Abstract
Hepatocyte nuclear factor–1β (HNF-1β) is a Pit-1, Oct-1/2, UNC-86 (POU)/homeodomain-containing transcription factor that regulates tissue-specific gene expression in the liver, kidney, and other organs. Humans with autosomal dominant mutations of HNF-1β develop maturity-onset diabetes of the young type 5 (MODY5) and congenital cystic abnormalities of the kidney. Autosomal recessive polycystic kidney disease (ARPKD) is an inherited cystic disorder that produces renal failure in infants and children and is caused by mutations of PKHD1. The proximal promoter of the mouse Pkhd1 gene contains an evolutionarily conserved HNF-1–binding site that is located near a region of deoxyribonuclease hypersensitivity. HNF-1β and the structurally related HNF-1α bind specifically to the Pkhd1 promoter and stimulate gene transcription. Mutations of the HNF-1 site or expression of a dominant-negative HNF-1β mutant inhibit Pkhd1 promoter activity in transfected cells. Transgenic mice expressing a dominant-negative HNF-1β mutant under the control of a kidney-specific promoter develop renal cysts, similarly to humans with MODY5. Pkhd1 transcripts are absent in the cells lining the cysts but are present in morphologically normal surrounding tubules. These studies identify a link between two cystic disease genes, HNF1β (MODY5) and PKHD1 (ARPKD). HNF-1β directly regulates the transcription of Pkhd1, and inhibition of PKHD1 gene expression may contribute to the formation of renal cysts in humans with MODY5.
Introduction
Hepatocyte nuclear factor–1β (HNF-1β, also known as vHNF-1, LFB3, or TCF2) belongs to a digenic family of homeodomain-containing transcription factors that play important roles in the liver-specific expression of genes such as albumin and α1-antitrypsin (1). HNF-1β and the structurally related HNF-1α (LFB1, TCF1) contain an amino-terminal dimerization domain, a (Pit-1,OCT1/2,UNC-86) POU domain and homeodomain that mediate sequence-specific DNA binding, and a carboxyl-terminal activation domain (2). HNF-1β and HNF-1α recognize an identical 15-bp consensus sequence, 5′-RGTTAATNATTAACM-3′, which is found in the proximal promoters of several liver-specific genes (3). The HNF-1 proteins bind to DNA as homodimers or heterodimers and usually function as transcriptional activators. Although HNF-1β and HNF-1α were first identified in the liver, they are also highly expressed in polarized epithelia in the kidney, intestine, pancreas, and other organs (1). Studies using in situ hybridization or HNF-1β/lacZ reporter genes have shown that, in the kidney, HNF-1β is expressed in tubular epithelial cells composing the nephrons and collecting ducts (4–6). During nephrogenesis, HNF-1β is expressed in comma-shaped and S-shaped bodies as well as developing proximal and distal tubules, which suggests that it may have a role in tubule differentiation. HNF-1β–deficient mice, however, are embryonic lethal prior to renal organogenesis, and few target genes that are regulated by HNF-1β in the kidney have been identified (4, 7, 8).
Mutations of the human HNF-1β gene (TCF2) are responsible for the autosomal dominant disorder MODY5 (maturity-onset diabetes mellitus of the young, type 5) (9). Humans with MODY5 present with early-onset type 2 diabetes mellitus due to an impairment in insulin secretion. In addition, many MODY5 patients have congenital abnormalities of the kidney and genitourinary tract (10, 11). The spectrum of renal abnormalities includes simple cysts, polycystic kidneys, cystic dysplasia, and glomerulocystic kidney disease. These disorders have in common the development of cysts in the tubular portion of the nephron (12). The renal abnormalities may appear before the onset of diabetes, which indicates that they are not due to diabetic nephropathy. Affected individuals may develop chronic kidney disease and progress to end-stage renal disease requiring dialysis or transplantation. Other renal manifestations of MODY5 include solitary kidney, enlarged nephrons (oligomeganephronia), and familial juvenile hyperuricemic nephropathy (13).
Autosomal recessive polycystic kidney disease (ARPKD) is a monogenic disorder that affects 1 in 20,000 births and produces renal failure in neonates and children (14–16). ARPKD is characterized by the association of cysts in the renal collecting ducts and biliary dysgenesis, a ductal plate malformation that produces aberrant intrahepatic bile ducts and portal fibrosis. Infants with the severe perinatal form of ARPKD have bilateral enlarged kidneys, intrauterine renal failure, and oligohydramnios. Up to 30–50% of affected neonates die shortly after birth due to sepsis or respiratory failure. Children who are diagnosed later in life have a better prognosis, but may develop complications of portal hypertension (15). ARPKD is caused by mutations of PKHD1 (polycystic kidney and hepatic disease 1) on chromosome 6p21.1-p12 (17, 18). More than 90 different mutations that are distributed throughout the PKHD1 gene have been identified (19–21). Amino acid substitutions are more commonly associated with a nonlethal presentation, whereas truncating mutations are associated with the severe perinatal form of the disease, suggesting a loss-of-function mechanism.
Consistent with the sites affected by the disease, the PKHD1 gene is expressed at high levels in the fetal and adult kidney and at lower levels in the liver and pancreas (17, 18, 22). The mouse orthologue, Pkhd1, is highly expressed in the kidney, with lower levels of expression in the liver, heart, stomach, intestine, skeletal muscle, uterus, and placenta (23). Within the kidney, Pkhd1 is abundantly expressed in the renal collecting ducts with lower levels of expression in the proximal and distal tubules (22, 23). In the developing kidney (metanephros), Pkhd1 is expressed in the branching ureteric bud and developing tubules. In the developing liver Pkhd1 is expressed in epithelial cells of the ductal plate, and expression continues in the intra- and extrahepatic bile ducts in the mature organ. Additional sites of embryonic expression include sympathetic ganglia, pancreatic ducts, trachea, and testis (23). The protein encoded by PKHD1, named polyductin or fibrocystin, is predicted to be an integral membrane protein containing a large extracellular domain, a single transmembrane segment, and a short carboxyl-terminal domain (17, 18). Fibrocystin/polyductin has been identified as a greater than 450-kDa protein that is expressed in the loops of Henle and collecting ducts in the kidney and bile ducts in the liver (24). Expression is also found in hepatocytes, pancreatic ducts, and the developing epididymis and testis. In ARPKD, no expression of fibrocystin/polyductin is detected in cystic collecting ducts (24).
Collectively, these studies indicate that the expression of Pkhd1 is tissue-specific and developmentally regulated and that the loss of Pkhd1 expression produces renal cysts. To begin understanding the regulation of Pkhd1 gene expression, we cloned and characterized the gene promoter. We identified an evolutionarily conserved HNF-1–binding site in the proximal Pkhd1 promoter and found that WT HNF-1β activated gene transcription. Expression of a dominant-negative HNF-1β (DN- HNF1β) mutant inhibited Pkhd1 gene expression and produced renal cysts in transgenic mice. These studies identify a possible mechanism of cystogenesis in humans with renal cysts and diabetes due to mutations of HNF-1β.
Methods
Library screening.
The RPCI-22 129S6/SvEvTac mouse bacterial artificial chromosome (BAC) library (Children’s Hospital Oakland Research Institute, Oakland, California, USA) was screened by hybridization of 32P-labeled DNA probes to high density filters. The probe was a 433-bp genomic fragment derived from intron 10 and intron 11 of mouse Pkhd1. BAC DNA was prepared using standard alkaline lysis protocols with Endo-Free kits (QIAGEN Inc., Valencia, California, USA). Eluted DNA was precipitated with isopropanol and dissolved in 10 mM Tris-Cl (pH 7.4) and 0.1 mM EDTA. Appropriate restriction fragments were subcloned into pBluescript II KS and sequenced using gene-specific primers.
Plasmids.
pBluescript II and pcDNA3 plasmids encoding WT HNF-1β and HNF-1α and an expression plasmid encoding a dominant-negative mutant form of mouse HNF-1β (DN-HNF1β) corresponding to the human A263insGG mutation have been described previously (8). In some experiments, V5 and FLAG epitope tags were added inframe to the carboxyl termini by PCR. Preliminary experiments verified that the presence of the epitope tags did not affect activity (not shown). Luciferase reporter plasmids containing the Pkhd1 promoter linked to the coding region of firefly luciferase were produced as follows: BAC 534 J18 was digested with EcoRI, and the 8.1-kb fragment was cloned into pBluescript. The resulting plasmid was digested with PstI, and the 519-bp fragment containing exon 1 and upstream sequence was cloned into pGL3-Basic. The noncoding region of Pkhd1 extending from exon 2 to 6.7 kb upstream to the transcription start site was amplified from the BAC using long-range PCR with Herculase (Stratagene, La Jolla, California, USA) and primers 5′-ACAAGTGGACATTCTTGGCACGGTTTAC-3′ and 5′-ATATTAGAAGCATTTCCCGGGTTGATTGGC-3′. The 9.4-kb product was cloned into pCR2.1-TOPO and digested with SmaI and BstZ171, EcoRV, BsaB1, or HindIII. The restriction fragments were cloned into pGL3-Basic to produce luciferase reporter plasmids containing 6.6 kb, 3.3 kb, 1.8 kb, and 1.1 kb of the promoter, respectively. The plasmid containing 3.3 kb of the promoter was digested with MluI and PstI and ligated to an MluI/PstI adapter to produce an intermediate plasmid. The plasmid was digested with PstI, and the 519-bp PstI fragment containing exon 1 was inserted to produce a luciferase reporter plasmid containing 444 bp of the promoter.
Site-directed mutagenesis.
Site-directed mutagenesis was performed using QuikChange kits (Stratagene) as described previously (8). The presence of the desired mutation was verified by DNA sequencing.
Cell culture, transfection, and reporter gene assays.
Mouse inner medullary collecting duct cells (mIMCD-3), HeLa cells, and C3H/10T1/2 cells were grown as described previously (8). HEK293 cells were grown in DMEM containing 10% FCS. Cells were plated in six-well dishes (1.2 × 105 to 3 × 105 cells/well) and transfected with 0.6 μg luciferase reporter plasmids using Effectene (QIAGEN Inc.). Cells were cotransfected with 20 ng pRL plasmid encoding Renilla luciferase to control for differences in transfection efficiency. After growth for 48 hours, the cells were lysed in 250 μl passive lysis buffer (Promega Corp., Madison, Wisconsin, USA), freeze-thawed once, and centrifuged. Supernatants (20 μl) were added to 96-well plates, and firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega Corp.), according to the manufacturer’s directions. Luciferase Assay Reagent II (100 μl) was added, and light output was measured for 10 seconds using a Wallac VICTOR V multilabel counter (Perkin Elmer, Wellesley, Massachusetts, USA). Firefly luciferase activity was normalized to Renilla luciferase activity, which was measured by adding 100 μl of Stop & Glo reagent and measuring light output for 5 seconds.
Electrophoretic mobility-shift assays.
Rabbit reticulocyte lysates programmed with HNF-1α or HNF-1β, and nuclear extracts from cultured cells were prepared as described previously (8). A 44-bp DNA fragment containing the sequence of the Pkhd1 promoter from –81 bp to –38 bp (Figure 1A) was end-labeled with 32P using T4 polynucleotide kinase. Binding reactions (20 μl) contained 25 mM NaCl, 10 mM Tris-Cl (pH 7.5), 1 mM MgCl2, 5 mM EDTA, 5% glycerol, 1 mM DTT, labeled probe (5 × 104 cpm), and 2–4 μg nuclear extracts or 5 μl programmed lysates. Some reactions included a 100-fold molar excess of unlabeled DNA fragment as specific competitors or Ab’s to HNF-1α, HNF-1β (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA), FLAG epitope tag (Sigma-Aldrich, St. Louis, Missouri, USA), or V5 epitope tag (Invitrogen Corp., Carlsbad, California, USA). All binding reactions contained 0.2–1 μg poly(dI·dC) as a nonspecific competitor. After incubation at room temperature for 20 minutes, samples were loaded on 4% polyacrylamide gels and electrophoresed at 8 V/cm for 2 hours in 0.5 × Tris-borate-EDTA. Gels were dried, and the radiolabeled bands were detected by autoradiography using Kodak BioMax MS film or a PhosphorImager (Storm 860; Molecular Dynamics, Sunnyvale, California, USA).
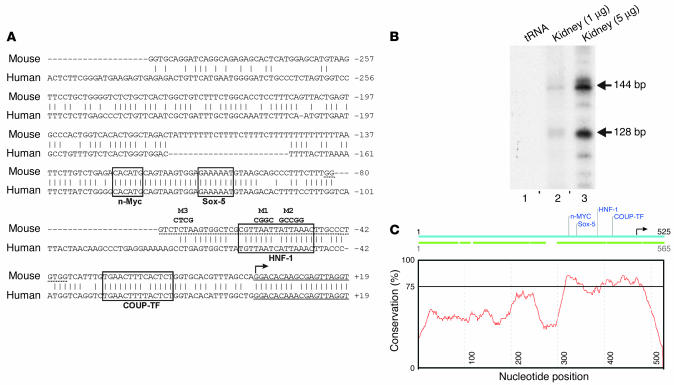
Figure 1.
Sequence of the Pkhd1 promoter. (A) Alignment of the mouse (upper) and human (lower) Pkhd1 promoters. Short vertical line indicates identity. Dash indicates gaps introduced to optimize the alignment. Bent arrow indicates the transcription initiation site at +1. Solid underlines indicate exon 1. Dashed underlines indicate the DNA fragment used for EMSA. Boxes indicate evolutionarily conserved transcription factor–binding sites identified using the ConSite program (conservation 75%, window 50, transcription factor threshold 80%). Nucleotides in bold type indicate the site-directed mutations M1–M3. The sequence of the mouse Pkhd1 promoter has been deposited in the GenBank database (accession number AY544205). (B) Mapping of the transcription initiation site by primer extension. Arrows indicate products corresponding to transcription initiation sites at +1 (144 bp) and +17 (128 bp) in kidney RNA (lanes 2 and 3) but not in yeast tRNA (lane 1). (C) Plot of the percentage of identity between the mouse (blue) and human (green) promoters. Horizontal line indicates 75% identity.
Northern blot analysis and real-time RT-PCR.
Poly(A)+ RNA (4 μg) was prepared from cultured cells, and Northern blot analysis was performed using methods similar to those described previously (25). Northern blots were hybridized with a 1.5-kb mouse Pkhd1 cDNA (nucleotides 11210–12723), 800-bp mouse HNF-1β cDNA (nucleotides 967–1766), and a cDNA encoding rat GAPDH as a positive control. Total RNA was isolated from kidneys using TRIzol (Invitrogen Corp.), and 2 μg RNA was used for cDNA synthesis. PCR reactions (50 μl) contained 1× Platinum Quantitative PCR SuperMix-UDG (Invitrogen Corp.) and 0.5× SYBR Green I (Molecular Probes Inc., Eugene, Oregon, USA). The following primers were used: Pkhd1 forward primer, 5′-AAGTCAAGGGCCATCACATC-3′; Pkhd1 reverse primer, 5′-ATGTTTCTGGTCAACAGCCC-3′; and Universal 18S Internal Standards (Ambion Inc., Austin, Texas, USA). Reactions were performed in 96-well dishes using an iCycler iQ machine (Bio-Rad Laboratories Inc., Hercules, California, USA). The PCR protocol was incubation at 95°C for 3 minutes followed by 35 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. Quality control was performed with melting curve analysis and sequencing of PCR products. Standard curves were produced to demonstrate linearity and determine efficiency. The expression of Pkhd1 relative to 18S rRNA was calculated using the method of Pfaffl (26).
Transcript mapping.
Primer extension was performed by annealing a 32P-labeled oligonucleotide derived from exon 1 (5′-GTGCCTCAAATGAGAGAAACCGTCCCCTTC-3′) to 1–5 μg poly(A)+ RNA from mouse kidney or yeast tRNA as a negative control. The primer was elongated with StrataScript reverse transcriptase (Stratagene), and the reaction products were resolved on 6% polyacrylamide-8 M urea gels. Gels were calibrated by sequencing reactions run in parallel lanes. Ribonuclease protection assays were performed as described previously (27) using an antisense riboprobe extending from exon 1 into the 5′ flanking region (nucleotides –196 to +145).
Immunoblot analysis and immunoprecipitation.
Kidney homogenates were analyzed by SDS-PAGE and transferred to nitrocellulose membranes as described previously (25). Immunoblots were incubated with a HRP-conjugated anti-FLAG Ab. Signal intensity was quantified with a FluorImager and expressed as a percentage relative to the highest signal measured in each group. HEK293 cells were transfected with expression plasmids encoding WT or mutant HNF-1β containing carboxyl-terminal V5 or FLAG epitope tags. Cell lysates were precipitated with anti-V5 Ab (Invitrogen Corp.) or anti-Myc Ab as a negative control. The immunoprecipitates were analyzed by immunoblot analysis with HRP-conjugated anti-FLAG Ab, as described previously (28).
Deoxyribonuclease I hypersensitive site mapping.
Deoxyribonuclease I (DNase I) hypersensitive site mapping was performed as described previously (8). Nuclei from mIMCD-3 cells or 10T1/2 cells were incubated with graded concentrations of DNase I (2–100 μg/ml) at 37°C for 5 minutes. After quenching the reaction, the samples were treated with ribonuclease A and proteinase K, and genomic DNA was purified by phenol/chloroform extraction and ethanol precipitation. DNase I “fadeout” was confirmed by agarose gel electrophoresis. DNA (10 μg) was digested with EcoRI or SpeI, and Southern blots were prepared. The blots were hybridized with 32P-labeled probes containing nucleotides –5 to +748 (3′ probe) or –4202 to –3740 (5′ probe).
Animals.
A cDNA encoding the DN-HNF1β mutant containing a carboxyl-terminal FLAG epitope tag was cloned into the unique SbfI site of a plasmid containing the 1.3-kb Ksp-cadherin promoter, β-globin TATA box, and bovine growth hormone polyadenylation signal. The insert was excised with NotI and XhoI and purified from the vector by gel electrophoresis. Transgenic mice were produced by the University of Texas Southwestern Transgenic Core Facility, as described previously (28). Microinjected C57BL/6 embryos were transferred to pseudopregnant foster mothers and permitted to develop to term. Progeny were genotyped by PCR analysis of tail biopsies and confirmed by Southern blot analysis. Studies were performed on 20- to 86-day-old transgenic founders and their age-matched nontransgenic littermates. Kidney-specific kinesin-II (Kif3a) KO mice were produced as described previously (25). All experiments involving animals were performed under the auspices of the Institutional Animal Care and Research Advisory Committee.
Ab and lectin staining.
Formalin-fixed, paraffin-embedded tissue sections and paraformaldehyde-fixed cryosections were stained with Ab’s to aquaporin-2 (AQP2), Na-K-Cl cotransporter (NKCC2), Na-Cl cotransporter (NCC), all gifts from M. Knepper (National Heart, Lung, and Blood Institute, Bethesda, Maryland, USA), or acetylated tubulin (Sigma-Aldrich), as described previously (28). Secondary Ab’s were conjugated to cyanine 3 (Cy3; Jackson ImmunoResearch Laboratories Inc., West Grove, Pennsylvania, USA), Alexa Fluor 594 (Molecular Probes Inc.), or Alexa Fluor 488. FLAG epitope–tagged proteins were detected using Cy3-conjugated anti-FLAG Ab (Sigma-Aldrich). Staining with FITC-coupled Lotus tetragonolobus agglutinin (LTA; Vector Laboratories, Burlingame, California, USA) was performed as described previously (28). Photomicrographs were obtained with a Zeiss Axioplan 2 microscope, and image deconvolution was performed using Openlab software (Improvision Inc., Lexington, Massachusetts, USA).
In situ hybridization.
Fluorescence in situ RNA hybridization was performed with tyramide signal amplification (TSA). Paraformaldehyde-fixed, paraffin-embedded tissue sections were dewaxed, dehydrated through graded ethanol, and air-dried. Sections were post-fixed with 4% paraformaldehyde, were treated with proteinase K for 15 minutes at 37°C, and acetylated with acetic anhydride for 10 minutes. A pCMVSport6 plasmid containing a 1.5-kb mouse Pkhd1 cDNA was linearized with NotI, and a digoxigenin-labeled antisense riboprobe was synthesized using SP6 RNA polymerase and Dig RNA Labeling Kits according to the manufacturer’s directions (Roche Applied Science, Indianapolis, Indiana, USA). As a negative control, a sense riboprobe was synthesized using T7 RNA polymerase after linearization with SalI. Sections were hybridized overnight at 60°C in buffer containing 50% formamide, 4× SSC, 0.25 mg/ml yeast tRNA, 0.5 mg/ml salmon sperm DNA, 5× Denhardt’s solution, and 200 ng/ml labeled probe. Slides were washed sequentially at 65°C in 5× SSC, 2× SSC containing 50% formamide, 2× SSC, 0.2× SSC, and 0.1× SSC, and then were incubated in blocking buffer (Roche Applied Science) containing 5% normal rabbit IgG. Digoxigenin-labeled probes were detected by incubation with HRP-conjugated anti-digoxigenin Ab (1:200; Dako Cytomation, Carpinteria, California, USA) followed by amplification with TSA-direct deposition of Cy3 (1:50; Perkin Elmer). The slides were then dehydrated and mounted with Vectashield (Vector Laboratories).
Statistical analysis.
Results of reporter gene assays are expressed as mean plus or minus standard error of six to nine independent transfections assayed in duplicate. The statistical significance of differences between the means was evaluated using Student paired t test with Bonferroni’s adjustment for multiple comparisons. Differences were considered to be significant at P values less than 0.05.
Results
Cloning of the mouse Pkhd1 gene promoter.
A mouse BAC library (RPCI-22 129S6/SvEvTac) was screened by filter hybridization with a 32P-labeled probe containing intron 10 and exon 11 of Pkhd1. Two positive clones were identified: 336J1 and 534J18. Clone 534J18 was found to contain exon 1 of the Pkhd1 gene and was characterized further. An 8.1-kb EcoRI fragment containing exon 1, a portion of intron 1, and the 5′ flanking sequence of the gene was subcloned into pBluescript II KS and sequenced. The mouse sequence was used in a BLAST search of the human genome (National Center for Biotechnology Information), and a chromosome 6–derived clone containing the homologous human PKHD1 sequence was identified (accession no. NT 007592). Figure 1A shows the sequence of the proximal 5′ flanking region of the mouse Pkhd1 gene aligned with the homologous human sequence.
The transcription initiation site of the mouse Pkhd1 gene was mapped by primer extension and ribonuclease protection assays. A 32P-labeled antisense oligonucleotide derived from exon 1 was annealed to poly(A)+ RNA from the mouse kidney. After elongation with reverse transcriptase, the reaction products were resolved on sequencing gels. Two major products were obtained that corresponded to transcription initiation sites located at nucleotides +1 and +17 (Figure 1B). The presence of more than one product suggested the existence of multiple transcription initiation sites. No extension products were produced from yeast tRNA, indicating that the products were specific. Ribonuclease protection assays produced three protected bands corresponding to multiple transcription initiation sites at +1, +5, and +17 (not shown). The transcription initiation site at +1 was located 18 bp upstream to the 5′ end of the cloned mouse Pkhd1 mRNA sequence in the GenBank database (accession no. AY130764). No TATA or CCAAT boxes were identified in their typical locations 5′ to the transcription initiation site. The absence of a TATA box often results in multiple transcription initiation sites, as was suggested by the primer extension and ribonuclease protection assays. The transcription initiation site at +1 was located within a weak initiator element (5′-YYANWYY-3′) that was evolutionarily conserved between mouse and human. A stronger initiator element was located 17 bp farther upstream.
The DNA elements that regulate gene expression are often evolutionarily conserved between different species. To identify evolutionarily conserved elements within the Pkhd1 promoter, the mouse and human sequences were compared using the ConSite program (Karolinska Institute, Stockholm, Sweden; http://mordor.cgb.ki.se/cgi-bin/CONSITE/consite). The overall sequence identity was 40–50% (Figure 1C). The region containing the transcription initiation site and extending 150 bp upstream was more than 75% identical, however. The ConSite program identified evolutionarily conserved bindings sites for N-myc at nucleotides –118 to –123, Sox-5 at nucleotides –100 to –106, COUP-TF at nucleotides –17 to –30, and Snail at nucleotides +15 to +20. At nucleotides –49 to –62 the mouse promoter contained the sequence 5′-CGTTAATTATTAAAC-3′, which matched a consensus HNF-1–binding site (5′-RGTTAATNATTAACM-3′) at 13 of 15 positions. The sequences of the mouse and human promoters were identical in this region except for a single nucleotide substitution at a redundant position (N). A similar analysis of the mouse Pkd1 and Pkd2 gene promoters did not identify any evolutionarily conserved HNF-1 sites within 1 kb of the transcription initiation site.
Identification of a DNase hypersensitive site in the proximal Pkhd1 promoter.
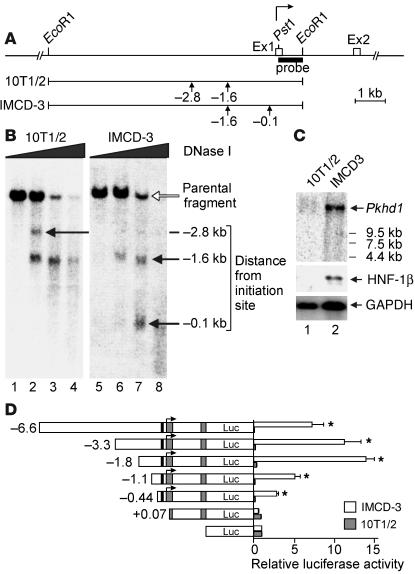
The binding of transcription factors to specific sites on DNA is accompanied by local disruptions of chromatin that render DNA in the region more susceptible to digestion by exogenous nucleases. Sites that are relatively sensitive to DNase (DNase hypersensitive sites) may contain binding sites for transcription factors that are involved in regulation of gene expression. To identify DNase hypersensitive sites in the Pkhd1 promoter, nuclei from mIMCD-3 renal epithelial cells, which endogenously express Pkhd1 (Figure 2C), were incubated with graded concentrations of DNase I. Genomic DNA was isolated, and an 8.1-kb EcoRI restriction fragment containing the first exon and 7.3 kb of 5′ flanking region, was visualized by indirect end labeling (Figure 2A). Incubation with increasing concentrations of DNase I resulted in the gradual disappearance of the 8.1-kb parental band and the appearance of sub-bands corresponding to hypersensitive sites located approximately 0.1 kb and 1.6 kb 5′ to the transcription initiation site (Figure 2B, right). The positions of the hypersensitive sites were confirmed by digestion with SpeI and hybridization to a 5′ probe (data not shown). The 10T1/2 cells, which do not endogenously express Pkhd1 (Figure 2C), contained the DNase hypersensitive site at –1.6 kb as well as an additional site at –2.8 kb, but did not contain the hypersensitive site at –0.1 kb (Figure 2B, left). These results demonstrate that the Pkhd1 promoter contains a DNase hypersensitive site located at or near the consensus HNF-1 site, the appearance of which correlates with gene transcription.
Figure 2.
DNase hypersensitive site mapping and deletion analysis of the mouse Pkhd1 promoter. (A) Structure of the 5′ end of the Pkhd1 gene. Boxes indicate exons. Bent arrow indicates the transcription initiation site at +1. Bar indicates the 3′ probe used for indirect end labeling. Vertical arrows indicate hypersensitive sites. (B) Southern blot of genomic DNA from mIMCD-3 cells (right) and 10T1/2 cells (left) after digestion with graded concentrations of DNase I. Open arrow indicates the parental 8.1-kb EcoRI fragment. Closed arrows indicate sub-bands corresponding to hypersensitive sites located at the positions indicated on the right. (C) Northern blot showing endogenous expression of Pkhd1 (upper panel) and HNF-1β (middle panel) in mIMCD-3 cells (lane 2) and absence of expression in 10T1/2 cells (lane 1). Lower panel shows expression of GAPDH as a loading control. (D) Deletion analysis of the Pkhd1 promoter. Left panel shows plasmids containing fragments of the Pkhd1 promoter linked to a promoterless luciferase (Luc) reporter gene. Bent arrow indicates the transcription initiation site at +1, gray boxes indicate exons, and black boxes indicate the consensus HNF-1 site. Right panel shows luciferase activity in transfected mIMCD-3 cells (white bars) and 10T1/2 cells (gray bars). Data are presented as mean ± SE of six to nine independent transfections. *P < 0.05 compared with empty pGL3-Basic.
The HNF-1 site is required for Pkhd1 promoter activity in transfected mIMCD-3 cells.
To verify that the proximal 5′ flanking region of Pkhd1 contained a functional gene promoter, and to evaluate the importance of the consensus HNF-1 site, reporter gene assays were performed in cultured cells. Plasmids containing different lengths of the Pkhd1 promoter linked to a firefly luciferase reporter gene were transfected into mIMCD-3 renal epithelial cells, and luciferase activity was measured 48 hours later. To correct for differences in transfection efficiency, the cells were cotransfected with a constant amount of pRL encoding Renilla luciferase, and promoter activity was inferred from the ratio of firefly and Renilla luciferase activities. Plasmids containing 6.6 kb, 3.3 kb, 1.8 kb, 1.1 kb, or 444 bp of the Pkhd1 promoter produced threefold to 14-fold stimulation of luciferase activity in transfected mIMCD-3 cells (Figure 2D). Promoter activity was abolished by truncation of the promoter from –444 bp to +76 kb, which deletes the consensus HNF-1 site and transcription initiation site. None of the plasmids produced significant luciferase activity when transfected into 10T1/2 cells that do not express Pkhd1 (Figure 2D).
Next, we determined the effects of mutations of the consensus HNF-1 site on Pkhd1 promoter activity. Two mutations (M1 and M2) that disrupted conserved nucleotides within the consensus HNF-1 site were introduced into the promoter by site-directed mutagenesis. A mutation outside the HNF-1 site was produced as a negative control (M3). (The sequences of the mutations are shown in Figure 1A.) WT and mutant plasmids were transfected into mIMCD-3 renal epithelial cells that endogenously expressed HNF-1β (Figure 2C), and luciferase activity was measured 48 hours later. Transfection of mIMCD-3 cells with a reporter plasmid containing 444 bp of the WT Pkhd1 promoter produced a threefold stimulation of luciferase activity (Figure 3A), similar to the results shown in Figure 2D. Mutations of the consensus HNF-1 site (M1 and M2) abolished promoter activity in transfected mIMCD-3 cells. In contrast, mutation M3 did not inhibit promoter activity but rather produced a small but reproducible stimulation. Similar results were obtained when the mutations were introduced into the 1.8-kb promoter (not shown). These results indicate that the consensus HNF-1 site is required for the activity of the Pkhd1 promoter in transfected mIMCD-3 cells.
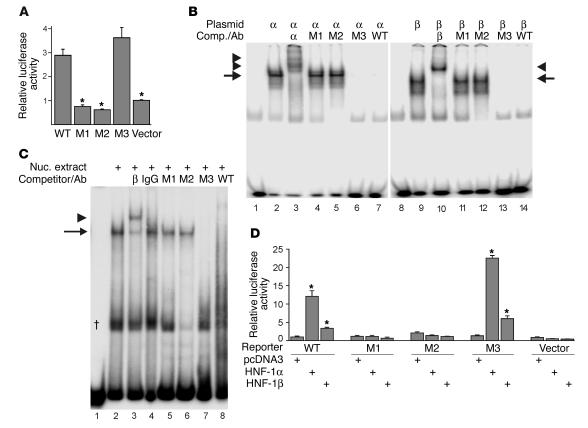
Figure 3.
Mutational analysis of the Pkhd1 promoter and binding of HNF-1. (A) Luciferase activity in mIMCD-3 cells transfected with reporter plasmids containing the WT 444-bp Pkhd1 promoter (WT), mutated promoter (M1–M3), or empty pGL3-Basic (Vector). Data are presented as mean ± SE of nine independent transfections. *P < 0.01 compared with WT promoter. (B) EMSA performed with a 44-bp DNA fragment containing the consensus HNF-1 site and reticulocyte lysates programmed with HNF-1α (lanes 2–7), HNF-1β (lanes 9–14), or unprogrammed lysates (lanes 1 and 8). Binding reactions were performed in the presence of anti–HNF-1α Ab (lane 3), anti–HNF-1β Ab (lane 10), or 100-fold excess unlabeled competitor (Comp.) DNA fragment (lanes 4–7, 11–14). (C) EMSA performed using the 44-bp DNA fragment and nuclear (Nuc) extracts from mIMCD-3 cells (lanes 2–8) or no protein (lane 1). Binding reactions were performed in the presence of anti–HNF-1β Ab (lane 3), irrelevant Ab (lanes 4), or 100-fold excess unlabeled DNA fragment (lanes 5–8). In B and C, arrows indicate retarded band, and arrowheads indicate supershifted band. †Complex that does not contain HNF-1β. (D) Luciferase activity in HeLa cells cotransfected with reporter plasmids containing the WT 444-bp Pkhd1 promoter (WT), mutated promoter (M1–M3), or empty pGL3-Basic (Vector) and expression plasmids encoding HNF-1α, HNF-1β, or empty pcDNA3. Data are presented as mean ±SE of six independent transfections. *P < 0.01 compared with cells cotransfected with empty expression plasmid.
HNF-1β and HNF-1α activate the Pkhd1 promoter directly.
To test whether HNF-1β or HNF-1α could bind to the Pkhd1 promoter, electrophoretic mobility-shift assays (EMSAs) were performed. HNF-1β and HNF-1α were translated in vitro using rabbit reticulocyte lysates and then were incubated with a 44-bp, 32P end-labeled DNA fragment containing the consensus HNF-1 site (sequence of the fragment is shown by the dashed underline in Figure 1A). The bound DNA was separated from unbound DNA by gel electrophoresis and was visualized by autoradiography. Figure 3B shows that DNA-binding activity was present in lysates programmed with HNF-1β or HNF-1α, but was not present in unprogrammed lysates. Addition of Ab’s directed against HNF-1β or HNF-1α produced a supershift, indicating the presence of the proteins in the complexes. In contrast, no supershift was observed after addition of an irrelevant Ab (not shown). The binding to HNF-1β and HNF-1α was specific since it could be inhibited with unlabeled DNA fragments containing WT sequence or a mutation outside the HNF-1 site (M3), but not with unlabeled DNA fragments containing mutations within the HNF-1 site (M1 and M2).
The mIMCD-3 cells endogenously express HNF-1β, but do not express HNF-1α, consistent with the pattern of expression of these genes in the renal collecting duct from which this cell line was derived (Figure 2C and data not shown). To determine whether endogenous HNF-1β could bind to the Pkhd1 promoter, nuclear extracts from mIMCD-3 cells were incubated with the 44-bp DNA fragment. Figure 3C shows that mIMCD-3 cells contained nuclear proteins that bound to the fragment containing the consensus HNF-1 site. The band indicated by the arrow could be supershifted with an Ab against HNF-1β, but not with an irrelevant Ab, indicating that HNF-1β was present in the DNA-protein complex. Binding was specific since it could be inhibited with unlabeled DNA fragments containing WT sequence or a mutation outside the HNF-1 site (M3), but not with unlabeled DNA fragments containing mutations within the HNF-1 site (M1 and M2). These results indicate that mIMCD-3 cells contain endogenous HNF-1β that binds specifically to the Pkhd1 promoter.
To test whether HNF-1β or HNF-1α could transactivate the Pkhd1 promoter, reporter gene assays were performed in HeLa cells that do not endogenously express either protein (8). HeLa cells were cotransfected with expression plasmids encoding HNF-1β or HNF-1α, or an equivalent amount of empty pcDNA3.1, together with a luciferase reporter plasmid containing the Pkhd1 promoter. Cotransfection with HNF-1α increased luciferase activity 12-fold, and cotransfection with HNF-1β produced a 3.3-fold increase (Figure 3D). The relative stimulation produced by HNF-1α and HNF-1β was similar to that observed previously on other HNF-1–responsive promoters (8). The stimulation of luciferase activity by HNF-1β and HNF-1α was abolished by mutations of the consensus HNF-1 site (M1 and M2), whereas a mutation outside the consensus HNF-1 site (M3) had no effect on the activation. These results demonstrate that either HNF-1β or HNF-1α can transactivate the Pkhd1 promoter. Mutations of the –49 bp site that prevented binding of HNF-1β and HNF-1α also prevented transactivation, indicating that HNF-1β and HNF-1α activated the promoter directly, rather than indirectly.
DN-HNF1β mutant inhibits DNA binding and promoter activity.
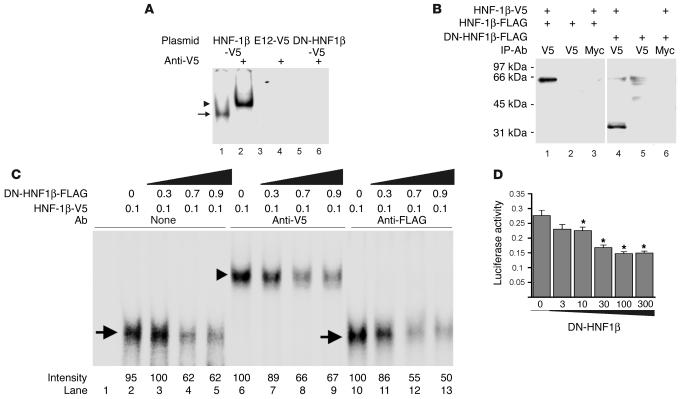
Since HNF-1β–deficient embryos die prior to gastrulation due to defects in visceral endoderm (4, 7), it was not possible to examine Pkhd1 gene expression in the kidneys of HNF-1β–deficient mice. As an alternative approach, we tested the effect of a dominant-negative mutation of HNF-1β on Pkhd1 promoter activity. The naturally occurring A263insGG mutation of human HNF-1β encodes a carboxyl terminally truncated protein that can dimerize with WT HNF-1β, but is unable to bind DNA, and therefore functions as a dominant-negative mutant (29). We produced the corresponding mutation of mouse HNF-1β and investigated its DNA-binding properties. EMSA assays showed that WT HNF-1β could bind to the Pkhd1 promoter and that an Ab to the carboxyl-terminal V5 epitope tag could supershift the complex (Figure 4A). In contrast, the DN-HNF1β mutant was unable to bind to the HNF-1 site. Likewise, no binding was observed to an irrelevant transcription factor, E12. Dimerization of HNF-1β is required for binding to DNA and is mediated by the amino terminus of the protein. Figure 4B (left) shows that WT HNF-1β formed homodimers that could be coimmunoprecipitated from transfected HEK293 cells. The DN-HNF1β mutant, which contains the amino-terminal domain, could heterodimerize with WT HNF-1β (Figure 4B, right).
Figure 4.
Effects of DN-HNF1β on DNA binding and promoter activity. (A) EMSA performed using a 44-bp DNA fragment containing the consensus HNF-1 site and nuclear extracts from HeLa cells transfected with expression plasmids encoding V5-tagged HNF-1β (lanes 1 and 2), E12 (lanes 3 and 4), or DN-HNF1β (lanes 5 and 6). Binding reactions were performed in the presence of anti-V5 Ab (lanes 2, 4, and 6) or no Ab (lanes 1, 3, and 5). Arrow indicates retarded band, and arrowhead indicates supershifted band. (B) HEK293 cells were transfected with plasmids encoding FLAG-tagged HNF-1β (lanes 1–3), V5-tagged HNF-1β (lanes 1, 3, 4, and 6), or FLAG-tagged DN-HNF1β (lanes 4–6). Cell lysates were immunoprecipitated (IP) with anti-V5 Ab (lanes 1, 2, 4, and 5) or anti-Myc Ab (lanes 3 and 6), and the immune complexes were subjected to immunoblot analysis with HRP-conjugated anti-FLAG Ab. (C) EMSA performed with the 44-bp DNA fragment and reticulocyte lysates programmed with the indicated amounts (nanograms) of plasmids encoding V5-tagged HNF-1β or FLAG-tagged DN-HNF1β. Binding reactions were performed in the presence of anti-V5 Ab (lanes 6–9), anti-FLAG Ab (lanes 10–13), or no Ab (lanes 1–5). Arrow indicates retarded band, and arrowhead indicates supershifted band. (D) Luciferase activity in mIMCD-3 cells cotransfected with a reporter plasmid containing the 444-bp Pkhd1 promoter and the indicated amounts (nanograms) of an expression plasmid encoding DN-HNF1β. Data presented are mean ± SE of nine independent transfections. *P < 0.05 compared with cells transfected with empty expression plasmid.
EMSA assays showed that the DN-HNF1β mutant inhibited the binding of WT HNF-1β to the Pkhd1 promoter (Figure 4C). Addition of increasing amounts of FLAG-tagged DN-HNF1β mutant to a constant amount of V5-tagged WT HNF-1β produced a dose-dependent decrease in DNA binding. Anti-V5 Ab’s supershifted the DNA-protein complexes, verifying that they contained WT HNF-1β, whereas anti-FLAG Ab’s did not supershift the complexes, indicating that they did not contain the DN-HNF1β mutant. To test the effects of the DN-HNF1β mutant on promoter activity, mIMCD-3 cells that endogenously express WT HNF-1β were cotransfected with an expression plasmid encoding the DN-HNF1β mutant and a luciferase reporter plasmid containing the Pkhd1 promoter. Transfection of DN-HNF1β produced a dose-dependent inhibition of Pkhd1 promoter activity (Figure 4D). The inhibition was specific since the DN-HNF1β mutant did not affect the activity of the promoter containing mutations of the HNF-1 site (not shown).
Transgenic mice expressing the DN-HNF1β mutant develop renal cysts.
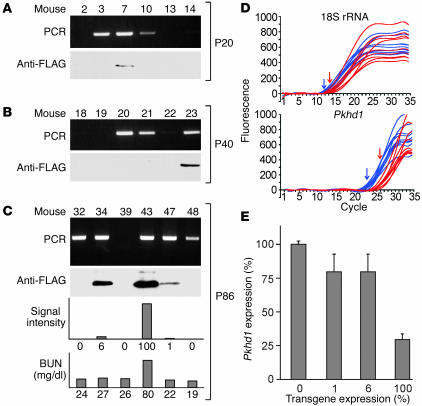
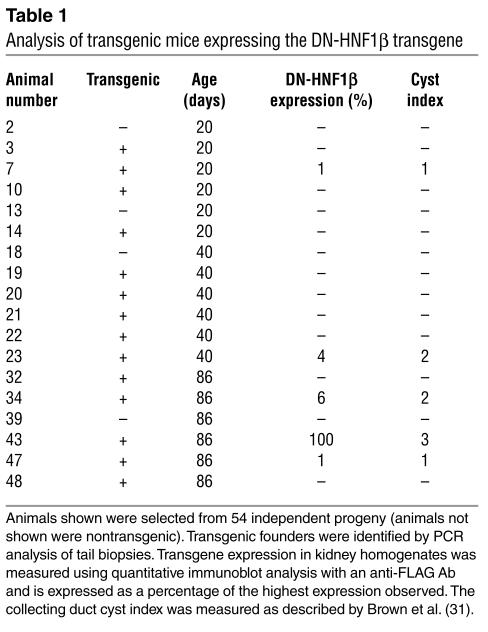
Humans that are heterozygous for the A263insGG mutation of HNF-1β develop renal cysts and diabetes mellitus (see below). To determine the effects of the mutation on kidney development in mice, transgenic mice expressing the corresponding mouse HNF-1β mutant were produced. To restrict the expression to the kidney and circumvent potential embryonic lethality, the transgene was expressed under the control of the kidney-specific Ksp-cadherin gene promoter. Previous studies have shown that this promoter directs transgene expression in all segments of the nephron, including Bowman’s capsule, proximal tubules, loops of Henle, distal tubules, and collecting ducts (28, 30). A cDNA encoding the DN-HNF1β mutant containing a carboxyl-terminal FLAG epitope tag was cloned into a plasmid downstream of the 1.3-kb Ksp-cadherin promoter and β-globin TATA box. The transgene DNA was microinjected into fertilized mouse oocytes, which were then transferred into pseudopregnant fosters. Fifty-four progeny were obtained, of which 14 (28%) carried the transgene (Figure 5, A–C, and Table 1). Immunoblot analysis using an anti-FLAG Ab showed that 5 of the 14 transgenic mice (36%) expressed the DN-HNF1β mutant in the kidney. Transgenic mice that expressed the DN-HNF1β mutant were infertile, probably due to transient expression in the sex ducts during embryonic development (28). Since permanent lines could not be established, the phenotype was analyzed in individual founders sacrificed at postnatal day 20 (P20) to P86. Nontransgenic, age-matched littermates were used as controls.
Figure 5.
Analysis of DN-HNF1β transgenic mice. (A–C) PCR analysis of tail biopsies (upper panels) and immunoblot analysis of kidney extracts (second panels) from mice sacrificed at P20 (A), P40 (B), or P86 (C). Lower panels in C show immunoblot signal intensity (expressed as percentage of the highest expression observed) and BUN in mice at P86. (D) Real-time RT-PCR tracings of Pkhd1 RNA and 18S rRNA in a transgenic mouse (red) and nontransgenic littermate (blue). Arrows indicate threshold cycles. (E) Pkhd1 RNA expression (percentage of WT) in four independent founders expressing the indicated amounts of DN-HNF1β mutant (percentage of highest expression). Data presented are mean ±SE of three separate measurements.
Table 1.
Analysis of transgenic mice expressing the DN-HNF1β transgene
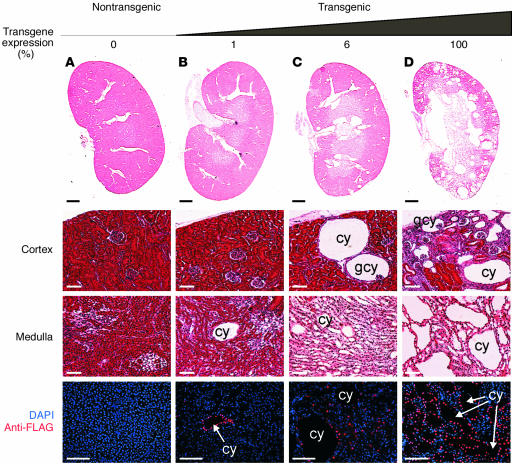
Examination of renal histology revealed that the DN-HNF1β transgenic mice developed renal cysts. Figure 6 shows H&E-stained sections of the kidneys from three independent transgenic founders at P86 compared with a nontransgenic littermate. Renal cysts were present in the transgenic mice but were absent in the age-matched control. The cysts were located in the cortex and medulla and included tubular cysts as well as glomerular cysts. They were lined by a single layer of cyst epithelial cells and were surrounded by areas of interstitial fibrosis. Two founders that had lower levels of expression of the DN-HNF1β mutant exhibited fusiform dilatation of collecting ducts and medullary cysts (Figure 6, B and C), whereas a founder with higher levels of expression showed multiple cysts throughout the cortex and medulla (Figure 6D). Collectively, renal cysts were observed in five independent founders, which indicates that the phenotype was not due to position effects, and were never observed in nontransgenic littermates. The magnitude of cyst formation in the collecting ducts was quantified using a cyst index (31) and correlated with the level of expression of the DN-HNF1β mutant (Table 1). Among the animals examined at P86, the highest-expressing founder had a blood urea nitrogen (BUN) of 79.2 mg/dl, consistent with renal failure, whereas the BUN of two lower-expressing founders was 22.2 mg/dl and 27.6 mg/dl, which was not significantly different from controls (23.1 ± 1.9 mg/dl) (Figure 5C).
Figure 6.
Formation of renal cysts in DN-HNF1β transgenic mice. (A–D) H&E-stained kidney sections from three P86 transgenic mice expressing the indicated amounts of the DN-HNF1β mutant (percentage of highest expression) and a nontransgenic littermate (A). Middle panels show higher magnification images of the renal cortex and medulla. Lower panels show localization of the DN-HNF1β mutant with an anti-FLAG Ab (red). Nuclei were counterstained with DAPI (blue). cy, cyst; gcy, glomerular cyst. Bars, 1 mm (top row), 50 μm (bottom three rows).
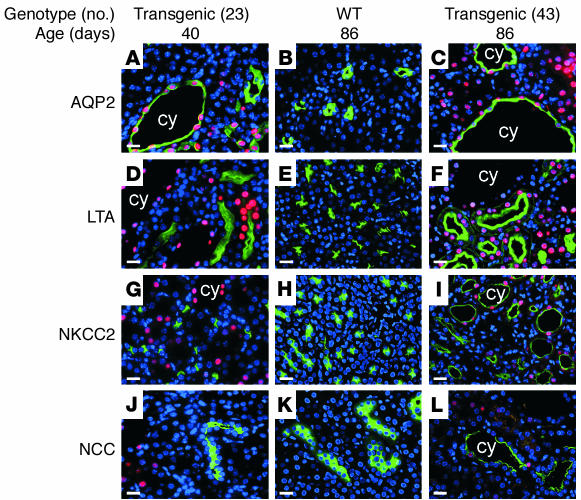
To localize the expression of the DN-HNF1β mutant in the cystic kidneys, paraffin sections were stained with an Ab directed against the carboxyl-terminal FLAG epitope. Figures 6 and 7 show that the Ab stained the nuclei of epithelial cells lining the cysts, verifying that cyst formation was dependent on the expression of the DN-HNF1β mutant. In some kidneys (Figure 7D), the DN-HNF1β mutant was also expressed in morphologically normal tubules that were identified as proximal tubules by colabeling with the lectin LTA. The absence of cysts in proximal tubules may be due to the high expression of HNF-1α in this nephron segment (1, 32). To identify the tubular origins of the cysts, the transgenic kidneys were costained with Ab’s or lectins that label specific nephron segments. Proximal tubules were labeled with LTA, thick ascending limbs of loops of Henle were labeled with an Ab to the NKCC2, distal tubules were labeled with an Ab to the NCC, and collecting ducts were labeled with an Ab to AQP2. Most kidneys showed the staining pattern shown in Figure 7 (A, D, G, and J) in which all the tubular cysts expressed AQP2, indicating that they were derived exclusively from renal collecting ducts. In a transgenic founder that highly expressed the DN-HNF1β mutant (Figure 7, C, F, I, and L), cysts originating from the collecting ducts, distal tubules, loops of Henle, and proximal tubules were observed.
Figure 7.
Origins of renal cysts in DN-HNF1β transgenic mice. Kidney sections from transgenic mice at P40 (A, D, G, J) and P86 (C, F, I, L) and a nontransgenic littermate (B, E, H, K) costained with anti-FLAG Ab (red) and Ab’s to AQP2 (A–C), NKCC2 (G–I), NCC (J–L), or FITC-coupled LTA (D–F) (green). Nuclei were counterstained with DAPI (blue). cy, cyst. Bar, 10 μm.
Mutations of HNF-1β inhibit Pkhd1 expression in vivo.
The effects of the DN-HNF1β mutant on Pkhd1 gene expression were measured using real-time RT-PCR. Total RNA from the kidneys of transgenic mice and nontransgenic littermates was transcribed into cDNA using reverse transcriptase. The cDNA encoding Pkhd1 was amplified using PCR and quantified by incorporation of SYBR green. Pkhd1 expression was calculated relative to 18S rRNA as an internal control. Figure 5D shows representative real-time RT-PCR tracings of Pkhd1 and 18S rRNA in a transgenic founder that highly expressed the DN-HNF1β mutant (red) and a nontransgenic littermate (blue). Figure 5E (right bar) shows that the overall expression of Pkhd1 in the transgenic kidney was decreased 3.3-fold. Animals with lower levels of transgene expression showed smaller decreases in the overall expression of Pkhd1 (middle two bars).
To examine the expression of Pkhd1 at higher resolution, fluorescence RNA in situ hybridization was performed. Figure 8 (A–C) shows the expression of Pkhd1 RNA transcripts in the kidneys from nontransgenic mice. As previously reported by others (22, 23), Pkhd1 transcripts were expressed in renal tubules, including collecting ducts that were identified by their expression of AQP2, but were not expressed in glomerular tufts. Hybridization to a sense riboprobe produced no significant signal (Figure 8D). Next, we examined the expression of Pkhd1 in the cystic kidneys from transgenic mice that expressed the DN-HNF1β mutant. To ensure that Pkhd1 expression was compared in the same nephron segments, the sections were costained with an Ab against AQP2. As shown in Figure 8 (E–K), five independent founders that expressed the DN-HNF1β mutant had no detectable expression of Pkhd1 in AQP2-positive cystic collecting ducts. Pkhd1 was expressed in the surrounding noncystic tubules. The observation of decreased Pkhd1 expression in multiple independent founders verified that the phenotype was not due to position effects. Costaining with an anti-FLAG Ab showed that Pkhd1 RNA transcripts were not detectable in the cytosol of cyst epithelial cells in which the DN-HNF1β mutant was highly expressed in the nucleus (Figure 8, L and M). Pkhd1 was expressed in the surrounding morphologically normal tubules that did not express the DN-HNF1β mutant. Pkhd1 was also expressed in noncystic proximal tubules that expressed the DN-HNF1β mutant, indicating that cyst formation was dependent on downregulation of Pkhd1 and not only transgene expression (not shown).
Figure 8.
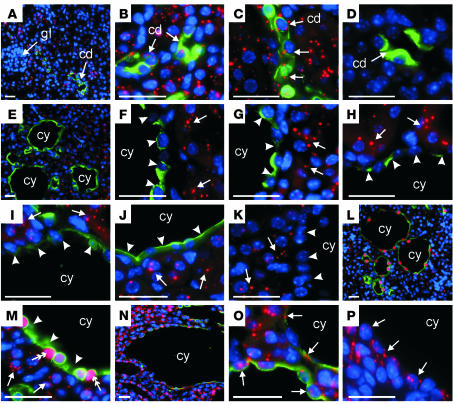
Inhibition of Pkhd1 expression in renal cysts of DN-HNF1β transgenic mice. (A–P) Fluorescence in situ hybridization of Pkhd1 antisense riboprobes (red) and (A–J, L–O) costaining with anti-AQP2 Ab (green) in kidney sections. (A–C) Nontransgenic mice show Pkhd1 expression in collecting duct (cd) (arrows) and absence in glomeruli (gl). (D) Hybridization to a sense riboprobe produces no significant signal. (E–K) Five independent DN-HNF1β transgenic founders, no. 23 (E and F), no. 7 (G), no. 43 (H), no. 47 (I), and no. 34 (J and K), show absence of Pkhd1 RNA expression (arrowheads) in the cells lining the renal cysts (cy). Arrows indicate expression in surrounding noncystic tubules. (L and M) Costaining of transgenic founder no. 23 with an anti-FLAG Ab (red) shows the absence of Pkhd1 RNA in the cytosol (arrowheads) of cyst epithelial cells in which the DN-HNF1β mutant was expressed in the nucleus (double arrow). Arrows indicate Pkhd1 expression in surrounding tubules that do not express the DN-HNF1β mutant. (N–P) Kidney-specific Kif3a KO mice show expression of Pkhd1 in the cells lining the renal cysts (arrows) as well as in surrounding tubules. (K and P) Costaining with anti-AQP2 was omitted to more clearly show the absence of Pkhd1 transcripts (arrowheads) in the cyst epithelial cells of DN-HNF1β transgenic mice (K) and the expression of Pkhd1 (arrows) in the cyst epithelial cells of Kif3a KO mice (P). Nuclei were counterstained with DAPI (blue). Bars, 20 μm.
As an additional test of specificity, we examined Pkhd1 expression in the renal cysts of kidney-specific Kif3a KO mice. KIF3A is a subunit of the kinesin-II motor protein that is required for intraflagellar transport and the synthesis of primary cilia. Previous studies have shown that kidney-specific inactivation of KIF3A using Cre/loxP recombination causes the loss of renal cilia and produces polycystic kidneys (25). Figure 8 (N–P) shows that Pkhd1 was expressed in the renal cysts of kidney-specific Kif3a KO mice. These results indicated that the inhibition of Pkhd1 expression in DN-HNF1β transgenic mice was specific and was not secondary to cyst formation itself.
Discussion
To begin understanding the transcriptional regulation of genes involved in maintaining proper tubule structure, we cloned and characterized the mouse Pkhd1 promoter. The proximal promoter contains an evolutionarily conserved consensus binding site for HNF-1 located at or near a region of DNase hypersensitivity. Mutations that disrupt the consensus HNF-1 site inhibit promoter activity in renal epithelial cells. HNF-1β and HNF-1α bind specifically to the Pkhd1 promoter and stimulate its activity directly. Expression of a DN-HNF1β mutant inhibits promoter activity in transfected cells. Taken together, these results demonstrate that HNF-1β and HNF-1α regulate the activity of the Pkhd1 promoter. One prediction of these findings is that naturally occurring mutations of the HNF-1 site in the human PKHD1 gene promoter may cause ARPKD. Mutations have been identified in the exons and introns of PKHD1 in only about 50–60% of ARPKD patients (19–21). Further studies will be required to determine whether some of the remaining patients carry mutations that disrupt the HNF-1 site in the promoter.
HNF-1β and HNF-1α constitute a family of homeodomain-containing transcription factors that regulate tissue-specific gene expression in the liver, kidney, pancreas, and other epithelial organs (1). In the kidney, the pattern of expression of HNF-1β overlaps with the expression of Pkhd1 (4–6). Like Pkhd1, HNF-1β is expressed exclusively in tubular epithelial cells in the kidney and is not expressed in glomerular tufts, blood vessels, or interstitial cells. HNF-1β is expressed in all segments of the nephron, especially the loops of Henle and distal tubules, and in renal collecting tubules, whereas HNF-1α is expressed only in proximal tubules (1, 32). In developing embryos, HNF-1β is expressed in the ureteric bud, mesonephros, and epididymis, which are also sites of Pkhd1 gene expression (5). In addition, both HNF-1β and Pkhd1 are expressed in bile ducts and pancreatic ducts. Taken together with the effects of HNF-1β mutations on Pkhd1 expression (see below), these studies suggest that HNF-1β is the principal member of the HNF-1 family that regulates Pkhd1 expression in the kidney. Since Pkhd1 is also expressed in nonepithelial tissues, such as sympathetic ganglia and blood vessels, other transcription factors besides HNF-1β must be responsible for expression at these sites.
Autosomal dominant mutations of HNF-1β produce MODY5, associated with congenital cystic abnormalities of the kidney. The A263insGG mutation of HNF-1β was first described in a Japanese family with early-onset type 2 diabetes (33). Five individuals from three generations were found to carry the mutation, and all had multiple bilateral renal cysts as determined by ultrasound. Four affected individuals had decreased renal function or end-stage renal disease. In addition, one individual had proteinuria prior to the diagnosis of diabetes. The A263insGG mutation consists of the insertion of a GG dinucleotide in the codon for Ala-263. This mutation causes a frameshift and produces a truncated protein that lacks the third helix of the homeodomain and the carboxyl-terminal activation domain. Biochemical studies have shown that the A263insGG mutant functions as a dominant-negative mutant when expressed in liver, pancreatic, and kidney cell lines, although the mechanism may depend on the promoter and cell type (8, 29, 34). Consistent with a dominant-negative mechanism, we found that the corresponding mutation of mouse HNF-1β produced a protein that dimerizes with WT HNF-1β, inhibits the binding of WT HNF-1β to the Pkhd1 promoter, and inhibits Pkhd1 promoter activity when expressed in cells that endogenously express WT HNF-1β. Expression of the DN-HNF1β mutant in transgenic mice under the control of a kidney-specific promoter led to the development of renal cysts and renal failure, similar to humans with the A263insGG mutation. These results confirm previous studies in zebra fish, showing that inactivation of HNF-1β produces renal cysts (35). Mutations of HNF-1β have also been shown to interfere with pronephric development in Xenopus embryos (34, 36). The expression of HNF-1β is inhibited in the cpk mouse model of polycystic kidney disease (37). Our studies demonstrate that a disease-causing mutation of HNF-1β produces kidney cysts in mammals and represent a mouse model of the congenital renal abnormalities in MODY5.
Similar to humans with MODY5, transgenic mice expressing the DN-HNF1β mutant develop cysts in renal tubules as well as glomerular cysts. Enlarged nephrons are observed in some MODY5 patients and in Xenopus embryos expressing the A263insGG mutant (34) but were not observed in transgenic mice expressing the DN-HNF1β mutant. Because the DN-HNF1β mutant was expressed from a kidney-specific promoter, our studies verify that mutations of HNF-1β are sufficient to produce kidney cysts and that the renal cystic disease is intrinsic to the kidney and is not secondary to diabetes. To determine the mechanism of cyst formation, we examined the expression of Pkhd1 in the DN-HNF1β transgenic kidneys. Consistent with the observations that HNF-1β regulates the Pkhd1 promoter and that DN-HNF1β inhibits Pkhd1 promoter activity, we found that Pkhd1 gene expression was decreased in the kidneys of transgenic mice. Cyst epithelial cells that highly expressed the DN-HNF1β mutant showed no detectable expression of Pkhd1. Taken together, these studies identify a link between two cystic disease genes, HNF1β and Pkhd1, and suggest that the mechanism of cyst formation in humans with autosomal dominant HNF-1β mutations involves inhibition of PKHD1 and other coordinately regulated genes.
Another prediction of our studies is that humans with MODY5 will also have decreased expression of Pkhd1 in the biliary tract, which is another site where the expression of HNF-1β and Pkhd1 overlap. Biliary dysgenesis and portal fibrosis are consistent findings in ARPKD due to mutations of PKHD1. Liver-specific inactivation of HNF-1β in transgenic mice produces a ductal plate malformation and biliary dysgenesis that resemble the lesions observed in the livers of ARPKD patients (38). Moreover, abnormalities in liver function and liver fibrosis have been seen in some humans with HNF-1β mutations (39). These observations suggest that HNF-1β may also be required for the expression of Pkhd1 in the developing biliary tract and that mutations of HNF-1β may produce biliary tract abnormalities through downregulation of Pkhd1.
Note added in proof.
Gresh et al. have shown that kidney-specific inactivation of HNF-1β produces renal cysts (40).
Acknowledgments
This work was supported by NIH grants R01 DK42921 and P50 DK57328 and grant 22a2R from the PKD Foundation. We thank Andrea Kaup and Baiju Sthapit for expert technical assistance, Bob Hammer for assistance with generation of transgenic mice, Ralph DiLeone for in situ hybridization protocols, J.T. Hsieh for assistance with real-time RT-PCR, Mark Knepper for Ab’s, Marco Pontoglio for helpful advice, and Fangming Lin for critically reviewing the manuscript.
Footnotes
Angus M. Sinclair’s present address is: Amgen Inc., Thousand Oaks, California, USA.
Nonstandard abbreviations used: aquaporin-2 (AQP2); autosomal recessive polycystic kidney disease (ARPKD); bacterial artificial chromosome (BAC); blood urea nitrogen (BUN); cyanine 3 (Cy3); deoxyribonuclease I (DNase I); dominant-negative HNF-1β (DN-HNF1β); electrophoretic mobility shift assay (EMSA); hepatocyte nuclear factor–1β (HNF-1β); Lotus tetragonolobus agglutinin (LTA); maturity-onset diabetes mellitus of the young, type 5 (MODY5); mouse inner medullary collecting duct (mIMCD); Na-Cl cotransporter (NCC); Na-K-Cl cotransporter (NKCC2); postnatal day (P); tyramide signal amplification (TSA).
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Pontoglio M. Hepatocyte nuclear factor 1, a transcription factor at the crossroads of glucose homeostasis. J. Am. Soc. Nephrol. 2000;11(Suppl. 16):S140–S143. [PubMed] [Google Scholar]
- 2.Mendel DB, Hansen LP, Graves MK, Conley PB, Crabtree GR. HNF-1α and HNF-1β (vHNF-1) share dimerization and homeo domains, but not activation domains, and form heterodimers in vitro. Genes Dev. 1991;5:1042–1056. doi: 10.1101/gad.5.6.1042. [DOI] [PubMed] [Google Scholar]
- 3.Baumhueter S, Courtois G, Crabtree GR. A variant nuclear protein in dedifferentiated hepatoma cells binds to the same functional sequences in the β fibrinogen gene promoter as HNF-1. EMBO J. 1988;7:2485–2493. doi: 10.1002/j.1460-2075.1988.tb03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbacci E, et al. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126:4795–4805. doi: 10.1242/dev.126.21.4795. [DOI] [PubMed] [Google Scholar]
- 5.Coffinier C, Barra J, Babinet C, Yaniv M. Expression of the vHNF1/HNF1β homeoprotein gene during mouse organogenesis. Mech. Dev. 1999;89:211–213. doi: 10.1016/s0925-4773(99)00221-x. [DOI] [PubMed] [Google Scholar]
- 6.Lazzaro D, de Simone V, de Magistris L, Lehtonen E, Cortese R. LFB1 and LFB3 homeoproteins are sequentially expressed during kidney development. Development. 1992;114:469–479. doi: 10.1242/dev.114.2.469. [DOI] [PubMed] [Google Scholar]
- 7.Coffinier C, Thepot D, Babinet C, Yaniv M, Barra J. Essential role for the homeoprotein vHNF1/HNF1β in visceral endoderm differentiation. Development. 1999;126:4785–4794. doi: 10.1242/dev.126.21.4785. [DOI] [PubMed] [Google Scholar]
- 8.Bai Y, Pontoglio M, Hiesberger T, Sinclair AM, Igarashi P. Regulation of kidney-specific Ksp-cadherin gene promoter by hepatocyte nuclear factor-1β. Am. J. Physiol. 2002;283:F839–F851. doi: 10.1152/ajprenal.00128.2002. [DOI] [PubMed] [Google Scholar]
- 9.Horikawa Y, et al. Mutation in hepatocyte nuclear factor-1β gene (TCF2) associated with MODY. Nat. Genet. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 10.Woolf AS, Feather SA, Bingham C. Recent insights into kidney disease associated with glomerular cysts. Pediatr. Nephrol. 2002;17:229–235. doi: 10.1007/s00467-001-0819-5. [DOI] [PubMed] [Google Scholar]
- 11.Woolf AS. Diabetes, genes, and kidney development. Kidney Int. 2000;57:1202–1203. doi: 10.1046/j.1523-1755.2000.00949.x. [DOI] [PubMed] [Google Scholar]
- 12.Bingham C, et al. Mutations in the hepatocyte nuclear factor-1β gene are associated with familial hypoplastic glomerulocystic kidney disease. Am. J. Hum. Genet. 2001;68:219–224. doi: 10.1086/316945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bingham C, et al. Atypical familial juvenile hyperuricemic nephropathy associated with a hepatocyte nuclear factor-1β gene mutation. Kidney Int. 2003;63:1645–1651. doi: 10.1046/j.1523-1755.2003.00903.x. [DOI] [PubMed] [Google Scholar]
- 14.Zerres K, Rudnik-Schoneborn S, Steinkamm C, Becker J, Mucher G. Autosomal recessive polycystic kidney disease. J. Mol. Med. 1998;76:303–309. doi: 10.1007/s001090050221. [DOI] [PubMed] [Google Scholar]
- 15.Guay-Woodford LM, Desmond RA. Autosomal recessive polycystic kidney disease; the clinical experience in North America. Pediatrics. 2003;111:1072–1080. doi: 10.1542/peds.111.5.1072. [DOI] [PubMed] [Google Scholar]
- 16.Igarashi P, Somlo S. Genetics and pathogenesis of polycystic kidney disease. J. Am. Soc. Nephrol. 2002;13:2384–2398. doi: 10.1097/01.asn.0000028643.17901.42. [DOI] [PubMed] [Google Scholar]
- 17.Ward CJ, et al. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat. Genet. 2002;30:259–269. doi: 10.1038/ng833. [DOI] [PubMed] [Google Scholar]
- 18.Onuchic LF, et al. PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription factor domains and parallel beta-helix 1 repeats. Am. J. Hum. Genet. 2002;70:1305–1317. doi: 10.1086/340448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furu L, et al. Milder presentation of recessive polycystic kidney disease requires presence of amino acid substitution mutations. J. Am. Soc. Nephrol. 2003;14:2004–2014. doi: 10.1097/01.asn.0000078805.87038.05. [DOI] [PubMed] [Google Scholar]
- 20.Bergmann C, et al. Spectrum of mutations in the gene for autosomal recessive polycystic kidney disease (ARPKD/PKHD1) J. Am. Soc. Nephrol. 2003;13:76–89. doi: 10.1097/01.asn.0000039578.55705.6e. [DOI] [PubMed] [Google Scholar]
- 21.Rossetti S, et al. A complete mutation screen of PKHD1 in autosomal-recessive polycystic kidney disease (ARPKD) pedigrees. Kidney Int. 2003;64:391–403. doi: 10.1046/j.1523-1755.2003.00111.x. [DOI] [PubMed] [Google Scholar]
- 22.Xiong H, et al. A novel gene encoding a TIG multiple domain protein is a positional candidate for autosomal recessive polycystic kidney disease. Genomics. 2002;80:96–104. doi: 10.1006/geno.2002.6802. [DOI] [PubMed] [Google Scholar]
- 23.Nagasawa Y, et al. Identification and characterization of Pkhd1, the mouse orthologue of the human ARPKD gene. J. Am. Soc. Nephrol. 2002;13:2246–2258. doi: 10.1097/01.asn.0000030392.19694.9d. [DOI] [PubMed] [Google Scholar]
- 24.Ward CJ, et al. Cellular and subcellular localization of the ARPKD protein, fibrocystin is expressed on primary cilia. Hum. Mol. Genet. 2003;12:2703–2710. doi: 10.1093/hmg/ddg274. [DOI] [PubMed] [Google Scholar]
- 25.Lin F, et al. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whyte DA, et al. Ksp-cadherin gene promoter. I. Characterization and renal epithelial cell-specific activity. Am. J. Physiol. 1999;277:F587–F598. doi: 10.1152/ajprenal.1999.277.4.F587. [DOI] [PubMed] [Google Scholar]
- 28.Shao X, Johnson JE, Richardson JA, Hiesberger T, Igarashi P. A minimal Ksp-cadherin promoter linked to a green fluorescent protein reporter gene exhibits tissue-specific expression in the developing kidney and genitourinary tract. J. Am. Soc. Nephrol. 2002;13:1824–1836. doi: 10.1097/01.asn.0000016443.50138.cd. [DOI] [PubMed] [Google Scholar]
- 29.Tomura H, et al. Loss-of-function and dominant-negative mechanisms associated with hepatocyte nuclear factor-1β mutations in familial type 2 diabetes mellitus. J. Biol. Chem. 1999;274:12975–12978. doi: 10.1074/jbc.274.19.12975. [DOI] [PubMed] [Google Scholar]
- 30.Shao X, Somlo S, Igarashi P. Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J. Am. Soc. Nephrol. 2002;13:1837–1846. doi: 10.1097/01.asn.0000016444.90348.50. [DOI] [PubMed] [Google Scholar]
- 31.Brown NE, Murcia NS. Delayed cystogenesis and increased ciliogenesis associated with the re-expression of polaris in Tg737 mutant mice. Kidney Int. 2003;63:1220–1229. doi: 10.1046/j.1523-1755.2003.00863.x. [DOI] [PubMed] [Google Scholar]
- 32.Pontoglio M, et al. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 33.Nishigori H, et al. Frameshift mutation, A263fsinsGG, in the hepatocyte nuclear factor-1β gene associated with diabetes and renal dysfunction. Diabetes. 1998;47:1354–1355. doi: 10.2337/diab.47.8.1354. [DOI] [PubMed] [Google Scholar]
- 34.Bohn S, et al. Distinct molecular and morphogenetic properties of mutations in the human HNF1β gene that lead to defective kidney development. J. Am. Soc. Nephrol. 2003;14:2033–2041. doi: 10.1097/01.asn.0000078808.70309.c4. [DOI] [PubMed] [Google Scholar]
- 35.Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wild W, et al. The mutated human gene encoding hepatocyte nuclear factor 1β inhibits kidney formation in developing Xenopus embryos. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4695–4700. doi: 10.1073/pnas.080010897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calvet JP. Injury and development in polycystic kidney disease. Curr. Opin. Nephrol. Hypertens. 1994;3:340–348. doi: 10.1097/00041552-199405000-00017. [DOI] [PubMed] [Google Scholar]
- 38.Coffinier C, et al. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1β. Development. 2002;129:1829–1838. doi: 10.1242/dev.129.8.1829. [DOI] [PubMed] [Google Scholar]
- 39.Montoli A, et al. Renal cysts and diabetes syndrome linked to mutations of the hepatocyte nuclear factor-1β gene: description of a new family with associated liver involvement. Am. J. Kidney Dis. 2002;40:397–402. doi: 10.1053/ajkd.2002.34538. [DOI] [PubMed] [Google Scholar]
- 40.Gresh, L., et al. 2004. A transcriptional network in polycystic kidney disease. EMBO J. In press. [DOI] [PMC free article] [PubMed]