Abstract
Background
In Plasmodium, the high level of genetic diversity and the interactions established by co-infecting parasite populations within the same host may be a source of selection on pathogen virulence and drug resistance. As different patterns have already been described in humans and mosquitoes, parasite diversity and population structure should be studied in both hosts to properly assess their effects on infection and transmission dynamics. This study aimed to characterize the circulating populations of Plasmodium spp and Plasmodium falciparum from a combined set of human blood and mosquito samples gathered in mainland Equatorial Guinea. Further, the origin and evolution of anti-malarial resistance in this area, where malaria remains a major public health problem were traced.
Methods
Plasmodium species infecting humans and mosquitoes were identified by nested-PCR of chelex-extracted DNA from dried blood spot samples and mosquitoes. Analysis of Pfmsp2 gene, anti-malarial-resistance associated genes, Pfdhps, Pfdhfr, Pfcrt and Pfmdr1, neutral microsatellites (STR) loci and Pfdhfr and Pfdhps flanking STR was undertaken to evaluate P. falciparum diversity.
Results
Prevalence of infection remains high in mainland Equatorial Guinea. No differences in parasite formula or significant genetic differentiation were seen in the parasite populations in both human and mosquito samples. Point mutations in all genes associated with anti-malarial resistance were highly prevalent. A high prevalence was observed for the Pfdhfr triple mutant in particular, associated with pyrimethamine resistance.
Analysis of Pfdhps and Pfdhfr flanking STR revealed a decrease in the genetic diversity. This finding along with multiple independent introductions of Pfdhps mutant haplotypes suggest a soft selective sweep and an increased differentiation at Pfdhfr flanking microsatellites hints a model of positive directional selection for this gene.
Conclusions
Chloroquine is no longer recommended for malaria treatment in Equatorial Guinea but sulphadoxine-pyrimethamine (SP) remains in use in combination with artesunate and is the only drug recommended in preventive chemotherapy in pregnancy. The high prevalence of point mutations in Pfdhfr and Pfdhps points to the danger of an eventual reduction in the efficacy of SP combined therapy in P. falciparum populations in Equatorial Guinea and to the essential continuous monitoring of these two genes.
Keywords: Malaria, Equatorial Guinea, Genetic diversity, Drug resistance, pfcrt, pfdhps, pfdhfr, pfmdr1, Microsatellites, Plasmodium falciparum
Background
Malaria continues to be one of the main public health problems in the world, affecting 106 countries, with approximately 216 million cases resulting in 650,000 yearly deaths [1]. This parasitic disease involves three living entities with complex interactions among them and transmission of Plasmodium parasites by their anopheline vectors is a crucial factor determining the epidemiology of malaria in endemic areas.
The level of genetic diversity of natural populations of Plasmodium is well demonstrated and both inter- and intra-specific mixed infections in the same host are common, especially in highly endemic areas [2]. The ecological interactions that these different and co-infecting parasite populations establish among them may be a source of selection on pathogen traits such as virulence and drug resistance.
Parasite genetic diversity and population structure in both humans and mosquitoes should be assessed in order to better determine the influence of different parasite populations on infection and transmission dynamics. In fact, both different associations of Plasmodium species as well as marked differences in the multiplicity of infection and allele diversity of Plasmodium falciparum populations were previously reported [3]. Furthermore, a recent analysis of both human peripheral blood samples and mosquitoes from the same location has revealed a completely unexpected picture related to the presence of Plasmodium vivax in an area where it had not yet been reported [4]. Differences have also been found in drug-resistant associated genes. In Gabon, Mharakurwa et al[5] reported that parasites in humans presented high levels of pyrimethamine (PYR)-resistant mutants, whereas parasites in Anopheles mosquitoes showed high levels of cycloguanil-resistant mutants.
For a period of time, the genetic diversity of P. falciparum populations has mainly been investigated through the analysis of mutation in polymorphic surface antigen coding genes [6,7]. However, this approach poses some limitations as it is impossible to know whether observed patterns reflect population history or natural selection [8]. Microsatellite sequences (STR), spread throughout the genome, are currently the neutral markers most commonly used to differentiate P. falciparum populations as these markers (short repeated nucleotide sequences) often present high levels of inter- and intra-specific polymorphism, particularly when the number of repetition is 10 or higher.
In Equatorial Guinea, malaria remains the major endemic disease and the leading cause of child mortality and morbidity. In recent years, the prevalence of infection has been reduced significantly on the Insular Region due to an effective vector control [1,9] whilst the prevalence of infection remains above 50% in children under five years old in mainland region [10]. Along with the high prevalence of infection, the dissemination of P. falciparum drug resistance still remains the main constraint to control malaria transmission in most endemic areas. Anti-malarial resistance has largely been studied through the analysis of mutations on several target genes associated with resistance to specific drugs, e g, Pfcrt[11] and Pfmdr1[12] linked to chloroquine (CQ) resistance; and Pfdhfr[13] and Pfdhps genes [14] associated with resistance to pyrimethamine (PYR) and sulphadoxine (SFX), respectively.
Increasing failure rates (40-50%) for CQ and around 25% resistance to sulphadoxine/pyrimethamine (SP) in under-five children was reported in 2003 in Malabo, the capital city of Equatorial Guinea located in the island of Bioko [15]. Nevertheless, CQ continued to be used in mainland region as the first-line treatment for uncomplicated malaria until 2009, and had been replaced by artesunate + sulphadoxine/pyrimethamine (AS+SP) combination on the island of Bioko in 2004 [16]. In 2009, artemisinin combination therapy (ACT) of artesunate/amodiaquine (AS/AQ) was adopted as first-line therapy based on the high levels of resistance to SP in neighbouring countries. More recently, a study conducted in Bata, the largest city in the mainland region, and Malabo revealed that AS/SP and AQ/SP combinations were both highly effective for the treatment of uncomplicated P. falciparum malaria [16]. SP is still prescribed alone for intermittent preventive therapy in pregnant women [17].
This study aimed to characterize the circulating populations of Plasmodium spp and P. falciparum from a combined set of human blood and mosquito samples collected in both coastal and inland villages from mainland Equatorial Guinea. Plasmodium falciparum diversity was analysed through the study of an antigen coding gene (Pfmsp2) as well as a set of neutral STR loci and four anti-malarial resistant associated genes (Pfcrt, Pfmdr1, Pfdhfr and Pfdhps). Finally, to trace the origin of anti-malarial resistance and its progression in this area, the presence of signatures of drug resistant selection in P. falciparum populations were investigated. The impact of these findings on control policies, especially the avoidance of dissemination of P. falciparum drug-resistant parasites in Equatorial Guinea, is discussed.
Methods
Sample collection
Peripheral blood samples from 97 inhabitants (zero to 78 years-old) were collected in 2005 in mainland Equatorial Guinea during the dry (February and August) and rainy (May) seasons from two villages, Miyobo (34 and 43 individuals in the dry and rainy seasons, respectively; 44 different individuals in total) and Ngonamanga (40 and 26 individuals in the dry and rainy seasons, respectively; 53 different individuals in total). Blood sampling has been performed in four consecutive days per individual, in order to better assess variations in the P. falciparum population’s composition. Further, 819 mosquito specimens were also collected during the same period and locations. Miyobo is located in a forested area on the riverbank of the Wele River, which crosses the mainland region from east to west. Ngonamanga is a coastal village surrounded by forest-savannah, 60 km north of Bata. In both, malaria is classified as hyperendemic. Both study areas and sample collection procedures have been described elsewhere [4].
Villagers were informed of the nature and aims of the study and voluntary participation of five households randomly selected by location was requested after approval of local authorities. Blood samples were collected after informed consent was received from all donors (parents or guardians responded on behalf of children). Mosquito collection was done after the approval of local authorities, the owner and occupants of the house. Written consent was obtained from the legal guardians of the recruited children and non-documented, oral consent was only requested from adults, due to the community-wide mistrust of signing official forms. The study was approved by the Ethical Committee of Equatorial Guinea’s Ministry of Health and Social Welfare, the National Malaria Control Programme, and local health authorities from the villages, which accepted the constraint and found no bio-ethical impediments to the study. Ethical clearance was also given by the Ethical Committees of the Instituto de Higiene e Medicina Tropical (Lisboa, Portugal) and the Instituto de Salud Carlos III (Madrid, Spain), according to EU regulations.
DNA extraction and molecular assays
Individual mosquitoes, dried on silica gel, and blood spot samples were stored at room temperature until DNA preparation. DNA was extracted using chelex according to Plowe et al[18] from blood spots and to Arez et al[19] from mosquitoes.
Detection of malaria infection and identification of Plasmodium species was carried out by nested-PCR amplification of the ssrRNA genes [20]. Plasmodium falciparum positive samples were further genotyped for:
a) Pfmsp2 gene by a nested-PCR as in Snounou et al[21];
b) Drug resistant associated genes by a nested PCR-RFLP analysis of the presence/absence of mutations at codons 75 and 76 of the Pfcrt gene, codons 86 and 1246 of the Pfmdr1 gene, codons 51, 59, 108 and 164 of the Pfdhfr gene and codons 436, 437, 540 and 581 of the Pfdhps gene [22],
c) Nine neutral microsatellite loci (STR) distributed throughout the genome of P. falciparum: TAA42, TAA81 (chromosome 5), TA1, TAA87, TAA109 (chromosome 6), ARA2 (chromosome 11), TA102, PfPK2 and Pfg377 (chromosome 12). Primer sequences and PCR conditions are described in Anderson et al[23] and Conway et al[24];
d) STRs flanking Pfdhfr and Pfdhps genes located 0.3 kb, 4.4 kb and 5.3 kb upstream of codon 108 of Pfdhfr (chromosome 4) and 0.8 kb, 4.3 kb and 7.7 kb downstream from codon 437 of Pfdhps (chromosome 8). Primer sequences and PCR conditions are described in Roper et al[25], Ndiaye et al[26] and Salgueiro et al[27]. Southeast Asian P. falciparum K1 laboratory strain was used as reference (at STRs flanking the Pfdhps gene, the allelic composition of the K1 strain matches that of the East African Pfdhps double mutant A437G/K540E haplotype lineage SGE 1 [28]).
Amplified fragments were run in an automatic sequencer (ABI 3730, Applied Biosystem) at Yale University, DNA Analysis Facility on Science Hill. The software GeneMarker (SoftGenetics) was used to measure allele sizes. Samples that failed the amplification in any of the loci or presented multiple STR peaks were excluded for the haplotype definition [25]. A new haplotype was considered when there was one or more allelic changes across all loci considered. For the remaining analyses, in cases where multiple peaks were present, only the value of the highest peak per locus was scored [8].
Statistical analysis
Pearson χ2 test was used to compare prevalence of infection between collection sites, seasons and hosts. Whenever Pearson χ2 test conditions were not satisfied, Fisher’s exact test was used (SPSS v.12 statistical software). Pearson’s χ2 test was also used to assess possible associations between Plasmodium species [29].
Prevalence of Pfmsp2 alleles and the minimum number of concurrent genotypes in the same isolate (multiplicity of infection (MOI): the largest number of alleles found in each sample) were calculated for all comparison groups; mosquitoes versus blood samples, Miyobo versus Ngonamanga and rainy season versus dry season.
STR data was analysed with FSTAT v. 2.9.3.2 [30] to obtain measures of genetic diversity [allelic richness Rs: a measure of the number of alleles independent of sample size, hence allowing to compare this quantity between different sample sizes; and expected heterozygosity He per locus and sample: this use an unbiased estimator Hs, which is calculated from individual allele frequencies and range from zero (no heterozygosity) to nearly 1.0 (for a system with a large number of equally frequent alleles)] and genetic differentiation using the Fst estimator. Linkage disequilibrium (LD) tests were performed with GENEPOP v.3.4 [31].
After the assessment of PYR- and SFX-associated wild type (or sensitive) and mutant alleles, comparisons were made between populations classified as “wild type”, “single mutant” (Pfdhfr: 51 or 59 or 108 or 164; Pfdhps: 436 or 437 or 540 or 581), “double mutant” (Pfdhfr: 51:108 or 59:108 or 51:59 or 51:164; Pfdhps: 436:581 or 437:581 or 540:581), “triple mutant” (Pfdhfr: 51:59:108 or 59:108:164 or 51:59:164, Pfdhps: 436:437:581 or 436:540:581), “quadruple mutant” (Pfdhfr: 51:59:108:164, Pfdhps 436:437:540:581). However this was not always possible due to the low number of samples in some groups, so that only the whole sample was subdivided and compared according to geographic collection sites.
In multiple tests, Bonferroni correction was applied by dividing 0.05 by the number of tests to minimize type I errors and obtain the actual cut-off for significance [32].
Results
Detection and identification of Plasmodium species
A total of 427 blood samples from 97 individuals were collected in both villages and seasons (44 individuals from Miyobo and 53 from Ngonamanga). A total of 819 mosquitoes were collected (509 from Miyobo and 310 from Ngonamanga), 536 belonging to Anopheles gambiae complex, 259 belonging to Anopheles nili complex (presumably Anopheles carnevalei), three to Anopheles funestus complex and 21 Anopheles moucheti moucheti.
In order to determine prevalence of infection, an individual was defined as infected if he/she had at least one positive sample among the multiple samples collected; therefore, only one sample was considered per individual and all calculations were performed having the number of individuals as denominator. Overall, prevalence of Plasmodium spp infection in humans was 93% in Miyobo and 81% in Ngonamanga, and was higher in the dry season (69%) than in the rainy (67%). In mosquitoes, the prevalence of infection was slightly higher in Ngonamanga (20%) than Miyobo (19%) and in the rainy season (22%) than in the dry season (16%). Although the four Plasmodium species were detected in both hosts, P. falciparum was the predominant species occurring in 90% of the isolates (both humans and mosquitoes) either in single or mixed infection (see Additional files 1 and 2). In humans, Plasmodium malariae was the second most prevalent species, occurring in 13% of individuals, followed by P. vivax (10%) and finally Plasmodium ovale (8%) (see Additional file 1). In mosquitoes, P. vivax was the second most prevalent species (9%), followed by P. malariae (4%) and P. ovale (2%) (see Additional file 2). A significantly higher number than expected of mixed infections with P. falciparum and P. malariae in both hosts (blood samples: χ2=8.973, p=0.003; mosquitoes: χ2=15.745, p<0.001) was found. No association was found for the pair P. falciparum and P. vivax.
Plasmodium falciparum genetic diversity
Pfmsp2
Plasmodium falciparum was detected in 302 out of the 427 samples collected and successful genotyping of Pfmsp2 gene was achieved in 73% (221/302) P. falciparum-positive blood samples and none in the 275 P. falciparum-positive mosquitoes. The unsuccessful amplification of Pfmsp2 in mosquitoes was probably due to degradation of parasite DNA in dried mosquitoes stored at room temperature for a long period of time.
No major differences in allelic diversity were detected between seasons or villages, which shared 11 out of 13 alleles; two unique alleles were detected in Ngonamanga in dry season (IC_400 and IC_700) and only one was observed in the rainy season (FC27_250) in both villages. The mean MOI was slightly higher in Miyobo than in Ngonamanga; 1.98 versus 1.83, respectively, and varied between 1.46 (Ngonamanga, rainy season) and 2.19 (Ngonamanga, dry season). When values are compared between villages without season distinction, mean MOI was slightly higher in Miyobo than in Ngonamanga (1.98 versus 1.83, respectively) and it was higher in the dry season in Ngonamanga (dry versus rainy: 2.19 versus 1.46), while the opposite occurred in Miyobo (dry versus rainy: 1.88 versus 2.07).
Neutral STRs loci
Ninety-nine per cent (299/302) P. falciparum-positive blood samples and 83% (228/275) P. falciparum-positive mosquitoes were successfully genotyped. The number of observed alleles (Na), allelic richness (Rs) and genetic diversity (uH) are shown in Table 1. All nine STR analysed were polymorphic and the number of alleles varied between seven in Pfg377 and 17 in TA109 in human samples, and six in TA42 and 18 in TA109 in mosquitoes. The majority of samples presented multiple P. falciparum genotypes but in general, the most common alleles are shared between parasite populations present in humans and mosquitoes. Genetic diversity (uH) also presented similar values; 0.75 versus 0.77, in humans and mosquitoes, respectively, as well as the number of alleles for each locus; 12 versus 11, in humans and mosquitoes respectively (see Table 1).
Table 1.
Neutral microsatellite diversity of Plasmodium falciparum populations from Ngonamanga and Miyobo in humans and mosquitoes
| n | TA1 | TA102 | ARA2 | TA87 | Pfk2 | TA81 | TA42 | Pfg377 | TA109 | Mean | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Humans |
TBs |
299 |
Na |
13 |
12 |
9 |
15 |
14 |
11 |
14 |
7 |
17 |
12 |
|
Rs |
12 |
11 |
8 |
12 |
13 |
10 |
10 |
5 |
13 |
10 |
|||
|
uH |
0.852 |
0.832 |
0.787 |
0.838 |
0.877 |
0.829 |
0.344 |
0.612 |
0.799 |
0.75 |
|||
|
BsM |
195 |
Na |
11 |
11 |
9 |
14 |
12 |
11 |
7 |
7 |
15 |
11 |
|
|
Rs |
10 |
9 |
8 |
9 |
10 |
9 |
5 |
5 |
9 |
8 |
|||
|
uH |
0.846 |
0.848 |
0.771 |
0.829 |
0.857 |
0.792 |
0.293 |
0.642 |
0.811 |
0.74 |
|||
|
BsN |
104 |
Na |
11 |
8 |
8 |
9 |
11 |
10 |
11 |
5 |
9 |
9 |
|
|
Rs |
10 |
7 |
8 |
8 |
11 |
10 |
7 |
5 |
7 |
8 |
|||
|
uH |
0.859 |
0.771 |
0.809 |
0.844 |
0.877 |
0.868 |
0.424 |
0.531 |
0.747 |
0.75 |
|||
|
BsW |
141 |
Na |
10 |
11 |
7 |
13 |
12 |
11 |
8 |
5 |
9 |
10 |
|
|
Rs |
9 |
10 |
7 |
10 |
11 |
10 |
6 |
5 |
8 |
8 |
|||
|
uH |
0.819 |
0.854 |
0.797 |
0.840 |
0.874 |
0.851 |
0.307 |
0.634 |
0.805 |
0.753 |
|||
|
BsD |
158 |
Na |
11 |
10 |
9 |
11 |
12 |
10 |
11 |
7 |
13 |
10 |
|
|
Rs |
10 |
8 |
8 |
9 |
11 |
9 |
8 |
5 |
9 |
9 |
|||
|
uH |
0.866 |
0.817 |
0.766 |
0.841 |
0.871 |
0.798 |
0.375 |
0.592 |
0.788 |
0.746 |
|||
|
Mosquitoes |
TMq |
228 |
Na |
13 |
12 |
8 |
11 |
12 |
10 |
6 |
8 |
18 |
11 |
|
Rs |
12 |
12 |
8 |
11 |
12 |
10 |
6 |
7 |
16 |
10 |
|||
|
uH |
0.88 |
0.874 |
0.810 |
0.849 |
0.833 |
0.779 |
0.365 |
0.712 |
0.869 |
0.77 |
|||
|
MqM |
130 |
Na |
12 |
11 |
7 |
9 |
12 |
9 |
6 |
6 |
15 |
10 |
|
|
Rs |
10 |
11 |
7 |
8 |
10 |
8 |
6 |
5 |
13 |
9 |
|||
|
uH |
0.851 |
0.870 |
0.780 |
0.845 |
0.845 |
0.753 |
0.462 |
0.713 |
0.848 |
0.77 |
|||
|
MqN |
98 |
Na |
9 |
7 |
7 |
8 |
8 |
8 |
3 |
6 |
14 |
8 |
|
|
Rs |
9 |
7 |
7 |
8 |
8 |
8 |
3 |
6 |
11 |
7 |
|||
|
uH |
0.862 |
0.815 |
0.760 |
0.814 |
0.816 |
0.747 |
0.191 |
0.718 |
0.837 |
0.73 |
|||
|
MqW |
86 |
Na |
11 |
11 |
5 |
9 |
12 |
9 |
6 |
7 |
8 |
9 |
|
|
Rs |
10 |
11 |
5 |
9 |
11 |
9 |
6 |
6 |
8 |
8 |
|||
|
uH |
0.882 |
0.904 |
0.741 |
0.823 |
0.846 |
0.774 |
0.428 |
0.717 |
0.808 |
0.769 |
|||
|
MqD |
142 |
Na |
10 |
7 |
8 |
9 |
10 |
8 |
4 |
5 |
17 |
9 |
|
|
Rs |
10 |
7 |
8 |
9 |
10 |
8 |
4 |
5 |
14 |
8 |
|||
| uH | 0.873 | 0.796 | 0.834 | 0.826 | 0.825 | 0.788 | 0.300 | 0.713 | 0.873 | 0.758 |
n: sample size; TBs: total of blood samples; BsM: Blood samples from Miyobo; BsN: Blood samples from Ngonamanga; BsW: Blood samples wet season; BsD: Blood samples dry season; TMq: total mosquitoes; MqM: Mosquitoes from Miyobo; MqN: Mosquitoes from Ngonamanga; MqW: Mosquitoes wet season; MqD: Mosquitoes dry season; Na: number of observed alleles; Rs: allelic richness; uH: unbiased estimation of genetic diversity.
MOI varied between 1.62 (Miyobo, dry season, mosquitoes) and 2.25 (Ngonamanga, dry season, humans) and tended to be higher in humans than in mosquitoes (2.09 versus 1.80, respectively). When values are compared between villages without season distinction, mean MOI in humans was slightly higher in Ngonamanga (2.07) than in Miyobo (2.11), and conversely in mosquitoes; 1.69 and 1.91, Ngonamanga and Miyobo respectively. No significant genetic differentiation was observed among all study groups.
Drug resistant associated genes
SNPs: Pfcrt, Pfmdr1, Pfdhfr and Pfdhps
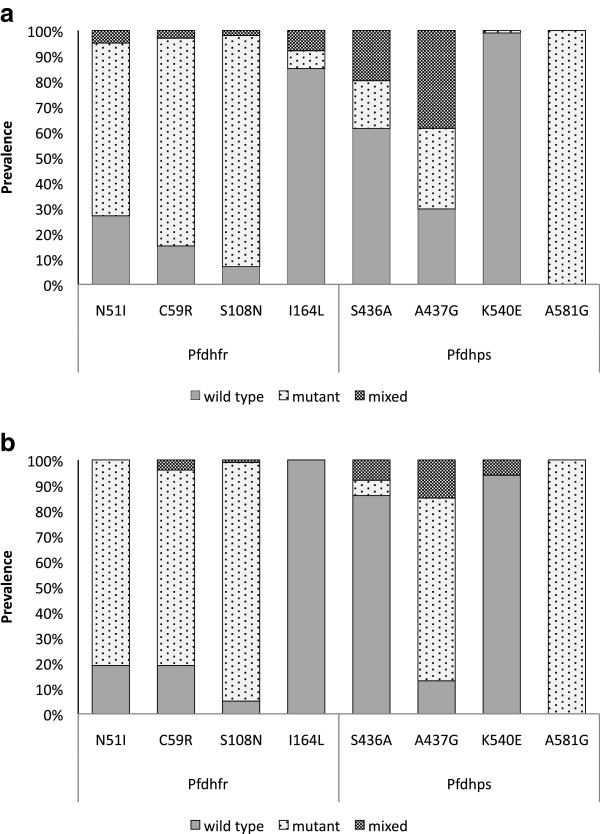
No major differences in the prevalence of mutant alleles were found among villages or seasons for any of the genes. Regarding Pfcrt and Pfmdr1 genes associated with CQ resistance, the prevalence of the Pfcrt mutant alleles (N75E and K76T), present in single or mixed infection, was 56% and 72% in humans and 64% and 54% in mosquitoes; and a much higher prevalence of mutation in codon N86Y (84% and 61%, in humans and mosquitoes, respectively) than in D1246Y (1% in both hosts) was found in Pfmdr1 gene (see Additional file 3). Regarding Pfdhfr gene, mutations N51I, C59R and S108N, associated with PYR resistance, presented prevalence, when in single or mixed infection, of 73%, 85%, 93% in humans and 81%, 81%, 95% in mosquitoes, respectively. The codon I164L was found in very low frequency (15% in humans and 0 in mosquitoes) (see Figure 1 and Additional file 3). While in Miyobo the double mutation (C59R/S108N) was the most prevalent in Ngonamanga most samples contained the triple mutation (N51I/C59R/S108N).
Figure 1.
Total prevalence of mutations in the eight codons of Pfdhfr and Pfdhps genes.Legend: Prevalence of mutations in the Pfdhfr (N51I, C59R, S108N, I164L) and Pfdhps (S436A, A437G, K540, 581G), in single (wild type or mutant) and mixed infections in both seasons and localities in humans (a) and mosquitoes (b).
Mutations A437G and A581G in Pfdhps gene associated with SFX resistance were detected at very high prevalence, the latter reaching 100%, whereas a very low prevalence of codon K540E was found (1% in humans and 6% in mosquitoes only in mixed infections) (see Figure 1 and Additional file 3). Mutation S436A occurred in 38% in humans and 14% in mosquitoes. When comparing the two villages, no major differences were found in mosquitoes, and the prevalence of mutant alleles were: 12% in Ngonamanga and 20% in Miyobo, unlike in humans where the prevalence of this mutation was higher in Ngonamanga (51%) than in Miyobo (31%).
Most samples presented the double mutation (A437G/A581G), but many samples (approximately 38%) containing the triple mutation (A436/G437/G581) were identified in Ngonamanga.
Despite the high prevalence of resistance-associated mutations in Pfdhfr and Pfdhps, no parasites containing the quintuple mutation (N51I/C59R/S108N/A437G/K540E) associated with the clinical failure of SP combination were found.
STR loci flanking Pfdhfr and Pfdhps genes
The analysis using the STR flanking Pfdhfr and Pfdhps genes was only conducted in human isolates, since the amplification rate of these loci was very low in mosquito samples.
Genetic diversity and linkage disequilibrium
The effect of SP selection on the P. falciparum population of Equatorial Guinea was evaluated by examining and comparing the levels of genetic diversity, LD and genetic differentiation between the Pfdhfr and Pfdhps flanking and neutral STR loci. Overall, genetic diversity estimated at neutral loci (He = 0.75; Rs = 14; N =244) was higher than at loci flanking both Pfdhfr (He = 0.15; Rs = 8; N =189) and Pfdhps (He = 0.80; Rs = 12; N = 189) genes (see Tables 2 and 3). In parasites holding Pfdhfr resistance associated alleles, triple mutants showed lower levels of genetic diversity (0.11) when compared to the single (0.36) and double mutants (0.21) (see Table 2). The mean He at three Pfdhfr loci was 0.22, which was much lower when compared to the mean He at 9 neutral loci (0.74) (see Table 2).
Table 2.
Statistics of the 15 STR loci of Plasmodium falciparum-positive individuals: mutants to PYR
|
Microsatellites |
Sampled populations - Miyobo |
Sampled populations - Ngonamanga |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
Single mutant |
Double mutant |
Triple mutant |
All samples |
Triple mutant |
||||||||
| |
( N =10) |
(N=37) |
(N=17) |
(N=64) |
(N=82) |
||||||||
| Rs | He | Rs | He | Rs | He | Rs | He | FST | P | Rs | He | ||
|
Loci flanking dhfr gene |
Dhfr 0.3 |
3 |
0.51 |
3 |
0.32 |
2 |
0.15 |
3 |
0.33 |
0.02 |
NS |
7 |
0.26 |
|
Dhfr 4.4 |
1 |
0.00 |
2 |
0.14 |
2 |
0.17 |
2 |
0.10 |
-0.04 |
NS |
3 |
0.10 |
|
|
Dhfr 5.3 |
3 |
0.56 |
2 |
0.18 |
1 |
0.00 |
2 |
0.24 |
0.10 |
0.03 |
3 |
0.08 |
|
| All loci |
2 |
0.36 |
2 |
0.21 |
2 |
0.11 |
2 |
0.22 |
0.03 |
0.05 |
4 |
0.15 |
|
|
Neutral loci |
TA1 |
4 |
0.82 |
5 |
0.83 |
6 |
0.89 |
6 |
0.85 |
0.04 |
0.04 |
12 |
0.87 |
| TA102 |
4 |
0.64 |
7 |
0.88 |
5 |
0.84 |
6 |
0.79 |
0.03 |
NS |
8 |
0.78 |
|
| ARA2 |
4 |
0.69 |
5 |
0.77 |
6 |
0.79 |
5 |
0.75 |
-0.03 |
NS |
9 |
0.81 |
|
| TA87 |
4 |
0.87 |
6 |
0.85 |
5 |
0.81 |
6 |
0.84 |
0.01 |
NS |
9 |
0.84 |
|
| PfPK2 |
5 |
0.93 |
6 |
0.86 |
5 |
0.85 |
6 |
0.88 |
-0.03 |
NS |
12 |
0.87 |
|
| TA81 |
4 |
0.87 |
5 |
0.81 |
4 |
0.71 |
5 |
0.80 |
-0.04 |
NS |
10 |
0.87 |
|
| TA42 |
3 |
0.60 |
2 |
0.11 |
2 |
0.28 |
2 |
0.32 |
0.06 |
NS |
8 |
0.36 |
|
| Pfg377 |
3 |
0.67 |
3 |
0.56 |
3 |
0.65 |
3 |
0.62 |
-0.04 |
NS |
7 |
0.55 |
|
| TA109 |
5 |
0.86 |
5 |
0.80 |
4 |
0.79 |
5 |
0.82 |
0.04 |
0.03 |
8 |
0.75 |
|
| All loci | 4 | 0.77 | 5 | 0.72 | 4 | 0.73 | 5 | 0.74 | <0.01 | NS | 9 | 0.74 | |
N: number of isolates genotyped, He: expected heterozygosity; Rs: allelic richness. All loci: mean over loci Rs and He and global Fst over loci as calculated by FSTAT. P:P-values of permutation tests to assess significance of Fst values. NS: non-significant (P>0.05).
Table 3.
Statistics of the 15 STR loci of Plasmodium falciparum-positive individuals: mutants to SFX
|
Microsatellites |
Sampled populations - Miyobo |
Sampled populations - Ngonamanga |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
Single mutant |
Double mutant |
All samples ( N =60) |
Double mutant |
Triple mutant |
All samples (N=85) |
|||||||||||
| |
|
|
|
(N=63) |
(N=22) |
|
|||||||||||
| |
(N=17) |
( N =43) |
|
||||||||||||||
| Rs | He | Rs | He | Rs | He | FST | P | Rs | He | Rs | He | Rs | He | FST | P | ||
|
Loci flanking dhps gene |
Dhps 0.8 |
4 |
0.60 |
5 |
0.75 |
5 |
0.67 |
0.02 |
NS |
9 |
0.81 |
5 |
0.64 |
9 |
0.73 |
0.06 |
0.01 |
|
Dhps 4.3 |
6 |
0.83 |
5 |
0.77 |
6 |
0.80 |
<0.01 |
NS |
7 |
0.78 |
4 |
0.71 |
7 |
0.75 |
0.13 |
<0.01 |
|
|
Dhps 7.7 |
6 |
0.79 |
7 |
0.84 |
8 |
0.82 |
0.09 |
<0.01 |
10 |
0.85 |
6 |
0.67 |
10 |
0.76 |
0.14 |
<0.01 |
|
| All loci |
5 |
0.74 |
6 |
0.79 |
6 |
0.76 |
0.04 |
<0.01 |
9 |
0.81 |
5 |
0.67 |
9 |
0.74 |
0.11 |
<0.01 |
|
|
Neutral loci |
TA1 |
5 |
0.79 |
8 |
0.87 |
8 |
0.83 |
0.04 |
NS |
9 |
086 |
8 |
0.88 |
9 |
0.87 |
<0.01 |
NS |
| TA102 |
5 |
0.78 |
9 |
0.87 |
9 |
0.83 |
0.04 |
0.01 |
6 |
0.75 |
7 |
0.86 |
7 |
0.80 |
<-0.01 |
NS |
|
| ARA2 |
6 |
0.79 |
6 |
0.69 |
7 |
0.74 |
0.06 |
<0.01 |
7 |
0.80 |
6 |
0.82 |
7 |
0.81 |
-0.01 |
NS |
|
| TA87 |
5 |
0.79 |
8 |
0.84 |
7 |
0.82 |
0.05 |
0.02 |
8 |
0.83 |
7 |
0.87 |
8 |
0.85 |
0.02 |
NS |
|
| PfPK2 |
5 |
0.83 |
8 |
0.80 |
8 |
0.81 |
0.11 |
0.02 |
10 |
0.87 |
8 |
0.87 |
11 |
0.87 |
0.02 |
<0.05 |
|
| TA81 |
5 |
0.77 |
6 |
0.78 |
7 |
0.78 |
-0.02 |
NS |
9 |
0.88 |
8 |
0.83 |
9 |
0.85 |
<0.01 |
NS |
|
| TA42 |
1 |
0.00 |
4 |
0.29 |
3 |
0.14 |
0.03 |
NS |
5 |
0.44 |
3 |
0.18 |
5 |
0.31 |
0.05 |
0.04 |
|
| Pfg377 |
3 |
0.56 |
4 |
0.59 |
3 |
0.57 |
<-0.01 |
NS |
5 |
0.59 |
3 |
0.40 |
5 |
0.50 |
0.01 |
NS |
|
| TA109 |
6 |
0.86 |
6 |
0.81 |
6 |
0.83 |
-0.02 |
NS |
5 |
0.72 |
5 |
0.60 |
6 |
0.66 |
0.16 |
<0.01 |
|
| All loci | 5 | 0.69 | 6 | 0.73 | 6 | 0.71 | 0.03 | <0.01 | 7 | 0.75 | 6 | 0.70 | 7 | 0.72 | 0.03 | 0.01 | |
N: number of isolates genotyped, He: expected heterozygosity; Rs: allelic richness. All loci: mean over loci Rs and He and global Fst over loci as calculated by FSTAT. P:P-values of permutation tests to assess significance of Fst values. NS: non-significant (P>0.05).
The reduction in the genetic diversity is not so marked in Pfdhps as in Pfdhfr. When double mutants and triple mutants are compared, there is a slight decrease in genetic diversity (He=0.81 for double versus He=0.67 for triple mutants) but values are still similar and high (see Table 3). Statistical tests for LD were conducted for all pairs of flanking STR on each of the mutant groups – single, double and triple mutants (105 possible tests for Pfdhfr and 316 for Pfdhps). Only two associations showed significant results (p <0.05), after Bonferroni’s correction was applied, in the Pfdhps double mutants group, involving loci 0.8 kb/4.3 kb and 4.3 kb/7.7 kb. No significant pairwise association was found involving the Pfdhfr gene.
Pfdhfr and Pfdhps haplotype characterization
Only samples with single infections and successful amplification of all loci were used for the haplotype characterization. Thus, haplotypes were reconstructed in 57 out of 298 human isolates genotyped for Pfdhfr and in 35 out of 296 human isolates genotyped for Pfdhps. For the Pfdhfr gene, nine distinct haplotypes were found (see Additional file 4). The haplotype H9, an exact match of the P. falciparum K1 strain used as a control (double mutation C59R/S108N and allele sizes of 113 bp, 183 bp and 210 bp to the 0.3 kb, 4.4 kb and 5.3 kb loci, respectively), was found in 10 samples from Miyobo. Most frequent haplotypes in 53 out of 57 samples (H1, H3, H5, H8) also matched STR sizes in K1 strain.
The majority of samples from Ngonamanga (97%) showed the triple-mutant IRNI (51I:59R:108N:164I – mutated codons appear underlined), while in Miyobo the most prevalent haplotype was the double-mutant NRNI (51N: 59R:108N:164I) (43%) followed by the triple-mutant IRNI with 26% (see Additional file 4).
Regarding the Pfdhps gene, 25 distinct haplotypes were found (see Additional file 5); 16 in Miyobo and nine in Ngonamanga, only one shared between the two villages. None of the haplotypes found matches with K1 strain (single mutation A437G and allele sizes of 131 bp, 103 bp and 108 bp to the 0.8 kb, 4.3 kb and 7.7 kb loci, respectively). The haplotypes found for the Pfdhps gene have multiple independent lineages since the majority of the haplotypes were unique. Nevertheless, the most prevalent haplotype in Ngonamanga was the triple-mutant AGKG (436A:437G:540K:581G) with 38%, whilst in Miyobo it was the double-mutant SGKG (436S:437G:540 K:581G) (63%).
Discussion
Malaria still is a major public health concern in Equatorial Guinea, especially in the mainland. In order to contribute to the update of the malaria situation in this area, a combined set of blood and mosquito samples from the same locations were analysed to characterize the genetic diversity of circulating populations of Plasmodium spp and especially of P. falciparum, in both hosts.
Plasmodium species diversity
This study presents a much higher prevalence of Plasmodium infection in mainland Equatorial Guinea (87%) than the one reported for the Insular Region in 2005 (32%) [33]. This difference is likely due to the fact that most malaria control activities have been deployed on the island of Bioko where the capital, Malabo is located. In 2004, the first stage of the project “The Bioko Island Malaria Control Project (BIMCP)” was launched and initial reports stated a significant decrease in the prevalence of infection, achieving an overall malaria prevalence of 18% in 2008 [10,34]. In the present study, although P. falciparum infections were the most frequent, P. vivax infections were detected for the first time both in humans and mosquitoes, which means that active transmission of this species not previously reported in this area is occurring. The apparent higher presence of P. vivax in mosquitoes might be due to its higher visibility in the vector, since in the human host this parasite can form dormant forms in the liver – hypnozoites - and go unnoticed, as discussed in [4].
Regarding mixed infections, P. falciparum and P. malariae are also associated in mainland Equatorial Guinea, as has been reported in other sub-Saharan countries [3,35-39]. This association was observed both in humans and mosquitoes, which suggests that no differing patterns of Plasmodium species association in the two hosts occurs as has formerly been reported in Guinea Bissau [3].
Plasmodium falciparum genetic diversity
Concerning P. falciparum genetic diversity, the analysis of both Pfmsp2 and neutral STR in humans showed similar levels of allelic diversity and MOI in both villages and seasons. No reduction of genotype diversity or MOI was observed with the decline of transmission, as seen in areas of lower endemicity, such as Sudan [40] or in areas with marked differences in malaria endemicity [41]. However, analogous results were obtained by Cano et al[42] in a study conducted on the island of Annobon, part of Equatorial Guinea Insular Region.
In mosquitoes, this analysis was only possible with the neutral STR and the results confirmed those obtained in humans, i. e., high levels of genetic diversity and no significant genetic differentiation between geographic locations, despite their different ecological differences or seasons. This is a sign of high malaria endemicity in mainland Equatorial Guinea and the similarity between population genetic structures is concordant with other studies in African highly malaria-endemic countries [8,40]. No significant genetic differentiation was seen between hosts, when comparisons between human blood samples and mosquitoes were made using neutral STR data. The most common alleles are found in both humans and mosquitoes, which may indicate consistency in the parasite populations that are being transmitted. Nevertheless, MOI values were higher in humans than in mosquitoes. As Arez et al[3] observed, a higher proportion of single-genotype infections in mosquitoes could point to a limited genetic diversity of the inocula and a high genetic diversity in humans resulting from superinfection phenomena.
Anti-malarial resistance evolution
The prevalence of the main point mutations associated with CQ resistance (75E and 76 T of Pfcrt gene and 86Y of Pfmdr1 gene) was nearly 71%. Although the mutation 1246Y in the Pfmdr1 gene has also been associated with reduced susceptibility to CQ [43], a very low frequency of this mutation was found in Equatorial Guinea (1%).
Nowadays, after the introduction of artemisinin-based combination therapy (ACT), decrease in prevalence of mutations associated with CQ resistance might be expected, due to the absence of drug pressure, as reported in Malawi, China, Kenya and Angola [44-47]. However, a recent study conducted in Equatorial Guinea [48] found higher prevalence of mutation in Pfcrt (codon 76) and in Pfmdr1 (codon 1246) (98% and 96%, respectively), than those found in this study (72% for Pfcrt codon 76 and 1% for Pfmdr1 codon 1246) in isolates collected in 2005, when CQ was still in use in mainland Equatorial Guinea. The increasing of these and other point mutants might be a result of selective pressure by AS-AQ combination, since AQ is a close Mannich base analogue of CQ, promoting the maintenance of CQ-resistant isolates with the mutant Pfcrt and Pfmdr1 genotypes. On the other hand, another possibility is the continuous use of CQ despite national therapeutic guidelines [49].
In Equatorial Guinea, SP has been used as a second-line therapy for many years and lately, though less intensely, as a first-line in combination with artemisinin derivatives and it is used in preventive chemotherapy in pregnancy. Although the failure rate of this combination has not suffered major variations since 1992, and in the late 1990s was still 10% [15], it was expected that the continuous use of this drug would rapidly lead to an increase of resistance levels as had happened in other countries such as Kenya [50] and Tanzania [51].
In fact, a high prevalence of mutation in genes associated with resistance to the SP combination (~70%) was observed in this study. PYR resistance seemed to be well established in mainland Equatorial Guinea and nearly 80% of parasite populations presented the triple mutant N51I/C59R/S108N in the Pfdhfr gene, both in humans and mosquitoes, as seen in other nearby countries such as Cameroon [52], Gabon [53] and São Tomé and Principe [27].
Regarding SFX resistance, a high prevalence of the mutation A437G in Pfdhps was detected. However the mutation K540E was practically non-existent, which is usual in West Africa [28]. The prevalence of S436A mutation was low, contrary to data from the neighbouring country Gabon, where this was the most frequent Pfdhps polymorphism [54]. The mutations S436A and A581G are less studied due to their low prevalence in some African countries, and the lack of knowledge of their role in treatment failure [55]. However, the prevalence of A581G mutation in this study reached 100%. Other recent studies conducted in different African countries showed an increase of the prevalence of A581G, during the last years [53,56].
The quintuple mutant, associated with SP clinical failure [57,58] and resulting from the combination of the Pfdhfr triple mutant N51I/C59R/S108N (linked to resistance to PYR) with the Pfdhps double mutant A437G/K540E (linked to resistance to SFX), was not detected since no samples containing the latter were found. No major differences in the prevalence of mutation between parasites in humans and mosquitoes occurred.
Analysis on Pfdhfr flanking STRs showed that the majority of haplotypes found were associated with triple mutants IRNI, especially in Ngonamanga, while the majority of isolates harboured double mutants NRNI in Miyobo. These two haplotypes have already been reported in Ghana [59] and the triple mutant IRNI was also found in Southeast Asia [60]. The majority of the haplotypes seems to have a single origin. In fact, the haplotypes found were very similar among them, with the majority of them corresponding to the H3 haplotype. This haplotype has arisen from H9 haplotype double mutant through an additional mutation occurring at position 59 of the Pfdhfr gene. Both H3 and H9 haplotypes share the same microsatellite profile.
The results of the present study suggest that PYR resistance was firstly established in Ngonamanga, probably due to the fact that Miyobo is more isolated and the introduction of the drug may have occurred later. It is likely that SP combination has been introduced first in Malabo (the capital of the country), and then its utilization was spread all over the country. Ngonamanga, being a coastal area (closer to the capital), may have started to use this drug earlier, and therefore to develop resistance sooner. The process of the addition of a single mutation in Pfdhfr alleles to double mutants, originating a high prevalence of triple mutants [61] was still occurring in Miyobo. The most common haplotype 113/183/210 with the triple mutant IRNI, already described in Senegal [26], should be related to the 109/183/210 background, found in Tanzania, South Africa, Southeast Africa [25] and, most recently, in Kenya [50].
It was expected that the extensive use of SP would lead to a rapid increase in resistance levels, leaving signatures of drug selective pressure, such as a reduction in genetic diversity around Pfdhps and Pfdhfr due to selective sweep; an increased genetic differentiation at the loci under selection; and, a significant LD between loci flanking Pfdhps and Pfdhfr genes [62].
Indeed, the reduction in heterozigosity in the loci flanking Pfdhfr gene with regard to the mean of heterozigosity in the neutral loci indicates that this gene has undergone strong selection in Equatorial Guinea. The higher mean of He around double mutant than the mean of He around triple mutant is consistent with a model of positive directional selection. The Fst values at STR loci linked to Pfdhfr gene were higher when compared with mean Fst at neutral loci, which supports this hypothesis. However, no significant LD values were found between flanking genes of interest.
According to the results herein presented, SFX resistance seems to have appeared more recently than PYR resistance in mainland Equatorial Guinea. Indeed, only mutations at codons A437G and A581G, from the Pfdhps polymorphic sites surveyed showed high prevalence. Point mutations at S436A and K540E codons were rarely seen. A wide diversity of haplotypes was detected, being the majority unique haplotypes, which is consistent with independent origins for those alleles. The most prevalent haplotype match with AGK1/SGK1 lineages of West African origin and a few others (436A:437G:540K/ 436S:437G:540 K) with probable independent origin. The double mutant lineage identified as SGE1 (436S:437G:540E), originated in East Africa [28], was not detected in this study. As occurred with PYR resistance, the resistance to SFX seems to have been established earlier in Ngonamanga, where the prevalence of triple mutants is higher than in Miyobo.
When double mutants (He=0.81) were compared with triple mutants (He=0.67), a reduction in the heterozigosity was seen. However the values found are remarkably higher when comparing to those found for the PYR resistance (mean He=0.22). These differences may be due to the presence of multiple lineages occurring within individual populations. Also, significant LD values were found between flanking Pfdhps gene, involving loci 0.8 kb/4.3 kb and 4.3 kb/7.7Kb. Overall, these results might be suggestive of soft selective sweep, where multiple lineages are superimposed within a single population causing higher He values than in populations where a single lineage is present [63].
The results gathered in this study suggests that the PYR resistance has been established for a while in mainland Equatorial Guinea leaving selection signatures as the decrease in genetic diversity and an increased genetic differentiation at the loci around Pfdhfr gene. In addition, the impact on genetic diversity was less clear at the loci flanking Pfdhps, with only evidence of a soft selective sweep effect. This agrees with a more recent introduction of resistance to SFX in Equatorial Guinea, which is in agreement with results obtained in a recent study [64].
Conclusions
CQ is no longer recommended for malaria treatment in Equatorial Guinea but SP remains in use in combination with artesunate and is the only drug recommended for intermittent preventive therapy in pregnancy [65]. Prevalence of infection in the mainland region, where most of the country’s population live, remains high despite the efforts undertaken to control malaria transmission mainly on the island of Bioko [10,34]. A close and continuous monitoring of point mutations frequency in the two genes associated with SP resistance, Pfdhfr and Pfdhps, is essential since there is the danger of an eventual reduction in the efficacy of SP combined therapy.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CM and VG carried out the laboratory analysis. JC carried out the sampling and field data collection. PS, PB, BdS, VER, AB and JC participated in the analysis and interpretation of data and helped to draft the manuscript. CM and APA drafted the paper. APA designed the study and participated in the analysis and interpretation of data. All authors read and approved the final manuscript.
Supplementary Material
Prevalence of Plasmodium infections in humans, in two villages of mainland Equatorial Guinea. n: sample size; F: P. falciparum; M: P. malariae; O: P. ovale; V: P. vivax; F+M: mixed infection by P. falciparum and P. malariae; F+O: mixed infection by P. falciparum and P. ovale; F+V: mixed infection by P. falciparum and P. vivax; F+M+O: mixed infection by P. falciparum, P. malariae and P. ovale; F+M+V: mixed infection by P. falciparum, P. malariae and P. vivax.
Prevalence of Plasmodium infections in mosquitoes, in two villages of mainland Equatorial Guinea. n: sample size; F: P. falciparum; M: P. malariae; O: P. ovale; V: P. vivax; F+M: mixed infection by P. falciparum and P. malariae; F+O: mixed infection by P. falciparum and P. ovale; F+V: mixed infection by P. falciparum and P. vivax; F+M+O: mixed infection by P. falciparum, P. malariae and P. ovale; F+M+V: mixed infection by P. falciparum, P. malariae and P. vivax.
Characterization of mutations in Pfcrt, Pfmdr1, Pfdhps and Pfdhfr genes, in humans and mosquitoes.
Pfdhfr point mutations and their respective STR haplotypes in allele size.
Pfdhps point mutations and their respective STR haplotypes in allele size.
Contributor Information
Cristina Mendes, Email: cristina.mendes@ihmt.unl.pt.
Patrícia Salgueiro, Email: psalgueiro@ihmt.unl.pt.
Vicenta Gonzalez, Email: vgonzalez@isciii.es.
Pedro Berzosa, Email: pberzosa@isciii.es.
Agustin Benito, Email: abenito@isciii.es.
Virgílio E do Rosário, Email: virgilio.rosario@ihmt.unl.pt.
Bruno de Sousa, Email: bruno.desousa@ihmt.unl.pt.
Jorge Cano, Email: jcano@isciii.es.
Ana Paula Arez, Email: aparez@ihmt.unl.pt.
Acknowledgements
We thank all families who participated in this study. We thank researchers and technicians from the National Malaria Control Program of the Equatorial Guinea’s Ministry of Health and Social Welfare and “Centro de Referencia para el Control de Endemias” (Instituto de Salud Carlos III, Equatorial Guinea).
This study was supported by PEst-OE/SAU/LA0018/2011 - Proj. Estratégico LA0018 2011/2012 (http://cmdt.ihmt.unl.pt/index.php/pt/) and PTDC/SAU-EPI/113326/2009, “Fundacão para a Ciência e Tecnologia/Ministério da Educação e Ciência”, FCT/MEC (http://alfa.fct.mctes.pt/index.phtml.pt), Portugal and by “Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación”, Madrid, Spain. C. Mendes and P. Salgueiro hold FCT grants (SRFH/BD/41473/2007 and SFRH/BPD/72532/2010, respectively).
References
- WHO. World malaria report 2011. Geneva: World Health Organization; 2011. http://www.who.int/malaria/world_malaria_report_2011/9789241564403_eng.pdf. [Google Scholar]
- Bruce MC, Donnelly CA, Alpers MP, Galinski MR, Barnwell JW, Walliker D, Day KP. Cross-species interactions between malaria parasites in humans. Science. 2000;287:845–848. doi: 10.1126/science.287.5454.845. [DOI] [PubMed] [Google Scholar]
- Arez AP, Pinto J, Palsson K, Snounou G, Jaensen TGT, Rosário V. Transmission of mixed Plasmodium species and Plasmodium falciparum genotypes. Am J Trop Med Hyg. 2003;68:161–198. [PubMed] [Google Scholar]
- Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, do Rosário VE, Benito A, Berzosa P, Arez AP. Duffy negative antigen is no longer a barrier to Plasmodium vivax – molecular evidences from the African West Coast (Angola and Equatorial Guinea) PLoS Negl Trop Dis. 2011;5:e1192. doi: 10.1371/journal.pntd.0001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mharakurwa S, Kumwenda T, Mkulama MAP, Musapa M, Chishimba S, Shiff CJ, Sullivan DJ, Thuma PE, Liu K, Agre P. Malaria antifolate resistance with contrasting Plasmodium falciparum dihydrofolate reductase (DHFR) polymorphisms in humans and Anopheles mosquitoes. Proc Natl Acad Sci USA. 2011;108:18796–18801. doi: 10.1073/pnas.1116162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arez AP, do Rosário VE. The relevance of molecular markers in the analysis of malaria parasite populations. Transbound Emerg Dis. 2008;55:226–232. doi: 10.1111/j.1865-1682.2008.01025.x. [DOI] [PubMed] [Google Scholar]
- Kiwanuka GN. Genetic diversity in Plasmodium falciparum merozoite surface protein 1 and 2 coding genes and its implications in malaria epidemiology: a review of published studies from 1997-2007. J Vector Borne Dis. 2009;46:1–12. [PubMed] [Google Scholar]
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite P. falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- Pardo G, Descalzo MA, Molina L, Custodio E, Lwanga M, Mangue C, Obono J, Nchama A, Roche J, Benito A, Cano J. Impact of different strategies to control Plasmodium infection and anaemia on the island of Bioko (Equatorial Guinea) Malar J. 2006;5:10. doi: 10.1186/1475-2875-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt I, Schwabe C, Benavente L, Torrez M, Ridl FC, Segura JL, Ehmer P, Nchama GN. Marked increase in child survival after four years of intensive malaria control. Am J Trop Med Hyg. 2009;80:882–888. [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naudé B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falciparum digestive vacuole transmembrane protein Pfcrt and evidence for their role in Chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/S1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DR, Wang P, Read M, Watkins WM, Sims PF, Hyde JE. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur J Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- Roche J, Guerra-Neira A, Raso J, Benito A. Surveillance of in vivo resistance of Plasmodium falciparum to antimalarial drugs from 1992 to 1999 in Malabo (Equatorial Guinea) Am J Trop Med Hyg. 2003;68:598–601. doi: 10.4269/ajtmh.2003.68.598. [DOI] [PubMed] [Google Scholar]
- Charle P, Berzosa P, Descalzo MA, de Lucio A, Raso J, Obono J, Lwanga M, Nlang N, Nchama A, Mangue C, Micha A, Nsee N, Mesie R, Benito A, Roche J. Efficacy of artesunate + sulphadoxine-pyrimethamine (AS + SP) and amodiaquine + sulphadoxine-pyrimethamine (AQ+ SP) for uncomplicated falciparum malaria in Equatorial Guinea (Central Africa) J Trop Med. 2009;2009:781865. doi: 10.1155/2009/781865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijk AMV, Hill J, Alegana VA, Kirui V, Gething PW, Kuile FO, Snow RW. Coverage of malaria protection in pregnant women in sub-Saharan Africa: a synthesis and analysis of national survey data. Lancet Infect Dis. 2011;11:190–207. doi: 10.1016/S1473-3099(10)70295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- Arez AP, Lopes D, Pinto J, Franco AS, Snounou G, do Rosário VE. Plasmodium sp.: Optimal protocols for PCR detection of low parasite numbers from mosquito (Anopheles sp.) samples. Exp Parasitol. 2000;94:269–272. doi: 10.1006/expr.2000.4496. [DOI] [PubMed] [Google Scholar]
- Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–292. doi: 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/S0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- PCR-ASRA Protocols for Plasmodium falciparum Drug-Resistance Mutation Analysis. 2002. http://medschool.umaryland.edu/cvd/plowe.html.
- Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers characterization of P. falciparum from finger-prick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/S0031182099004552. [DOI] [PubMed] [Google Scholar]
- Conway DJ, Machado RL, Singh B, Dessert P, Mikes ZS, Povoa MM, Oduola AM, Roper C. Extreme geographical fixation of variation in the Plasmodium falciparum gamete surface protein gene Pfs48/45 compared with microsatellite loci. Mol Biochem Parasitol. 2001;115:145–156. doi: 10.1016/S0166-6851(01)00278-X. [DOI] [PubMed] [Google Scholar]
- Roper C, Pearce R, Bredenkamp B, Gumede J, Drakeley C, Mosha F, Chandramohan D, Sharp B. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet. 2003;361:1174–1181. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- Ndiaye D, Daily JP, Sarr O, Ndir O, Gaye O, Mboup S, Roper C, Wirth DF. Defining the origin of Plasmodium falciparum resistant dhfr isolates in Senegal. Acta Trop. 2006;99:106–111. doi: 10.1016/j.actatropica.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgueiro P, Vicente JL, Ferreira C, Teófilo V, Galvão A, Do Rosário VE, Cravo P, Pinto J. Tracing the origins and signatures of selection of antifolate resistance in island populations of Plasmodium falciparum. BMC Infect Dis. 2010;10:163. doi: 10.1186/1471-2334-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RJ, Pota H, Evehe MS, Bâ e-H, Mombo-Ngoma G, Malisa AL, Ord R, Inojosa W, Matondo A, Diallo DA, Mbacham W, van den Broek IV, Swarthout TD, Getachew A, Dejene S, Grobusch MP, Njie F, Dunyo S, Kweku M, Owusu-Agyei S, Chandramohan D, Bonnet M, Guthmann JP, Clarke S, Barnes KI, Streat E, Katokele ST, Uusiku P, Agboghoroma CO, Elegba OY, Cissé B, A-Elbasit IE, Giha HA, Kachur SP, Lynch C, Rwakimari JB, Chanda P, Hawela M, Sharp B, Naidoo I, Roper C. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009;6:e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitenbroek DG. SISA Binomial. 1997. http://www.quantitativeskills.com/sisa/
- Goudet J. FSTAT (version 1.2): A program to estimate and test gene diversities and fixation indices. J Hered. 2002;86:485–486. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenism. J Hered. 1995;86:248–249. [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.2307/2409177. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt I, Torrez M, Schwabe C, Benavente L, Seocharan I, Jituboh D, Nseng G, Sharp B. Factors influencing the effectiveness of malaria control in Bioko Island, Equatorial Guinea. Am J Trop Med Hyg. 2007;76:1027–1032. [PMC free article] [PubMed] [Google Scholar]
- Overgaard HJ, Reddy VP, Abaga S, Matias A, Reddy MR, Kulkarni V, Schwabe C, Segura L, Kleinschmidt I, Slotman MA. Malaria transmission after five years of vector control on Bioko Island, Equatorial Guinea. Parasit Vectors. 2012;5:253. doi: 10.1186/1756-3305-5-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineaux L, Storey J, Cohen JE, Thomas A. A longitudinal study of human malaria in the West African Savanna in the absence of control measures: relationships between different Plasmodium species, in particular P. falciparum and P. malariae. Am J Trop Med Hyg. 1980;29:725–737. doi: 10.4269/ajtmh.1980.29.725. [DOI] [PubMed] [Google Scholar]
- Richie TL. Interactions between malaria parasites infecting the same vertebrate host. Parasitology. 1988;96:607–639. doi: 10.1017/S0031182000080227. [DOI] [PubMed] [Google Scholar]
- Boudin C, Robert V, Verhave JP, Carnevale P, Ambroise-Thomas P. Plasmodium falciparum and P. malariae epidemiology in a West African village. Bull World Health Organ. 1991;69:199–205. [PMC free article] [PubMed] [Google Scholar]
- Marques PX, Saúte F, Pinto VV, Cardoso S, Pinto J, Alonso PL, do Rosário VE, Arez AP. Plasmodium species mixed infections in two ecologically different areas of Manhiça District, Mozambique. Int J Biol Sci. 2005;1:96–102. doi: 10.7150/ijbs.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce MC, Macheso A, Kelly-Hope LA, Nkhoma S, McConnachie A, Molyneux ME. Effect of transmission setting and mixed species infections on clinical measures of malaria in Malawi. PLoS One. 2008;3:e2775. doi: 10.1371/journal.pone.0002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiker HA, Satti G, Walliker D. Genetic changes in the population of Plasmodium falciparum in a Sudanese village over a three-year period. Am J Trop Med Hyg. 1995;53:7–15. [PubMed] [Google Scholar]
- Mobegi VA, Loua KM, Ahouidi AD, Satoguina J, Nwakanma DC, Amambua-Ngwa A, Conway DJ. Population genetic structure of Plasmodium falciparum across a region of diverse endemicity in West Africa. Malar J. 2012;11:223. doi: 10.1186/1475-2875-11-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano J, Berzosa P, de Lucio A, Descalzo MA, Bobuakasi L, Nzambo S, Ondo M, Buatiche JN, Nseng G, Benito A. Transmission of malaria and genotypic variability of Plasmodium falciparum on the Island of Annobon (Equatorial Guinea) Malar J. 2007;6:141. doi: 10.1186/1475-2875-6-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA. 2009;106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimdé AA, Kouriba B, Taylor TE, Plowe CV. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, Sasi P, Marsh K, Borrmann S, Mackinnon M, Nzila A. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mu J, Li G, Chen P, Guo X, Fu L, Chen L, Su X, Wellems TE. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76 T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People’s Republic of China. Am J Trop Med Hyg. 2005;72:410–414. [PubMed] [Google Scholar]
- Fançony C, Gamboa D, Sebastião Y, Hallett R, Sutherland C, Sousa-Figueiredo JC, Nery SV. Various pfcrt and pfmdr1 genotypes of Plasmodium falciparum cocirculate with P. malariae, P. ovale spp., and P. vivax in Northern Angola. Antimicrob Agents Chemother. 2012;56:5271–5277. doi: 10.1128/AAC.00559-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor A, Toro C, Fernández-Martínez A, Baquero M, Benito A, Berzosa P. Molecular markers in Plasmodium falciparum linked to resistance to anti-malarial drugs in samples imported from Africa over an eight-year period (2002-2010): impact of the introduction of artemisinin combination therapy. Malar J. 2012;11:100. doi: 10.1186/1475-2875-11-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowe CV, Wellems TE. Molecular approaches to the spreading problem of drug resistant malaria. Adv Exp Med Biol. 1995;390:197–209. doi: 10.1007/978-1-4757-9203-4_17. [DOI] [PubMed] [Google Scholar]
- McCollum AM, Poe AC, Hamel M, Huber C, Zhou Z, Shi YP, Ouma P, Vulule J, Bloland P, Slutsker L, Barnwell JW, Udhayakumar V, Escalante AA. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J Infect Dis. 2006;194:189–197. doi: 10.1086/504687. [DOI] [PubMed] [Google Scholar]
- Trigg JK, Mbwana H, Chambo O, Hills E, Watkins W, Curtis CF. Resistance to pyrimethamine/sulfadoxine in Plasmodium falciparum in 12 villages in northeast Tanzania and a test of chlorproguanil/dapsone. Acta Trop. 1997;63:185–189. doi: 10.1016/S0001-706X(96)00617-1. [DOI] [PubMed] [Google Scholar]
- Tahar R, Basco LK. Molecular epidemiology of malaria in Cameroon. XXII. Geographic mapping and distribution of Plasmodium falciparum dihydrofolate reductase (dhfr) mutant alleles. Am J Trop Med Hyg. 2006;75:396–401. [PubMed] [Google Scholar]
- Aubouy A, Jafari S, Huart V, Migot-Nabias F, Mayombo J, Durand R, Bakary M, Le Bras J, Deloron P. DHFR and DHPS genotypes of Plasmodium falciparum isolates from Gabon correlate with in vitro activity of pyrimethamine and cycloguanil, but not with sulfadoxine-pyrimethamine treatment efficacy. J Antimicrob Chemother. 2003;52:43–49. doi: 10.1093/jac/dkg294. [DOI] [PubMed] [Google Scholar]
- Mawili-Mboumba DP, Ekala MT, Lekoulou F, Ntoumi F. Molecular analysis of DHFR and DHPS genes in P. falciparum clinical isolates from the Haut-Ogooué region in Gabon. Acta Trop. 2001;78:231–240. doi: 10.1016/S0001-706X(01)00084-5. [DOI] [PubMed] [Google Scholar]
- Alker AP, Kazadi WM, Kutelemeni AK, Bloland PB, Tshefu AK, Meshnick SR. dhfr and dhps genotype and sulfadoxine-pyrimethamine treatment failure in children with falciparum malaria in the Democratic Republic of Congo. Trop Med Int Health. 2008;13:1384–1391. doi: 10.1111/j.1365-3156.2008.02150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MD, Eyase FL, Akala HM, Bedno SA, Prigge ST, Coldren RL, Moss WJ, Waters NC. Increased prevalence of the pfdhfr/phdhps quintuple mutant and rapid emergence of pfdhps resistance mutations at codons 581 and 613 in Kisumu, Kenya. Malar J. 2010;9:338. doi: 10.1186/1475-2875-9-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, Mukadam RA, Rogerson SJ, Lescano AG, Molyneux ME, Winstanley PA, Chimpeni P, Taylor TE, Plowe CV. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- Talisuna AO, Nalunkuma-Kazibwe A, Langi P, Mutabingwa TK, Watkins WW, Van Marck E, Egwang TG, D’Alessandro U. Two mutations in dihydrofolate reductase combined with one in the dihydropteroate synthase gene predict sulphadoxine-pyrimethamine parasitological failure in Ugandan children with uncomplicated falciparum malaria. Infect Genet Evol. 2004;4:321–327. doi: 10.1016/j.meegid.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Alam MT, de Souza DK, Vinayak S, Griffing SM, Poe AC, Duah NO, Ghansah A, Asamoa K, Slutsker L, Wilson MD, Barnwell JW, Udhayakumar V, Koram K. Selective sweeps and genetic lineages of Plasmodium falciparum drug-resistance in Ghana. J Infect Dis. 2011;203:220–227. doi: 10.1093/infdis/jiq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maïga O, Djimdé AA, Hubert V, Renard E, Aubouy A, Kironde F, Nsimba B, Koram K, Doumbo OK, Le Bras J, Clain J. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J Infect Dis. 2007;196:165–172. doi: 10.1086/518512. [DOI] [PubMed] [Google Scholar]
- Malisa AL, Pearce RJ, Abdulla S, Mshinda H, Kachur PS, Bloland P, Roper C. Drug coverage in treatment of malaria and the consequences for resistance evolution - evidence from the use of sulphadoxine/pyrimethamine. Malar J. 2010;9:190. doi: 10.1186/1475-2875-9-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Cornejo OE, Rojas A, Udhayakumar V, Lal AA. Assessing the effect of natural selection in malaria parasites. Trends Parasitol. 2004;20:388–395. doi: 10.1016/j.pt.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Nair S, Nash D, Sudimack D, Jaidee A, Barends M, Uhlemann AC, Krishna S, Nosten F, Anderson TJ. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol Biol Evol. 2007;24:562–573. doi: 10.1093/molbev/msl185. [DOI] [PubMed] [Google Scholar]
- Naidoo I, Roper C. Following the path of most resistance: dhps K540E dispersal in African Plasmodium falciparum. Trends Parasitol. 2010;26:447–456. doi: 10.1016/j.pt.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt I, Sharp B, Benavente LE, Schwabe C, Torrez M, Kuklinski J, Morris N, Raman J, Carter J. Reduction in infection with Plasmodium falciparum one year after the introduction of malaria control interventions on Bioko Island, Equatorial Guinea. Am J Trop Med Hyg. 2006;74:972–978. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prevalence of Plasmodium infections in humans, in two villages of mainland Equatorial Guinea. n: sample size; F: P. falciparum; M: P. malariae; O: P. ovale; V: P. vivax; F+M: mixed infection by P. falciparum and P. malariae; F+O: mixed infection by P. falciparum and P. ovale; F+V: mixed infection by P. falciparum and P. vivax; F+M+O: mixed infection by P. falciparum, P. malariae and P. ovale; F+M+V: mixed infection by P. falciparum, P. malariae and P. vivax.
Prevalence of Plasmodium infections in mosquitoes, in two villages of mainland Equatorial Guinea. n: sample size; F: P. falciparum; M: P. malariae; O: P. ovale; V: P. vivax; F+M: mixed infection by P. falciparum and P. malariae; F+O: mixed infection by P. falciparum and P. ovale; F+V: mixed infection by P. falciparum and P. vivax; F+M+O: mixed infection by P. falciparum, P. malariae and P. ovale; F+M+V: mixed infection by P. falciparum, P. malariae and P. vivax.
Characterization of mutations in Pfcrt, Pfmdr1, Pfdhps and Pfdhfr genes, in humans and mosquitoes.
Pfdhfr point mutations and their respective STR haplotypes in allele size.
Pfdhps point mutations and their respective STR haplotypes in allele size.