapolis, IN, USA). The cells were lysed via sonication and centrifuged at 40,000 × g for 25 minutes. The supernatants were filtered through 0.45-μm polyethersulfone membranes and applied to a 30 mL Ni-NTA Superflow affinity column (Qiagen, Valencia, CA, USA) equilibrated with buffer A. For each run, the column was washed with five column volumes of buffer A, and proteins were eluted with a linear gradient from 25 to 250 mM imidazole (pH 7.4). The His6-MBP-ExsD protein was digested with 5 mg His-tagged TEV (S219V) protease [37] while being dialyzed overnight in 150 mM NaCl, 50 mM Tris-HCl (pH 7.4), 11.6 mM imidazole (pH 7.4), and 1 mM DTT. The sample was then passed through a second Ni-NTA column to remove both the His6-MBP tag and the protease, using the same buffers as the first Ni-NTA column. The protein sample was collected in the flow through. The sample was diluted with 50 mM Tris-HCl (pH 7.4) and 2 mM DTT in order to lower the NaCl concentration to 50 mM. The ExsD sample was loaded onto a HiTrap Q HP column (GE Healthcare, Waukesha, WI, USA) that had been equilibrated with 50 mM NaCl, 25 mM Tris-HCl (pH 7.4), and 2 mM DTT, and elution was achieved by applying a linear gradient of NaCl from 50 mM to 1 M. Finally, gel filtration was performed using 150 mM NaCl, 25 mM Tris-HCl (pH 7.4), and 2 mM TCEP (ExsD storage buffer). ExsDM59R was purified in the same manner as wild-type ExsD, except that no anion exchange was performed. All purification steps were performed at 4°C. After each purification step, fractions were analyzed via SDS-PAGE and pooled accordingly. ExsD and ExsDM59R were concentrated to 4.5 mg/mL and 6.8 mg/mL, respectively. Protein samples were flash-frozen using liquid nitrogen and stored at -80°C.
The His6-MBP-ExsA fusion protein was treated differently. Following the initial Ni-NTA affinity purification step, the fusion protein was dialyzed against a buffer of 50 mM NaCl, 25 mM Tris-HCl (pH 7.4), and 2 mM DTT and loaded onto a HiTrap Q HP column (GE Healthcare) that had been equilibrated with the same buffer. The His6-MBP-ExsA fusion protein was eluted using a linear NaCl gradient from 0.05 M to 1 M. The sample was dialyzed against 2 L of 45 mM NaCl, 25 mM Tris-HCl (pH 7.15), and 2 mM DTT (buffer B) overnight. The sample was then loaded onto a HiTrap Heparin HP column (GE Healthcare) equilibrated in buffer B and eluted with a 0.05 M to 1 M gradient of NaCl. The NaCl concentration in the His-MBP-ExsA sample was adjusted to 0.5 M, and the fusion protein was digested with 3 mg of His-tagged TEV(S219V) protease at 4°C overnight. Next, ExsA was run through a second Ni-NTA Superflow affinity column, this time collecting ExsA in the flow through. Finally, gel filtration using a HighLoad 26/60 Superdex 200 prep grade column (GE Healthcare) was performed with the ExsA sample using 500 mM NaCl, 25 mM Tris-HCl (pH 7.4), and 2 mM TCEP (ExsA storage buffer). The sample was concentrated to 1 mg/mL, flash-frozen using liquid nitrogen, and stored at -80°C.
RNA polymerase purification and specific activity determination
RNA polymerase (RNAP) was purified from P. aeruginosa PAO1 cells following the original procedure of Allan and Kropinski [38]. However, changes were made to the later chromatographic steps. All purification steps were performed at 4°C. P. aeruginosa PAO1 cultures were grown in LB broth to an OD600 of 0.8, harvested by centrifugation at 6,000 × g, then lysed by sonication. The cell debris was removed by centrifugation at 35,000 × g for 30 minutes, and 25% polyethyleneimine (pH 7.5) was added to the supernatant to a final concentration of 0.5% in order to precipitate the RNAP. The supernatant was centrifuged at 35,000 × g for 30 minutes. The polyethyleneimine precipitate was washed with 10 mM Tris-HCl (pH 8.0), 250 mM NaCl, 5% glycerol, 0.05 mM EDTA, 1 mM DTT, and 0.1 mM PMSF (wash buffer) and centrifuged at 35,000 × g for 30 minutes. RNAP was released by resuspending the pellet in 10 mM Tris-HCl (pH 8.0), 800 mM NaCl, 5% glycerol, 0.05 mM EDTA, 1 mM DTT, and 0.1 mM PMSF (release buffer) and centrifuged at 25,000 × g for 30 minutes. Ammonium sulfate was added to the supernatant to a final concentration of 30%, followed by gentle stirring for one hour, and centrifugation at 35,000 × g for 30 minutes. Additional ammonium sulfate was then added to bring the supernatant to 60% saturation. After a second centrifugation at 35,000 × g for 30 minutes, the pellet was resuspended in 1 mL wash buffer per liter of original culture. The suspension was dialyzed versus 2 L wash buffer overnight. The dialyzed RNAP sample was centrifuged, and the supernatant was filtered in preparation for gel filtration. The sample was run through a Sephacryl S-300 HR column (GE Healthcare) using wash buffer. The fractions were analyzed by SDS-PAGE and collected to run on a Hi-Trap Heparin HP column (GE Healthcare) using a loading buffer composed of 10 mM Tris-HCl (pH 8.0), 250 mM NaCl, 5% glycerol, 0.05 mM EDTA, and 1 mM TCEP. RNAP was eluted using a linear gradient of 0.25 M to 1 M NaCl. Fractions were analyzed via SDS-PAGE, pooled, and concentrated to 1 mg/mL of total protein. Glycerol was added to a final concentration of 50%. RNAP was aliquoted and stored at -20°C.
The specific activity of the purified P. aeruginosa RNAP was determined by comparing its activity to a standard curve generated with different amounts of E. coli RNA Polymerase Holoenzyme (Epicentre Biotechnologies, Madison, WI, USA) using an ExsA-independent RNA-1 promoter which produces a 108 base transcript [29].
Site-directed mutagenesis
The ExsDM59R variant was generated by site-directed mutagenesis using Quik-Change (Stratagene) and the manufacturer's suggested protocol. The following primers were used: 5′-CTGCAGCGGCGGCTGCCGCGCCTGCGGCTGGAGC-3′ 5′-GGCGCGGCAGCCGCCGCTGCAGCAACGCCAG-3′.
ExsA-dependent in vitro transcription assays
The linear DNA template used in each assay encompassed positions -207 to 94 of the PexsD promoter, relative to the transcription start site; and from this template, RNA polymerase synthesizes an 82 base mRNA transcript. The template was produced by PCR using forward primer 5′-CATCAGTTGCTGCTCAACAGCG-3′ and reverse primer 5′-CACCGCTTCTCGGGAGTACTGC-3′. The PCR product was run on a 2% agarose gel and purified using the Wizard SV Gel and PCR Clean-up System (Promega, Madison, WI, USA). Each 30 μL transcription assay reaction contained 4.4 fM of promoter template, 50.4 μM bovine serum albumin (to eliminate non-specific protein-protein interactions), 10 U purified RNA polymerase from P. aeruginosa (see above), 1 U RiboGuard RNase Inhibitor (Epicentre Biotechnologies), 15 ng/μL poly(deoxyinosinic-deoxycytidylic) acid (to prevent non-specific transcription initiation), 133 mM NaCl, 32 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 25 μM EDTA, 0.9 mM TCEP, 0.2 mM DTT, and 15.5% glycerol. The time-course experiments contained 64 nM ExsA and either no ExsD or 50 μM ExsD (no ExsA was added for the RNA-1 control experiments). Samples were mixed and allowed to equilibrate at room temperature for five minutes. Samples were then pre-incubated for 10 minutes at either 30°C or 37°C, depending on the experiment. Next, 3 μL NTPs (stock concentrations of 200 μM ATP, CTP, GTP and 40 μM UTP) mixed with 0.2 μL (0.2 μCi) of 3.3 mM P32-alpha UTP was added to each sample to start the reaction, and samples were incubated at either 30°C or 37°C, depending on the experiment. After the reactions were stopped by adding 12 μL 1X stop solution (3M ammonium acetate, 50 mM EDTA, 0.11 mg/mL glycogen), 170 μL 100% cold ethanol was added, and the samples were incubated at -20°C for one hour. Following centrifugation at 12,000 × g for 15 minutes, the supernatant was discarded and pellets were resuspended in 12 μL 1X TBE (Tris/Borate/EDTA)-urea sample buffer and heated at 70°C for five minutes. After a brief centrifugation, the samples were loaded onto a 10% TBE-urea gel and run at 200 mV for 60 minutes. Gels were exposed to a storage phosphor screen (GE Healthcare) for 16 hours. The phosphor screen was scanned using a Typhoon Trio Variable Mode Imager (GE Healthcare), and gel bands were quantified using Image Quant TL v2005 (Amersham Biosciences, Piscataway, NJ, USA). Each experiment was performed in triplicate, and curve fits were analyzed with XLfit (IDBS, Bridgewater, NJ, USA).
Analytical size exclusion chromatography
100 μL samples of 2.1 μM purified wild-type ExsD and ExsDM59R, each containing 150 mM NaCl, 25 mM Tris-HCl (pH 7.4), and 2 mM TCEP, were separately loaded onto a Superdex 200 10/300 GL column (GE Healthcare). The proteins were eluted with 150 mM NaCl, 25 mM Tris-HCl (pH 7.4), and 2 mM TCEP. The absorbance at UV280 was plotted on the y-axis and Ve/Vo was plotted on the x-axis, where Ve is the elution volume and Vo is the void volume (7.93 mL). Cytochrome C, carbonic anhydrase, bovine serum albumin, and β-amylase standards were individually run using the same elution buffer. These standards were plotted using the log of their known molecular weights on the y-axis and Ve/Vo on the x-axis; from this, a best fit line was determined. The molecular weights of wild-type ExsD and ExsDM59R were subsequently estimated using the fitted linear equation.
Circular dichroism (CD) measurements
Circular dichroism measurements using far-UV (200-240 nm) of wild-type ExsD and ExsDM59R, each at a concentration of 5.3 μM, were separately measured at 4°C using a JASCO J-815 Circular Dichroism Spectrometer (JASCO, Easton, MD, USA) with a 1 mm pathlength cuvette. Each protein sample contained 150 mM NaCl, 25 mM Tris-HCl (pH 7.4), and 2 mM TCEP. The spectra obtained represent an average of three scans that were corrected for the buffer baseline.
Differential scanning fluorimetry (DSF)
The DSF experiments were performed utilizing an ABI 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Each 30 μL reaction contained 5x Sypro Orange (Ex. 490 nm, Em. 530 nm) (Invitrogen), 10 μM wild-type ExsD or ExsDM59R, 150 mM NaCl, 8.3 mM Tris-HCl (pH 7.4), and 0.67 mM TCEP. Starting at 10°C, the temperature was incrementally increased to 68°C at a rate of 1°C per minute. Data analysis was conducted using XLfit (IDBS).
LexA mono-hybrid assay
Quik-Change (Stratagene) mutagenesis was used to modify the previously constructed arabinose-inducible vector pJNexsDΔα [27] by introducing an M59R substitution in exsD using primer ExsDM59R (5′-CGTTGCTGCAGCGGCGCCTGCCGCGCCTGC-3′). The resulting plasmid was designated pAM102. E. coli strain SU101, which carries a LexA-repressible PsulA-lacZ reporter, was transformed with pAM102 and the IPTG-inducible pSR658-exsA vector [27] and selected on LB agar with gentamicin (15 μg/mL) and tetracycline (12 μg/mL). Self-association of ExsA in the presence of ExsD or ExsDM59R was measured as previously described [27, 39]. Briefly, strains containing the appropriate expression vectors were grown overnight at 30°C with shaking in 5 mL LB with 50 μM IPTG, 0.5% arabinose, and appropriate antibiotics. The following day, cultures were diluted to OD600 = 0.1 in 5 mL trypticase soy broth supplemented with 100 mM monosodium glutamate and 1% glycerol, as well as 50 μM IPTG, 0.5% arabinose, and antibiotics and grown at 30°C with shaking to OD600 = 1.0. β-galactosidase levels were measured as previously described [27]. Miller units were reported as the average of three replicates with error bars representing the standard error of the mean (SEM). Immunoblots were performed with α-ExsD and α-LexA1-87 to confirm the stability of ExsD and ExsA-LexA1-87, respectively.
Supplementary Material
Acknowledgements
This study was supported by the National Institutes of Health (1R21AI101774-01 to FDS and RO1- AI055042 to TLY) and the American Heart Association (09SDG2260401 to FDS).
Abbreviations
- CD
circular dichroism
- DSF
differential scanning fluorimetry
- ITC
isothermal titration calorimetry
- DTT
dithiothreitol
- EMSA
electrophoretic mobility shift assay
- ExsACDE
ExsA-ExsC-ExsD-ExsE
- IPTG
isopropyl-β-D-thiogalacto-pyranoside
- MBP
maltose binding protein
- TCEP
Tris(2-carboxyethyl)phosphine
- TEV
tobacco etch virus
- T3SS
type III secretion system
References
- 1.Church D, Elsayed S, Reid O, Winston B, Lindsay R. Burn wound infections. Clin Microbiol Rev. 2006;19:403–34. doi: 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Depuydt P, Benoit D, Vogelaers D, Claeys G, Verschraegen G, Vandewoude K, Decruyenaere J, Blot S. Outcome in bacteremia associated with nosocomial pneumonia and the impact of pathogen prediction by tracheal surveillance cultures. Intensive Care Med. 2006;32:1773–81. doi: 10.1007/s00134-006-0354-8. [DOI] [PubMed] [Google Scholar]
- 3.Garau J, Gomez L. Pseudomonas aeruginosa pneumonia. Curr Opin Infect Dis. 2003;16:135–43. doi: 10.1097/00001432-200304000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Gaynes R, Edwards JR. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41:848–54. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 5.Johnson LE, D'Agata EM, Paterson DL, Clarke L, Qureshi ZA, Potoski BA, Peleg AY. Pseudomonas aeruginosa bacteremia over a 10-year period: multidrug resistance and outcomes in transplant recipients. Transpl Infect Dis. 2009;11:227–34. doi: 10.1111/j.1399-3062.2009.00380.x. [DOI] [PubMed] [Google Scholar]
- 6.Trautmann M, Lepper PM, Haller M. Ecology of Pseudomonas aeruginosa in the intensive care unit and the evolving role of water outlets as a reservoir of the organism. Am J Infect Control. 2005;33:S41–9. doi: 10.1016/j.ajic.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Wine JJ. The genesis of cystic fibrosis lung disease. J Clin Invest. 1999;103:309–12. doi: 10.1172/JCI6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hauser AR, Cobb E, Bodi M, Mariscal D, Valles J, Engel JN, Rello J. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med. 2002;30:521–8. doi: 10.1097/00003246-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Hsu DI, Okamoto MP, Murthy R, Wong-Beringer A. Fluoroquinolone-resistant Pseudomonas aeruginosa: risk factors for acquisition and impact on outcomes. J Antimicrob Chemother. 2005;55:535–41. doi: 10.1093/jac/dki026. [DOI] [PubMed] [Google Scholar]
- 10.Roy-Burman A, Savel RH, Racine S, Swanson BL, Revadigar NS, Fujimoto J, Sawa T, Frank DW, Wiener-Kronish JP. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J Infect Dis. 2001;183:1767–74. doi: 10.1086/320737. [DOI] [PubMed] [Google Scholar]
- 11.Angus AA, Evans DJ, Barbieri JT, Fleiszig SM. The ADP-ribosylation domain of Pseudomonas aeruginosa ExoS is required for membrane bleb niche formation and bacterial survival within epithelial cells. Infect Immun. 2010;78:4500–10. doi: 10.1128/IAI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dacheux D, Toussaint B, Richard M, Brochier G, Croize J, Attree I. Pseudomonas aeruginosa cystic fibrosis isolates induce rapid, type III secretion-dependent, but ExoU-independent, oncosis of macrophages and polymorphonuclear neutrophils. Infect Immun. 2000;68:2916–24. doi: 10.1128/iai.68.5.2916-2924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans DJ, Frank DW, Finck-Barbancon V, Wu C, Fleiszig SM. Pseudomonas aeruginosa invasion and cytotoxicity are independent events, both of which involve protein tyrosine kinase activity. Infect Immun. 1998;66:1453–9. doi: 10.1128/iai.66.4.1453-1459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finck-Barbancon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish JP, Fleiszig SM, Wu C, Mende-Mueller L, Frank DW. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Molecular microbiology. 1997;25:547–57. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 15.Maresso AW, Deng Q, Pereckas MS, Wakim BT, Barbieri JT. Pseudomonas aeruginosa ExoS ADP-ribosyltransferase inhibits ERM phosphorylation. Cell Microbiol. 2007;9:97–105. doi: 10.1111/j.1462-5822.2006.00770.x. [DOI] [PubMed] [Google Scholar]
- 16.Saliba AM, Filloux A, Ball G, Silva AS, Assis MC, Plotkowski MC. Type III secretion-mediated killing of endothelial cells by Pseudomonas aeruginosa. Microb Pathog. 2002;33:153–66. [PubMed] [Google Scholar]
- 17.Sato H, Frank DW. ExoU is a potent intracellular phospholipase. Molecular microbiology. 2004;53:1279–90. doi: 10.1111/j.1365-2958.2004.04194.x. [DOI] [PubMed] [Google Scholar]
- 18.Zolfaghar I, Evans DJ, Ronaghi R, Fleiszig SM. Type III secretion-dependent modulation of innate immunity as one of multiple factors regulated by Pseudomonas aeruginosa RetS. Infect Immun. 2006;74:3880–9. doi: 10.1128/IAI.01891-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008;451:485–8. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- 20.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of flagellar type III secretion. Nature. 2008;451:489–92. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- 21.Shen DK, Filopon D, Kuhn L, Polack B, Toussaint B. PsrA is a positive transcriptional regulator of the type III secretion system in Pseudomonas aeruginosa. Infection and Immunity. 2006;74:1121–9. doi: 10.1128/IAI.74.2.1121-1129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urbanowski ML, Lykken GL, Yahr TL. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proceedings of the National Academy of Sciences. 2005;102:9930–5. doi: 10.1073/pnas.0504405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences. 2005;102:8006–11. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hovey AK, Frank DW. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol. 1995;177:4427–36. doi: 10.1128/jb.177.15.4427-4436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCaw ML, Lykken GL, Singh PK, Yahr TL. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Molecular microbiology. 2002;46:1123–33. doi: 10.1046/j.1365-2958.2002.03228.x. [DOI] [PubMed] [Google Scholar]
- 26.Thibault J, Faudry E, Ebel C, Attree I, Elsen S. Anti-activator ExsD forms a 1:1 complex with ExsA to inhibit transcription of type III secretion operons. J Biol Chem. 2009;284:15762–70. doi: 10.1074/jbc.M109.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brutinel ED, Vakulskas CA, Yahr TL. ExsD inhibits expression of the Pseudomonas aeruginosa type III secretion system by disrupting ExsA self-association and DNA binding activity. J Bacteriol. 2010;192:1479–86. doi: 10.1128/JB.01457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vakulskas CA, Brady KM, Yahr TL. Mechanism of Transcriptional Activation by Pseudomonas aeruginosa ExsA. Journal of Bacteriology. 2009;191:6654–6664. doi: 10.1128/JB.00902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finney AH, Blick RJ, Murakami K, Ishihama A, Stevens AM. Role of the C-terminal domain of the alpha subunit of RNA polymerase in LuxR-dependent transcriptional activation of the lux operon during quorum sensing. J Bacteriol. 2002;184:4520–8. doi: 10.1128/JB.184.16.4520-4528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernhards RC, Jing X, Vogelaar NJ, Robinson H, Schubot FD. Structural evidence suggests that antiactivator ExsD from Pseudomonas aeruginosa is a DNA binding protein. Protein Sci. 2009;18:503–13. doi: 10.1002/pro.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lykken GL, Chen G, Brutinel ED, Chen L, Yahr TL. Characterization of ExsC and ExsD self-association and heterocomplex formation. J Bacteriol. 2006;188:6832–40. doi: 10.1128/JB.00884-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogelaar NJ, Jing X, Robinson HH, Schubot FD. Analysis of the crystal structure of the ExsC.ExsE complex reveals distinctive binding interactions of the Pseudomonas aeruginosa type III secretion chaperone ExsC with ExsE and ExsD. Biochemistry. 2010;49:5870–9. doi: 10.1021/bi100432e. [DOI] [PubMed] [Google Scholar]
- 33.Dasgupta N, Lykken GL, Wolfgang MC, Yahr TL. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Molecular microbiology. 2004;53:297–308. doi: 10.1111/j.1365-2958.2004.04128.x. [DOI] [PubMed] [Google Scholar]
- 34.Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S. The Single-Nucleotide Resolution Transcriptome of Pseudomonas aeruginosa Grown in Body Temperature. PLoS pathogens. 2012;8:e1002945. doi: 10.1371/journal.ppat.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dotsch A, Eckweiler D, Schniederjans M, Zimmermann A, Jensen V, Scharfe M, Geffers R, Haussler S. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PloS one. 2012;7:e31092. doi: 10.1371/journal.pone.0031092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nallamsetty S, Austin BP, Penrose KJ, Waugh DS. Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 2005;14:2964–71. doi: 10.1110/ps.051718605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 38.Allan B, Kropinski AM. DNA-dependent RNA polymerase from Pseudomonas aeruginosa. Biochem Cell Biol. 1987;65:776–82. doi: 10.1139/o87-101. [DOI] [PubMed] [Google Scholar]
- 39.Daines DA, Granger-Schnarr M, Dimitrova M, Silver RP. Use of LexA-based system to identify protein-protein interactions in vivo. Methods in enzymology. 2002;358:153–61. doi: 10.1016/s0076-6879(02)58087-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.