Summary
Mobile introns home to allelic sites, but how they are stimulated to invade ectopic locations on an evolutionary time-scale is unclear [1]. Here we show that a group I intron can move to degenerate sites under oxidizing conditions. The phage T4 td intron endonuclease, I-TevI is responsible for this infidelity. We demonstrate here that I-TevI, which promotes mobility and is subject to autorepression [2] and translational control [3], is also regulated post-translationally by a redox mechanism. Redox regulation is exercised by a zinc finger (ZF), in a linker that connects the catalytic domain of I-TevI to the DNA binding domain (Fig. 1A). Four cysteines (Cys4) coordinate Zn2+ in the ZF (Fig. 1B), which ensures that I-TevI cleaves its DNA substrate at a fixed distance, 23–25 nucleotides upstream of the intron insertion site [4]. In addition to providing structural diversification, Cys4 ZFs can be molecular switches [5]. Redox signaling provides a rapid response, with switching orchestrated through reversible thiol coordination of Zn2+ [6–8]. We show that the fidelity of I-TevI cleavage is controlled by redox-responsive Zn2+ cycling. When the ZF is mutated, or after exposure of the wild-type I-TevI to H2O2, intron homing to degenerate sites is increased, likely due to indiscriminate DNA cleavage. These results suggest a mechanism for rapid intron dispersal, joining recent descriptions of the activation of biomolecular processes by oxidative stress through cysteine chemistry [9, 10].
Results
Redox-dependent switch in activity of I-TevI
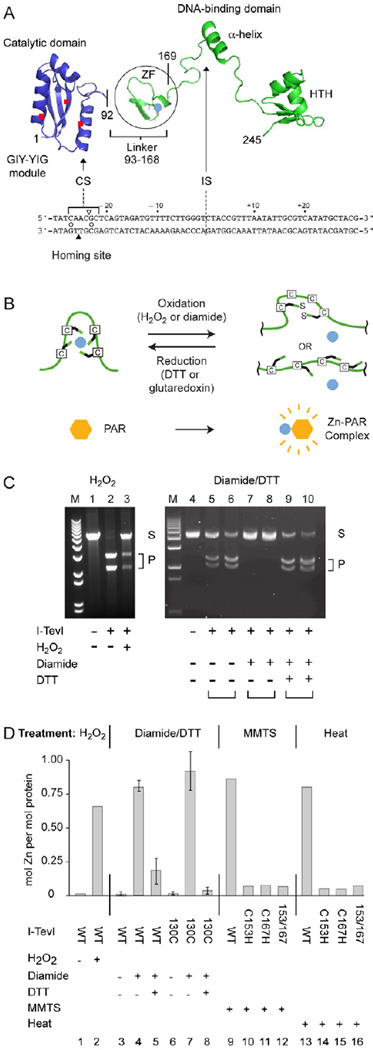
We first investigated the role of the zinc finger (ZF) of I-TevI (Fig. 1A and B) after in vitro treatment with H2O2. H2O2, a natural by-product of oxidative metabolism, inhibited I-TevI, presumably by oxidation of the ZF (Fig. 1C, lanes 1–3). To test the reversibility of the response, we treated I-TevI with diamide, an oxidizer that allows cycling, and then dithiothreitol (DTT), to reduce the ZF (Fig. 1C, lanes 4–10). Incubation of I-TevI with diamide again prevented cleavage (Fig. 1C, lanes 7 and 8), whereas DTT restored activity (Fig. 1C, lanes 9 and 10).
Figure 1. I-TevI cleavage is redox sensitive.
(A) Modular endonuclease I-TevI with zinc finger (ZF). The catalytic domain of the GIY-YIG endonuclease family is separated by a lengthy linker, which contains the ZF (Zn2+, turquoise dot), from the DNA binding domain with an α-helix and helix-turn-helix (HTH). I-TevI binds to the DNA homing site (below structure). The triangles indicate the cleavage on the top (open) and bottom (filled) strands, 23 and 25 nt from the intron insertion site (IS). The open circles between the strands indicate the two consensus nucleotides for cleavage.
(B) Redox scheme for ZF. The liberation and re-sequestration of a zinc ion (turquoise) under oxidative and reductive conditions is shown, with complexing of released Zn2+ by PAR reagent.
(C) Cleavage assays under different redox conditions. Cleavage of pBS tdΔIn substrate (250 ng) at 37°C with wild type I-TevI (25 ng) after treatment with 1 mM H2O2 for 5 min (lanes 1–3) or with I-TevI-H40Y (1.25 µg or 2.5 µg for each condition tested; I-TevI:DNA molar ratio 6:1 and 3:1, respectively), after treatment with 2.5 mM diamide in the absence and presence of 10 mM DTT for 15 min (lanes 4–10) is shown. S = substrate, P = cleavage products, M = size markers. The ability of T4 thioredoxin, Grx1, plus glutathione to provide reducing equivalents is shown in Supplemental Figure 1.
(D) Abstraction of Zn2+ from the I-TevI ZF and redox-dependent Zn2+ cycling. Zn2+ release from I-TevI-H40Y was measured by the PAR assay. Treatment (5 min) was with 0.5 mM H2O2, or 2.5 mM diamide and 10 mM DTT or 10 mM MMTS at 25°C or heat (100°C). WT was wild-type I-TevI in columns 1 and 2, and I-TevI-H40Y derivatives in columns 3–16. Truncated derivative C130 contains linker sequence (from residue 130) and the DNA binding domain, but not the catalytic domain [36]. Redox cycling (15-min incubation for each condition) is in columns 3–8. Error bars represent the results from three independent experiments.
Two biologically relevant molecules, phage T4 thioredoxins Grx-1 and Grx-2 might provide the reducing equivalents. Grx-1 alone could not restore the cleavage activity to diamide-treated I-TevI (Supplemental Fig. 1, lane 5), but in the presence of glutathione, Grx-1 was able to function in a similar manner to DTT (Supplemental Fig. 1 lane 8). However, I-TevI was not a substrate for Grx-2 (data not shown), suggesting that there may be glutaredoxin specialization.
Zn2+ content of I-TevI changes under oxidizing and reducing conditions
I-TevI was incubated with the Zn2+-complexing reagent 4-(2-pyridylazo) resorcinol (PAR) to determine Zn2+ levels by spectroscopic measurement (500 nm). H2O2 treatment resulted in Zn2+release (Fig. 1D, columns 1 and 2). Likewise, the reversible oxidant, diamide, led to Zn2+ release, at a molar excess of 4:1 over I-TevI (Fig. 1D, columns 3 and 4). When DTT was added at an 8:1 molar excess to counteract the diamide and re-reduce I-TevI, free Zn2+ dropped >3-fold, indicating that I-TevI re-sequestered the Zn2+ (Fig. 1D, column 5), and that there was Zn2+ cycling. Up to 1 mole Zn2+/mole I-TevI was released suggesting that almost all of the ZF pockets were occupied with Zn2+.
To demonstrate that the source of the Zn2+ is from the segment of I-TevI that contains the ZF and unrelated to the I-TevI catalytic domain, parallel experiments were carried out with deletion derivative I-TevI-130C [11], which lacks the catalytic domain. The data indicate similar Zn2+ cycling (Fig. 1D, columns 6–8). Furthermore, cysteine-to-histidine mutants of the ZF (C153H, C167H and C153H/C167H) could not coordinate Zn2+, as shown with the sulfhydryl-reactive reagent methyl methanethiosulfate (MMTS) (Fig. 1D, columns 9–12) or heat (Fig. 1D, columns 13–16). Collectively, these results indicate that the metal ion is reversibly coordinated by the ZF.
ZF mutants have altered cleavage activity on degenerate homing sites
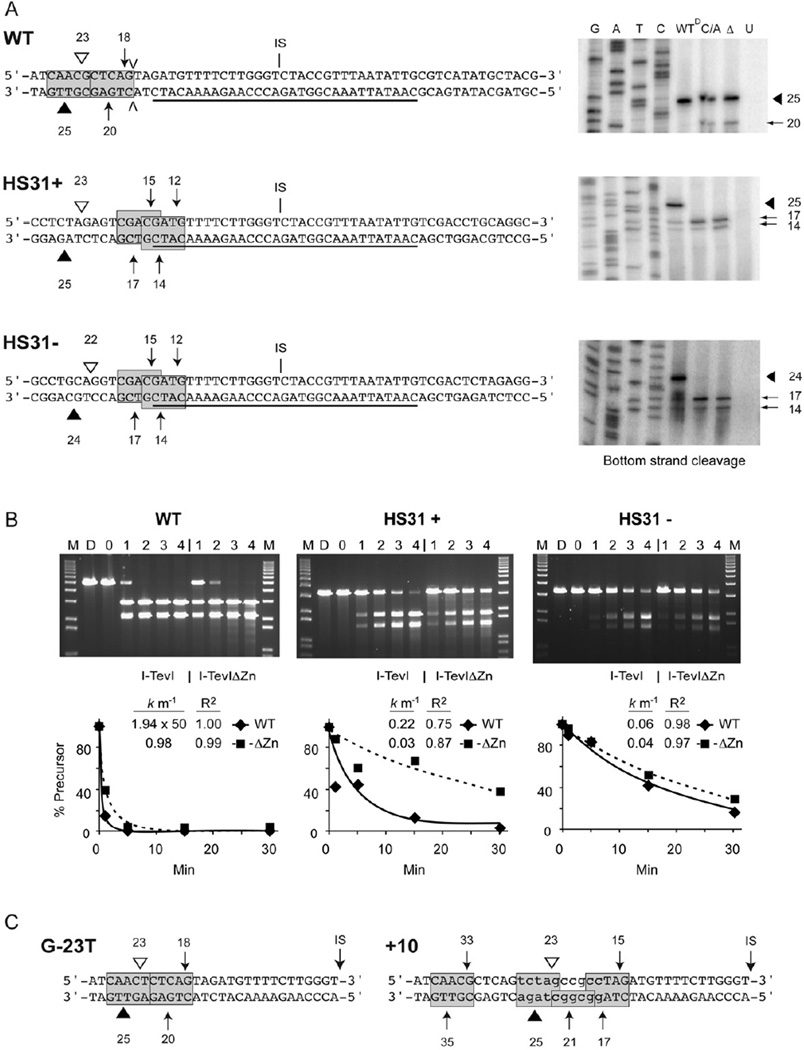
We had shown that mutants from which the ZF was deleted (ΔZn) or which contained four Cys-to-Ala mutations (CZnA) cleaved the operator site, which lacks the preferred cleavage sequence, more efficiently than did the wild-type enzyme [4, 12]. To test the effect of Zn2+depletion on enzyme fidelity we used those mutants, because H2O2 and diamide decrease cleavage to a point that defies measurement of cleavage accuracy. We thus measured cleavage of the wild-type homing site and two non-native sites that lack the natural cleavage sequence. These are the HS31+ and HS31− homing sites, with a synthetic duplex spanning the intron insertion site (15 bp upstream and 16 bp downstream), but lacking the cleavage site, cloned bidirectionally into pBSM13− (Fig. 2A). I-TevI binds the synthetic duplex and cleaves plasmid sequences, at a distance of −23/−25 for HS31+, as for the wild type, and −22/−24 for HS31− (Fig. 2A, Supplemental Fig. 2A) [13].
Figure 2. Cleavage with I-TevI ZF mutants on different substrates.
(A) Homing-site sequences and cleavage-site mapping. Wild-type (WT) tdΔIn, HS31+ and HS31− DNA substrates are shown, with the central 31-bp homing-site duplex (underscored) inserted into pBSM13-sequence in HS31+ and HS31−. Cleavage sites by WT I-TevI (triangles) and I-TevI-ΔZn (arrows) are shown on the top/bottom strands. IS = intron insertion site. V = insertion referred to in panel C. Gels for bottom and top strand cleavage are to the right and in Supplemental Figure 2A, respectively. Lanes: GATC = sequencing ladder; WT = wild type, C/A = CZnA I-TevI mutant and Δ = ΔZn mutant; U = unprogrammed wheat germ extract. Extracts are undiluted except for wild-type I-TevI on wild-type substrate, which was diluted 50-fold (WTD). Arrows point to cleavages, with Δs representing cleavage by wild-type enzyme and the tailed arrows representing cleavage by the ZF mutants. Cleavage sites on HS31+ and HS31− substrates are shown on the left. The 5'-CX↑XX↓G-3' consensus cleavage sequences are boxed and shaded.
(B) Cleavage kinetics. Assay compares wild-type I-TevI and I-TevI-ΔZn mutant synthesized in vitro on WT tdΔIn and on HS31+ and HS31− mutant substrates. Top band = DNA substrate and lower two bands are cleavage products. M = 1-kb DNA ladder. D = DNA only, 0 = unprogrammed lysate. Lanes 1, 2, 3 and 4 represent 1, 5, 15 and 30 min incubation with enzyme, respectively. Reaction mixtures contained equal amounts of protein except for WT DNA with WT I-TevI, which was diluted 1:50. Plots below each gel show precursor loss, fit to a first-order decay. Rate constants and R2 values are indicated.
(C) Cleavage sites on G-23T and +10 substrates. Ten-nucleotide insertion in +10 is in lower case (see V in panel A). Experiments were performed and cleavages represented as in panel B (Supplemental Fig. 2B).
I-TevI and I-TevI-ΔZn cleavage on the different substrates was quantitated and the data fit to a first order decay (Fig. 2B). Rate constants indicated that wild-type I-TevI cleaves the wild-type substrate ~100-fold more rapidly than does I-TevI-ΔZn. In contrast, on the HS31+ and HS31− substrates, I-TevI is only 7-fold and 1.5-fold faster, respectively, than I-TevI-ΔZn. These results indicate a relatively higher level of cleavage by enzymes with mutated ZFs on the artificial substrates (with non-native cleavage sites) by factors of 14- to 65-fold, suggesting that the mutant enzyme cleaves less specifically than the wild-type I-TevI.
ZF mutants are less discriminate than wild-type I-TevI
To test the fidelity of I-TevI ZF mutants, we mapped the cleavages of wild-type I-TevI, I-TevI-ΔZn and I-TevI-CZnA on the HS31+ and HS31− substrates. Interestingly, multiple cleavages were apparent with the mutant proteins (Fig. 2A). The I-TevI ZF constrains the catalytic domain and mutating the ZF allows the enzyme to stretch for the preferred cleavage site [4, 14], the sequence 5′-CX↑XX↓G-3′ [15]. This sequence occurs twice in each artificial homing substrate (Fig. 2A). Accordingly, whereas the wild-type enzyme cut the wild-type substrate at the normal sites, 23 and 25 bp from the intron insertion site (IS), the ZF mutants cleaved both at this native site and at a CXXXG sequence 18 and 20 bp from the IS (Fig. 2A, Supplemental Fig. 2A). Moreover, the wild-type enzyme cleaved predominantly at 23 and 25 bp on HS31+ and at 22 and 24 bp on HS31− (Fig. 2A). In both cases cleavage was after a G on the top strand, shortening the customary cleavage distance on the HS31− site by 1 bp. In contrast, the ZF mutants barely cleaved at the customary distance, but rather, cleaved at CXXXG sequences 12 and 14 bp and 15 and 17 bp from the IS on both HS31+ and HS31− (Fig. 2A). Apparently, the absence of a ZF relaxes the distance constraint of I-TevI, and enables the enzyme to perform promiscuous cleavage.
To test other examples of degenerate homing sites with mutants lacking the ZF, we selected a G-23T mutant of the CXXXG sequence [14], and a 10-nucleotide insertion within the homing site [4]. Again, cleavage by the wild-type enzyme was at −23 and −25 from the insertion site, whereas I-TevI-ΔZn yielded multiple cleavages at CXXXG sequences at different distances from IS (Fig. 2C; Supplemental Fig. 2B), indicating increased infidelity of ZF mutants on degenerate substrates.
Relaxed cleavage by I-TevI-ΔZn elevates in vivo homing on mutant homing sites
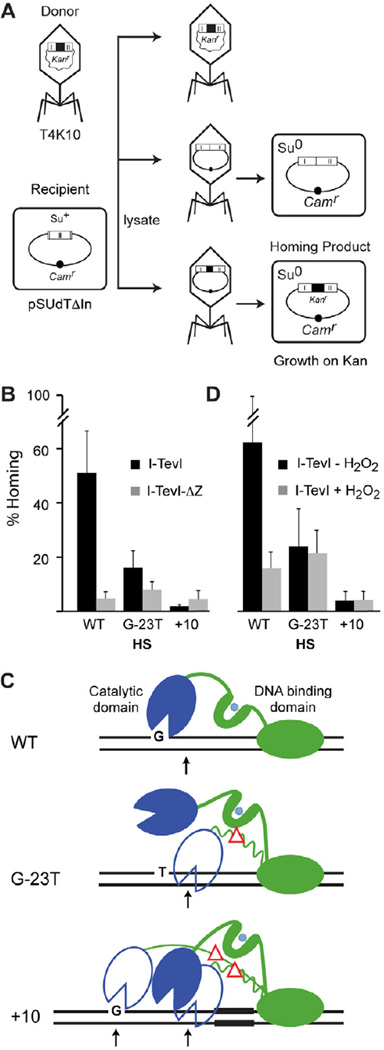
To determine if promiscuous cleavage by I-TevI ZF variants would provide double-strand breaks for intron homing, we used a phage-to-plasmid transduction assay [16, 17] (Fig. 3A). The kanr-marked donor intron was delivered by T4K10 phage carrying the wild-type intron, or T4K10-ΔZn, with an I-TevI ZF deletion, to cells containing a homing-site plasmid. The intron mobility frequency into the Camr recipient pSUdTΔIn, with the wild-type homing site, measured as CamrKanr/Camr transductant colonies, was >11-fold depressed with the ΔZn donor, relative to the wild-type donor (Fig. 3B left). In contrast, when the homing site was mutated at G-23, a key residue for cleavage by wild-type I-TevI and not by the ΔZn mutant (Fig 2C), there was only a two-fold depression of homing with the ΔZn mutant relative to the wild-type phage (Fig. 3B and C). Moreover, when the homing site contains a 10-bp insertion, which places the cleavage site 10-bp further upstream, homing by the ΔZn donor is elevated >2-fold relative to the wild-type intron donor (Fig. 3B and C). Homing events were accurate for each of the six crosses (data not shown). These results suggest that the cleavages introduced by the ΔZn mutant are substrates for intron homing and that a ΔZn mutant donor intron can mobilize relatively more efficiently than the wild-type donor into degenerate sites, to generate bona fide homing products. These results are assay-independent, with similar frequencies for intron movement from a wild-type or ΔZn intron donor plasmid to variant homing sites in the phage [18] (data not shown).
Figure 3. Elevated homing of ΔZn mutant and H2O2-treated introns into variant homing sites.
(A) Schematic of transduction assay for homing. The T4K10 phage donor provides the kanr-marked intron to the plasmid recipient (pSUdTΔIn) containing a wild-type or mutant homing site. After infection of a suppressor-containing cell (Su+), the lysate was used to transduce Suo cells and Camr recipients and CamrKanr homing products were selected on appropriate media.
(B) Homing frequencies in the presence and absence of the ZF. The frequency was calculated for the different donor phage with wild-type (WT) or mutant recipients (G-23T and +10) by determining % homing products (CamrKanr) relative to recipient plasmids (Camr), with n≥4. HS = homing site. The difference in homing frequencies between WT and ΔZn donors was significantly greater, by a two-tailed paired T-test, on the wild-type substrate (~11-fold; P<0.05) than on the G-23T (~2-fold decrease; P<0.05) or +10 substrates (no decrease), indicating an enhanced ability of ΔZn over wild-type donors to mobilize to variant substrates.
(C) Interpretive diagrams of I-TevI on mutant homing sites. Wild-type I-TevI containing the ZF (ZF) is shown in solid cartoon, whereas ΔZF mutant is represented in outline, with a red triangle in place of the ZF. The preferred G residue preceding the top-strand cleavage is indicated.
(D) Homing frequencies in the presence and absence of hydrogen peroxide. Results are represented as in panel B, with n = 6. With the wild-type substrate, homing frequency significantly decreased upon treatment with H2O2 (~4-fold; P<0.05), whereas with the mutant substrates, H2O2 had no significant effect on homing frequencies (P>0.05), again suggesting increased mobility upon ablation of the zinc finger on mutant substrates.
Oxidative stress as a stimulus to intron promiscuity
Given the redox-responsiveness of the ZF in vitro (Fig. 1C and D), the cleavage infidelity of I-TevI-ΔZn mutants in vitro, and the relatively enhanced homing of the ΔZn intron into variant homing sites in vivo, we wished to test if oxidative stress might enhance mobility to variant homing sites. We therefore measured in vivo homing in the presence of the natural oxidizing agent H2O2, which we showed previously results in Zn2+ release and reduces cleavage efficiency on wild-type substrate (Fig. 1C and D). We used the wild-type T4K10 intron donor phage on wild-type and mutant homing sites, as before, but with cells treated with H2O2, or left untreated. Whereas homing was reduced ~4-fold in the presence of 0.3 mM H2O2 with the wild-type recipient, the levels were similar in the presence of the oxidizing agent, with both the G-23T and +10 homing sites (Fig. 3D), indicating a relative stimulation of homing in the presence of H2O2 on mutant substrates. In contrast, there was no elevation in homing in the presence compared with the absence of H2O2 with the T4K10-ΔZn intron donor on mutant substrates (data not shown), consistent with the ZF mediating the H2O2 effect. These results suggest that H2O2 decreases the enzyme specificity by disassembling the ZF (Fig. 1D), leading to the relative stimulation of mobility on degenerate substrates.
Discussion
Here we showed how a modular intron endonuclease, I-TevI, has relaxed cleavage fidelity with ablation of its ZF. H2O2, which leads to life-threatening oxidative stress, results in the potentially adaptive release of the metal ion from the ZF. We propose that oxidation of the ZF in turn leads to spurious DNA cleavage, thereby creating the potential for the observed intron movement to degenerate sites. On an evolutionary time-scale, such stress likely contributes to intron mobility to ectopic sites.
Post-translational control as a third level of regulation of I-TevI
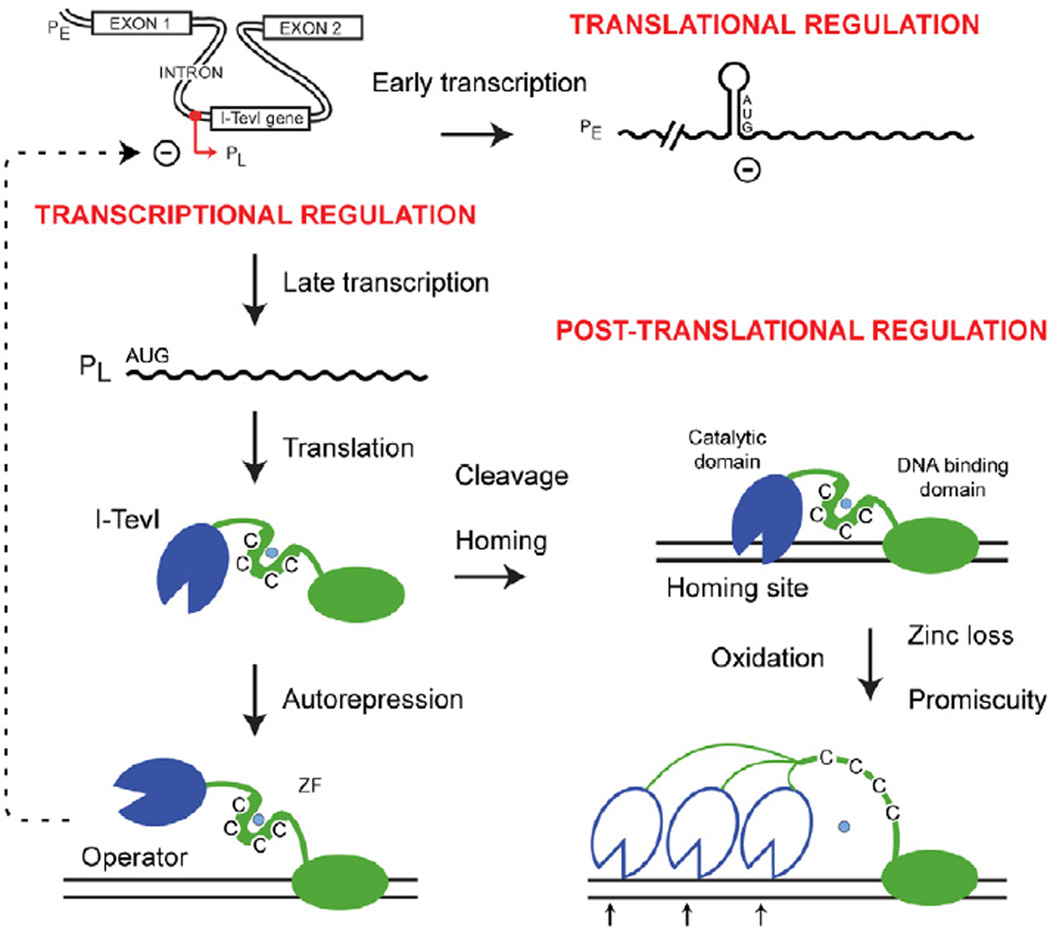
Expression of I-TevI is a tightly regulated process, occurring at both the transcriptional and translational levels. Transcription is temporally controlled by middle and late promoters [3] and negatively regulated by the autorepression activity of I-TevI, [2] (Fig. 4). Additionally, expression of I-TevI from the precursor RNA is repressed by sequestration of the translational start site [3, 19]. In this study we expand this complex regulatory scheme to include post-translational control, by proposing a redox-dependent mechanism, which promotes I-TevI infidelity, increasing the sampling of variant homing sites (Fig. 3C and 4). We postulate that this post-translational control of I-TevI in response to oxidative stress is an immediate regulator of protein function that requires no feedback loops such as those involved in transcription and translation. Thus, tight negative regulation over I-TevI expression is programmed into the phage developmental cycle, in contrast to instantaneous responsiveness of I-TevI fidelity and intron dissemination to cellular stress.
Figure 4. I-TevI regulation includes redox control.
I-TevI is regulated at three levels: transcription, translation and post-translation. The primary transcript from PE is translationally silenced, whereas the late transcript from PL is translated [3]. I-TevI can then act as an autorepressor at PL or as an endonuclease at the homing site [2]. We propose that post-translationally, I-TevI is regulated by oxidative stress, which abstracts Zn2+ (turquoise dot) from the ZF, resulting in promiscuous cleavage, and homing.
I-TevI is subject to a redox fidelity switch via its ZF
One of the principal control mechanisms within redox biology is the Zn2+ switch, where thiol coordination of the zinc ion is reversible and dependent upon the redox status of the cell, thereby regulating protein function [20]. Here we demonstrated that the four cysteines in the ZF in I-TevI [21] can cycle Zn2+ in a redox dependent manner to impact I-TevI cleavage activity (Fig. 1 B–D). Furthermore, reduction of the I-TevI ZF could be accomplished with T4 phage Grx-1 and glutathione but not with the T4 Grx-2 (Supplemental Fig. 1). Multiple glutaredoxins have been observed in other viral systems [22], suggesting that specialization could be occurring within these proteins to maximize viral proliferation. It is possible that I-TevI is utilizing Grx-1’s reducing potential to maintain the ZF for faithful DNA cleavage and intron homing.
Artificial control over a homing endonuclease utilizing an engineered ZF has been demonstrated for PI-SceI. The inserted cysteines allowed for redox-dependent, reversible cleavage by the nuclease [23]. Here, we report that a natural redox switch controls the function of a ZF, whose thiol coordinating residues in I-TevI are positioned to provide a distance sensor for accurate homing-site cleavage [4]. Oxidation switches off the distance determinant and enhance the enzyme's ability to explore sequence for ectopic cleavage sites (Fig. 3C and 4).
I-TevI is more active in promoting homing into a mutant site when its ZF is destroyed by mutation or oxidation (H2O2). This infidelity may provide I-TevI the chance to sample altered substrates in phage or host genes and provide a mechanism for ectopic homing [24, 25]. On an evolutionary time-scale, the naturally relaxed binding specificity of I-TevI [26] might combine with stress-induced cleavage infidelity to allow for repair with limited exon homology. We have previously demonstrated that DSB-repair can occur with regions of microhomology [27]. Such an illegitimate repair mechanism might be a way for the intron to move to non-native substrates.
Homing endonucleases must balance two seemingly opposite requirements to persist within a population. They need to be highly sequence specific to invade a particular site, while they must be promiscuous to permit ectopic movement to new sequence space [28]. Control of cleavage specificity in the modular I-TevI allows exactly such dichotomy of function: site-specific cleavage when the enzyme is structurally intact, and alternative cleavage in response to oxidative stress when the ZF ceases to function as a distance determinant.
Mobile genetic elements are stimulated to move when the host is exposed to stressors such as DNA damage, starvation and osmotic shock [29]. This dynamic not only ensures their propagation when the host is threatened, but also can generate genetic diversity, by ectopic movement that may be advantageous to the host. It is therefore not entirely surprising that transposases, of which intron endonucleases can be considered a functional equivalent, are the most abundant and ubiquitous genes in nature [30]; not only do they promote movement, but they do so to the potential selective advantage of the host, particularly in response to metabolic or environmental stressors. In this context, it is noteworthy that group II introns were recently shown to be stimulated to retrotranspose in reaction to nutrient depletion [31]. Now it appears that group I introns, which splice and mobilize by different pathways, are impacted by oxidative stress to increase their promiscuity.
Experimental Procedures
Plasmids and phages
Strains, plasmids and phage are listed and described in Supplemental Table S1. Oligonucleotides are listed in Supplemental Table S2. Description of the construction of T4K10td-Kan and T4K10tdΔZn-Kan phage is in Supplemental Methods. Homing-site mutants for homing assays were constructed by transferring the tdΔIn EcoRI fragment containing the mutations into the desired plasmid vector.
Expression and purification of proteins
Three different enzyme preparations were used as indicated in figure legends. The low-activity but faithful I-TevI-H40Y mutant of I-TevI allows for overexpression and purification of this otherwise toxic protein in vivo [11] as described in Supplemental Methods. Second, wild-type high-activity I-TevI was prepared by intein-mediated purification to minimize toxicity [32]. Third, given this toxicity of I-TevI and ZF mutant derivatives, these proteins were also synthesized in catalytically active form in vitro using wheat-germ extracts with mRNA synthesized from the SP6 promoter of an I-TevI clone, as described previously [4, 11]. Plasmids used for in vitro-synthesized I-TevI are listed in Supplemental Table 1.
Determination of protein-bound Zn2+
A spectroscopic technique, based on 4-(2-pyridylazo) resorcinol (PAR), was utilized to determine Zn2+ content of I-TevI derivatives [33, 34] as described in Supplemental Methods.
Cleavage assays and mapping
I-TevI was incubated with 250 ng of DNA substrate (tdΔIn) as described in Supplemental Methods. Cleavage was quantitated with an Alpha Innotech FluoroChem 8900 imager. To determine whether redox conditions influence cleavage activity, H2O2, diamide or DTT were added to a final concentration of 1 mM, 2.5 mM or 10 mM, respectively. Cleavage assays were performed as described [35]. Cleavage-site mapping was carried out with I-TevI synthesized in vitro using a Thermosequenase Cycling-Sequencing 2.0 kit (USB) as described [15]. Sequence ladders of the homing site were generated as described in Supplemental Methods. Substrates for cleavage and cleavage-site mapping experiments are listed in Supplemental Table 1.
Phage-to-plasmid homing assay
We used phage-to-plasmid T4-mediated transduction [17] to test mobility of wild-type I-TevI and ΔZn mutants of T4K10 on wild-type and variant substrates, as described in Supplemental Methods. H2O2 was added at 0.3 mM at time of infection.
Supplementary Material
Highlights.
Zn 2+ content of I-TevI intron endonuclease is redox sensitive
Cleavage fidelity of I-TevI is lost upon ablation of the zinc finger
Intron mobility to degenerate sites is increased when zinc finger is dysfunctional
Oxidative stress stimulates intron promiscuity
Acknowledgements
The authors thank members of the Belfort lab, especially Gil Amitai, Brian Callahan, and Venkata Chalamcharla for useful discussions and data analysis; Brian Callahan, Venkata Chalamcharla, Tao Huang, Ingrid Hahn and Natalya Topilina for comments on the manuscript and analysis; Maryellen Carl for expert manuscript preparation; and John Dansereau for help with the figures. We acknowledge use of Wadsworth Center’s DNA sequencing core. This work was supported by NIH grants GM44844 and GM39422 to M.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Justin B. Robbins, Wadsworth Center, New York State Department of Health, State University of New York, Center for Medical Science, 150 New Scotland Avenue, Albany, NY 12208, USA.
Dorie Smith, Wadsworth Center, New York State Department of Health, State University of New York, Center for Medical Science, 150 New Scotland Avenue, Albany, NY 12208, USA.
Marlene Belfort, Department of Biomedical Sciences, State University of New York, Center for Medical Science, 150 New Scotland Avenue, Albany, NY 12208, USA.
References
- 1.Lambowitz AM, Belfort M. Introns as mobile genetic elements. Ann. Rev. Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 2.Edgell DR, Derbyshire V, Van Roey P, LaBonne S, Stanger MJ, Li Z, Boyd TM, Shub DA, Belfort M. Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor. Nat. Struct. Mol. Biol. 2004;11:936–944. doi: 10.1038/nsmb823. [DOI] [PubMed] [Google Scholar]
- 3.Gott JM, Zeeh A, Bell-Pedersen D, Ehrenman K, Belfort M, Shub DA. Genes within genes: Independent expression of phage T4 intron open reading frames and the genes in which they reside. Genes & Devel. 1988;2:1791–1799. doi: 10.1101/gad.2.12b.1791. [DOI] [PubMed] [Google Scholar]
- 4.Dean AB, Stanger MJ, Dansereau JT, Van Roey P, Derbyshire V, Belfort M. Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI. Proc. Natl. Acad. Sci. USA. 2002;99:8554–8561. doi: 10.1073/pnas.082253699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ilbert M, Graf PC, Jakob U. Zinc center as redox switch--new function for an old motif. Antioxidants & redox signaling. 2006;8:835–846. doi: 10.1089/ars.2006.8.835. [DOI] [PubMed] [Google Scholar]
- 6.Jakob U, Eser M, Bardwell JC. Redox switch of hsp33 has a novel zinc-binding motif. J. Biol. Chem. 2000;275:38302–38310. doi: 10.1074/jbc.M005957200. [DOI] [PubMed] [Google Scholar]
- 7.Webster KA, Prentice H, Bishopric NH. Oxidation of zinc finger transcription factors: physiological consequences. Antioxidants & redox signaling. 2001;3:535–548. doi: 10.1089/15230860152542916. [DOI] [PubMed] [Google Scholar]
- 8.Jakob U, Muse W, Eser M, Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 9.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 10.Zhou A, Carrell RW, Murphy MP, Wei Z, Yan Y, Stanley PLD, Stein PE, Pipkin F, Read RJ. A redox switch in angiotensinogen modulates angiotensin release. Nature. 2010;468:108–111. doi: 10.1038/nature09505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derbyshire V, Kowalski JC, Dansereau JT, Hauer CR, Belfort M. Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J. Mol. Biol. 1997;265:494–506. doi: 10.1006/jmbi.1996.0754. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q-Q, Derbyshire V, Belfort M, Edgell DR. Distance determination by GIY-YIG intron endonucleases: discrimination between repression and cleavage functions. Nucleic Acids Res. 2006;34:1755–1764. doi: 10.1093/nar/gkl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell-Pedersen D, Quirk SM, Bryk M, Belfort M. I-TevI, the endonuclease encoded by the mobile td intron, recognizes binding and cleavage domains on its DNA target. Proc. Natl. Acad. Sci. USA. 1991;88:7719–7723. doi: 10.1073/pnas.88.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryk M, Belisle M, Mueller JE, Belfort M. Selection of a remote cleavage site by I-TevI, the td intron-encoded endonuclease. J. Mol. Biol. 1995;247:197–210. doi: 10.1006/jmbi.1994.0133. [DOI] [PubMed] [Google Scholar]
- 15.Edgell DR, Stanger MJ, Belfort M. Coincidence of cleavage sites of intron endonuclease I-TevI and critical sequences of the host thymidylate synthase gene. J. Mol. Biol. 2004;343:1231–1241. doi: 10.1016/j.jmb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Kreuzer KN, Alberts BM. Characterization of a defective phage system for the analysis of bacteriophage T4 DNA replication origins. J. Mol. Biol. 1986;188:185–198. doi: 10.1016/0022-2836(86)90303-7. [DOI] [PubMed] [Google Scholar]
- 17.Clyman J, Belfort M. Trans and cis requirements for intron mobility in a prokaryotic system. Genes and Devel. 1992;6:1269–1279. doi: 10.1101/gad.6.7.1269. [DOI] [PubMed] [Google Scholar]
- 18.Quirk SM, Bell-Pedersen D, Belfort M. Intron mobility in the T-Even phages: High frequency inheritance of group I introns promoted by intron open reading frames. Cell. 1989;56:455–465. doi: 10.1016/0092-8674(89)90248-1. [DOI] [PubMed] [Google Scholar]
- 19.Gibb EA, Edgell DR. Better late than early: Delayed translation of intron-encoded endonuclease I-TevI is required for efficient splicing of its host group I intron. Mol. Microbiol. 2010;78:35–46. doi: 10.1111/j.1365-2958.2010.07216.x. [DOI] [PubMed] [Google Scholar]
- 20.Maret W. Zinc and sulfur: a critical biological partnership. Biochemistry. 2004;43:3301–3309. doi: 10.1021/bi036340p. [DOI] [PubMed] [Google Scholar]
- 21.Van Roey P, Waddling CA, Fox KM, Belfort M, Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gvakharia BO, Koonin EK, Mathews CK. Vaccinia virus G4L gene encodes a second glutaredoxin. Virology. 1996;226:408–411. doi: 10.1006/viro.1996.0669. [DOI] [PubMed] [Google Scholar]
- 23.Posey KL, Gimble FS. Insertion of a reversible redox switch into a rarecutting DNA endonuclease. Biochemistry. 2002;41:2184–2190. doi: 10.1021/bi015944v. [DOI] [PubMed] [Google Scholar]
- 24.Edgell DR, Belfort M, Shub DA. Barriers to intron promiscuity in bacteria. J. Bacteriol. 2000;182:5281–5289. doi: 10.1128/jb.182.19.5281-5289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J. Mol. Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryk M, Quirk SM, Mueller JE, Loizos N, Lawrence C, Belfort M. The td intron endonuclease makes extensive sequence tolerant contacts across the minor groove of its DNA target. EMBO J. 1993;12:2141–2149. doi: 10.1002/j.1460-2075.1993.tb05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker MM, Belisle M, Belfort M. Intron homing with limited exon homology: illegitimate double-strand-break repair in intron acquisition by phage T4. Genetics. 1999;153:1513–1523. doi: 10.1093/genetics/153.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 29.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 30.Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coros CJ, Piazza CL, Chalamcharla VR, Smith D, Belfort M. Global regulators orchestrate group II intron retromobility. Mol. Cell. 2009;34:250–256. doi: 10.1016/j.molcel.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu W, Wood DW, Belfort G, Derbyshire V, Belfort M. Intein-mediated purification of cytotoxic endonuclease I-TevI by insertional inactivation and pH-controllable splicing. Nucleic Acids Res. 2002;30:4864–4871. doi: 10.1093/nar/gkf621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robbins JB, Murphy MC, White BA, Mackie RI, Ha T, Cann IK. Functional analysis of multiple single-stranded DNA-binding proteins from Methanosarcina acetivorans and their effects on DNA synthesis by DNA polymerase BI. J. Biol. Chem. 2004;279:6315–6326. doi: 10.1074/jbc.M304491200. [DOI] [PubMed] [Google Scholar]
- 34.Robbins JB, Stapleton M, Smith D, Dansereau JT, Derbyshire V, Belfort M. Homing endonuclease I-TevIII: dimerization as a means to a double strand break. Nucleic Acids Res. 2007;35:1589–1600. doi: 10.1093/nar/gkl1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu QQ, Dansereau JT, Puttamadappa SS, Shekhtman A, Derbyshire V, Belfort M. Role of the interdomain linker in distance determination for remote cleavage by homing endonuclease I-TevI. J. Mol. Biol. 2008;379:1094–1106. doi: 10.1016/j.jmb.2008.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalski JC, Belfort M, Stapleton MA, Holpert M, Dansereau JT, Pietrokovski S, Baxter SM, Derbyshire V. Configuration of the catalytic GIY-YIG domain of intron endonuclease I-Tev I: coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.