Abstract
The objective of the study was to identify the microbiological spectrum and drug-sensitivity pattern of peritonitis in patients on continuous ambulatory peritoneal dialysis. This was a prospective study done over a period of a year-and-a-half at a tertiary-care hospital in a hilly state of India. The effluent dialysate bags from 36 consecutive patients with peritonitis were studied. One hunderd ml dialysate fluid was processed under aseptic conditions by lysis centrifugation method. Microscopy and culture was done from the deposits for bacteriological, fungal, and mycobacterial isolates. They were identified by colony morphology and their biochemical reactions. Drug susceptibility testing was done by Kirby-Bauer disc diffusion method. In 36 dialysates, 33 (91.6%) dialysates were culture-positive and in 3 (8.4%), the culture was negative. A total of 36 microorganisms were isolated in 33 cultures. Among the 36 microorganisms, 19 (52.8%) isolates were gram-positive, 10 (27.8%) were gram-negative, 5 (13.9%) were fungi, and 2 (5.6%) were mycobacterial isolates. All gram-positive organisms were sensitive to ampicillin, amoxi-clavulanic acid, cefazolin, clindamycin, and vancomycin. Neither a methicillin-resistant Staphylococci aureus nor a vancomycin-resistant Enterococcus was isolated in gram-positive isolates. Gram-negative organisms were sensitive to ciprofloxacin, ceftriaxone, ceftazidime, cefepime, gentamicin, piperacillin–tazobactam and imipenem. One of the gram-negative isolate was an extended spectrum beta-lactamase producer. Gram-positive peritonitis was more frequent than gram-negative peritonitis in our continuous ambulatory peritoneal dialysis patients. Mycobacterial causes were responsible for peritonitis in patients with culture-negative peritonitis which was not responding to the conventional antimicrobial therapy.
Keywords: Continuous ambulatory peritoneal dialysis, drug sensitivity, microbiology, peritonitis
Introduction
Peritonitis is still the leading cause of technique failure in continuous ambulatory peritoneal dialysis (CAPD) patients.[1] The incidence of peritonitis depends on the factors such as age, race, educational background, environment, and type of dialysis system used.[2] But the outcome depends on the organisms isolated.[3,4] Several studies have shown a decreasing trend in the gram-positive peritonitis and an increasing trend in the incidence of gram-negative peritonitis.[4] The microbiological spectrum of PD-related peritonitis seen in patients from developing countries such as India may be different from that observed in patients from developed countries, and it may be attributed to the difference in social, environmental, educational, and financial background, and surrounding climate of the patients.[1,5,6] The main modality of chronic dialysis for the patients of end-stage renal disease is chronic peritoneal dialysis (PD) at our center which is the only tertiary care hospital in the state of Himachal Pradesh providing dialysis services. We analyzed our data regarding the incidence and outcome of the peritonitis caused by different types of microorganisms.
Materials and Methods
It was a prospective study involving patients undergoing CAPD at our center who developed peritonitis over a period of a year-and-a-half (January 1, 2009 to June 30, 2010). The study was approved by the institutional ethics committee and an informed consent was obtained from the patients before their inclusion in the study. Peritonitis was defined according to the International Society of Peritoneal Dialysis recommendations.[7]
The patients’ exchange bags containing effluent dialysate were delivered to the microbiology laboratory for culturing on the same day that they were collected from the patients. The bags not processed immediately, were refrigerated at 4°C. From these exchange bags, 100 ml of fluid was withdrawn with a sterile needle and syringe under aseptic conditions. This fluid was centrifuged in sterile tubes at a rate of 3000 g for 15 min and supernatant was discarded, leaving 0.5 ml of deposit. In the centrifuged deposit, 10 ml of sterile distilled water was added and the mixture was shaken vigorously for 30 s.[8,9] After vigorous shaking, the deposit was centrifuged at 3,000 g for 15 min and supernatant was discarded. The deposit was divided into three parts, the first part of the deposit was used for gram staining, Ziehl-Neelsen (ZN) staining, and 10% KOH mount to detect the presence of yeast cells or fungal hyphae.[10] The second part of the deposit was used for culturing the bacteria which was done on Blood agar (BA) and MacConkey agar at a temperature of 37°C for 24-48 h. Culturing for fungi was done on Sabouraud-Dextrose agar with and without antibiotics at temperatures of 25°C and 37°C for 4 weeks, and culturing for mycobacteria was done on Lowenstein Jensen medium at 37°C for 8-12 weeks. The third part of the deposit was inoculated into Brain-Heart Infusion (BHI) broth and incubated at 37°C. BHI broth was observed daily for the development of turbidity. After the development of turbidity, the fluid was gram-stained and plated on appropriate media for isolation and identification of the microorganisms. BHI broths showing no growth were discarded after seven days of incubation.
The drug sensitivity was done by Kirby-Bauer disc diffusion method on Mueller Hinton agar. For gram-positive organisms, ampicillin, amoxi-clavulanic acid, cefazolin, clindamycin, and vancomycin discs were tested. The cefoxitin discs were used to detect methicillin-resistant Staphylococcus aureus (MRSA). For gram-negative organisms, ciprofloxacin, cefotaxime, ceftriaxone, ceftazidime, cefepime, gentamicin, piperacillin–tazobactam, and imipenem discs were tested as per Clinical and Laboratory Standard Institute guidelines for antimicrobial susceptibility testing.[11]
Results
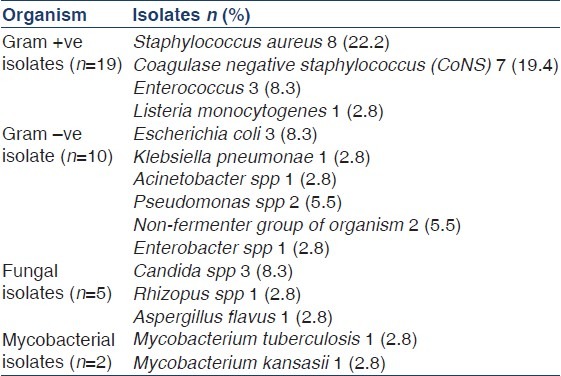
There were 36 episodes of peritonitis in 25 patients during the study period (15 male patients, 11 diabetics, and mean age 58.4 ± 9.1 years). The mean duration of PD in these patients was 29.6 ± 21.6 (range 6-92) patient-months. The peritonitis rate was 0.59 episodes per patient-year. A total of 36 effluent dialysate bags from 36 consecutive patients of CAPD peritonitis were submitted for microbiological evaluation. Initial smear showed gram-positive cocci in 2, gram-negative bacilli in 1, and 1 showed yeast cells [Table 1]. No acid-fast bacillus was detected with ZN staining. Out of a total of 36 dialysate cultures, 33 (91.6%) dialysate were culture-positive and the culture was sterile in 3 (8.4%) [Figure 1]. A total 36 microorganisms were isolated in 33 CAPD dialysate cultures. Among the 36 microorganisms, 19 (52.8%) isolates were gram-positive, 10 (27.8%) isolates were gram-negative, 5 (13.9%) isolates were fungi and 2 (5.6%) mycobacterial spp. were isolated [Table 2]. Among the 19 gram-positive microorganisms, Staphylococcus spp. 15 (78.9%) were the most common group of pathogens involved in CAPD peritonitis [Table 3]. Staphylococcus aureus accounted for 8 (22.2%), coagulase-negative staphylococcus (CoNS) 7 (19.4%), and 3 (8.33%) isolates were Enterococcus fecalis and one isolate was Listeria monocytogenes. Among the 10 (27.8%) gram-negative organisms: The microorganisms of Enterobacteriaceae were isolated in 5: E. coli 3, Klebsiella pneumoniae and Enterobacter spp. 1 each, and environmental Gram-negative bacilli were cultured in other 5: Pseudomonas spp. 2, Acinetobacter spp. 1 and Non-fermenter spp. 2. Among the 5 (13.9%) fungal isolates: Candida albicans 1, non-candida albicans spp 2, and 1 each of Rhizopus spp. and Aspergillus flavus were recovered. The culture also showed the growth of 1 (2.8%) isolate each of Mycobacterium tuberculosis and Mycobacterium kansasii. All these organisms were identified by colony morphology and their biochemical reactions.
Table 1.
Result of smear staining of the centrifuged dialysate (Total=36)
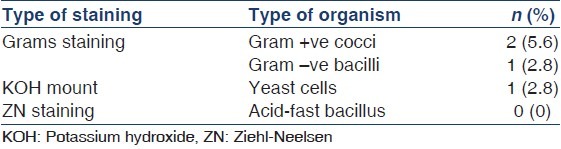
Figure 1.
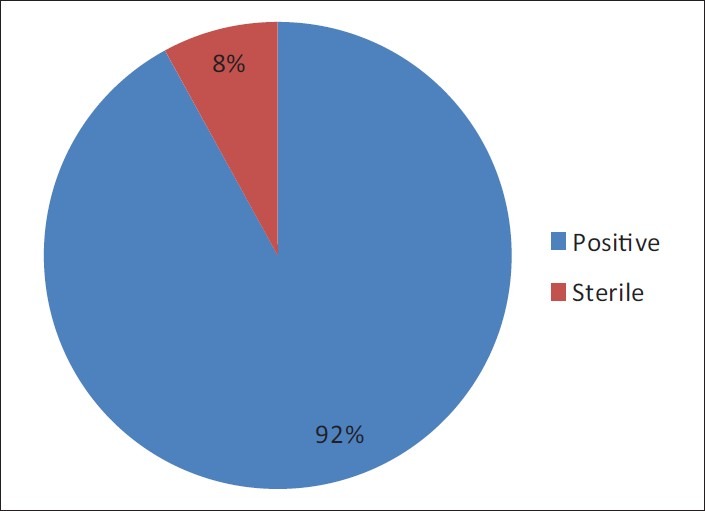
Result of continuous ambulatory peritoneal dialysis dialysate culture
Table 2.
Type of organisms isolated (Total=36)

Table 3.
Spectrum of microorganisms isolated in culture (Total=36)
When we analyzed the organisms according to their origin [Figure 2], the organisms of skin origin were more frequent (50%) than the organisms of environmental origin (25%), followed by those of fecal origin (22%). We considered Staphylococcus aureus, CoNS, and Candida spp. to be of skin origin.[1] E. coli, Klebsiella pneumoniae, Enterobacter spp., and Enterococcus spp. were considered to be of fecal origin. Pseudomonas spp, Non-fermenter spp., Acinetobacter spp., Rhizopus spp., Aspergillus flavus, and Mycobacterium kansasii were of environmental origin.[12] Mycobacterium tuberculosis was of endogenous in origin.
Figure 2.
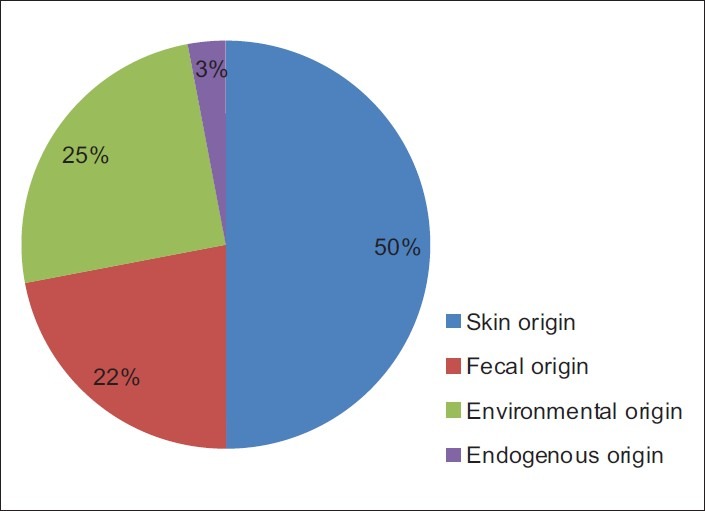
Origin of microorganisms isolated in culture
Gram-positive organisms were sensitive to all antibiotics which were tested. Neither an MRSA nor a vancomycin-resistant Enterococcus was isolated. Listeria monocytogenes was sensitive to amoxi-clavulanic acid. All gram-negative organisms were sensitive to ciprofloxacin, ceftriaxone, cefataxime, cefepime, gentamicin, but Klebsiella pneumoniae was resistant to beta- lactam antibiotics (Extended spectrum beta-lactamase producers [ESBL]) and was sensitive to imipenem.
There was an episode of recurrent peritonitis caused by Staphylococcus aureus in one of the CAPD patient and when nasal swab of CAPD patient and dialysis-assistant was cultured, Staphylococcus aureus was isolated from the dialysis-assistant and had the same antibiotics profile as the CAPD dialysate culture. Among the 5 (14%) fungal isolates, 3 (8.3%) were associated with CoNS (Aspergillus flavus, Rhizopus spp and non-candida albicans each in combination with CoNS).
A total of 31 (86.1%) episodes of peritonitis were cured. In 3 (8.3%) episodes of peritonitis of polymicrobial cause, the catheters were removed and patients were switched over to hemodialysis and in 2 (5.5%) episodes of peritonitis caused by mycobacterial spps, the patients died. One patient died shortly after the start of treatment and in the other, the diagnosis was made postmortem.
Discussion
PD-related peritonitis could be caused by touch contamination, catheter-related problems, bowel pathology, gynecological disease, or systemic bacteremia.[13,14] Despite the advances in PD system connectology, contamination at the time of the PD exchange remains a major cause of peritonitis. Worldwide, gram-positive peritonitis followed by the gram-negative peritonitis are the leading causes of PD-related peritonitis.[15] Likewise, our study also observed that the main causes of peritonitis are gram-positive (53%) and gram-negative (28%) microorganisms and a lesser percentage of other agents.
Among the gram-positive organisms, the Staphylococcus aureus and the CoNS accounted for 15 (41.7%) episodes of the CAPD peritonitis. The Staphylococcus aureus was isolated in 8 (22.2%) and the CoNS in 7 (19.4%) CAPD dialysates. In our study, one nasal carrier of Staphylococcus aureus was detected in the dialysis-assistant which had the same antibiotic profiles as Staphylococcus aureus isolated from the patient's CAPD fluid. The nasal carriers of Staphylococcus aureus had significantly higher frequencies of exit-site infections and peritonitis than a non-carrier.[16] The CoNS is an important pathogen in CAPD peritonitis. About one-fifth of the cases of peritonitis was observed to be caused by CoNS in this study. Also, there are studies which have reported that the majority of CAPD peritonitis is caused by CoNS.[17] This may primarily be due to touch contamination or due to the formation of a biofilm.
Enterococci may cause between 2% and 6% of PD-related peritonitis episodes and their identification is a hallmark of a gastrointestinal origin of the infection. Overall, outcomes in enterococcus peritonitis are similar to those in CoNS peritonitis and are better than those of Escherichia coli peritonitis.[18] In our study, 3 (8.3%) Enterococcus fecalis were isolated. They were sensitive to tested antibiotics and none of them were resistant to vancomycin. Listeria monocytogenes was isolated in one dialysate culture and identified by biochemical reactions and motility at 22°C temperatures. The organisms were sensitive to ampicillin, amoxi-clavulanic acid, and vancomycin and peritonitis was cured using these antibiotics. A study has reported CAPD peritonitis caused by Listeria monocytogenes isolated in a female patient which was sensitive to beta-lactam antibiotics.[19]
Advances in connectology have significantly reduced the overall incidence of peritonitis, particularly that caused by gram-positive organisms. However, the incidence of gram-negative peritonitis remains at a steady level, and therefore, it has become proportionally more important. Gram-negative organisms now account for 20-30% of all PD-related peritonitis. Moreover, gram-negative peritonitis is often more severe and associated with worse outcomes.[20] Gram-negative organisms were responsible for 28% of the peritonitis in this study which were equally divided between microorganisms of fecal origin and environmental origin. Gram-negative peritonitis usually occurs either because of fecal origin or transmural migration of the infecting organisms. Gram-negative peritonitis episodes attributable to transmural migration of bacteria across the bowel wall are usually associated with multiple gram-negative organisms and anaerobic organisms.[1] In our patients, transmural migrations were an unlikely route of peritonitis, because none of our cultures isolated multiple gram-negative and anaerobic organisms. We think that poor hand-washing technique and lack of access to fresh running water for hand washing may have been responsible for contamination during peritoneal dialysis exchange procedure. Among the gram-negative organisms, Pseudomonas aeruginosa, Non-fermenter spp., and Acinetobacter spp. were the cause of peritonitis in 5 (13.9%) episodes of peritonitis. The origin may be the skin or a contaminated water bath used to heat the dialysis bag.[21] Our study revealed that E. coli causes more peritonitis episodes as compared to the other gram-negative organisms, which is similar to the pattern observed in other parts of the world. A study from India has reported resistance in two-third of the gram-negative organisms to the third generation cephalosporin possibily due to availability of these antimicrobials without prescriptions and misuse of antimicrobials by primary-care physicians.[22] In our study, an important observation was made from the antimicrobial sensitivity profile that all the gram-negative pathogens were sensitive to quinolones, gentamicin and third-generation cephalosporins except Klebsiella pneumoniae which was ESBL-producer and was sensitive to imipenem.
In the recent years, fungal peritonitis complicating CAPD is being increasingly recognized. Recent antibiotic therapy, frequent episodes of bacterial peritonitis, and immunosuppression are the major risk factors of fungal peritonitis which accounts for 1-15% of episodes of peritonitis in various studies.[23] The majority of these fungal peritonitis episodes are caused by Candida species. Candida albicans has historically been reported to be a more common cause than non-albicans species, but in recent reports, a shift has been observed. Now non-albicans Candida may be more common and there is increasing recognition of filamentous fungi as pathogens in CAPD patients.[23] The incidence of fungal peritonitis was 14% in the present study. There were 2 isolates of non-candida albicans and 1 isolate each of candida albicans, Rhizopus spp. and Aspergillus flavus in a total of 5 fungal isolates. The origin of fungal isolates may be from the patients’ skin, environment, or from the mucous membranes.[24]
In our study, Mycobacterium tuberculosis (1) and Mycobacterium kansasii (1) were isolated in two dialysate cultures. The pathogenesis of peritoneal tuberculosis (TB) in patients undergoing CAPD remains speculative. Chronic renal failure may lead to a defect in the cellular arm of immune response and predispose to Mycobacterium tuberculosis infection. The exact portal of entry of Mycobacterium tuberculosis into the peritoneum remains unclear. Some studies proposed that infection with Mycobacterium tuberculosis is acquired by direct contamination via peritoneal dialysate, whereas other researchers believe that infection is spread from another focus of TB in the body.[25]
The clinical presentation of TB peritonitis can be very nonspecific. A triad of abdominal pain, abdomen distension, and fever has been reported to occur in <60% of the patients.[26] In our patients, abdominal pain, distension, and cloudy fluid were the symptoms and peritonitis was not resolving in spite of the conventional broad-spectrum antimicrobial treatment. The diagnosis of TB peritonitis requires a high index of suspicion but early diagnosis is often difficult to make.[27,28] Radiologic imaging techniques are not sensitive or specific for diagnostic purposes for tuberculosis. Some studies observed that despite the presence of molecular diagnostic techniques, we will depend upon conventional microbiological cultures for the identification of mycobacterial species.[29] However, direct microscopic smear detection of acid-fast bacilli in the ascitic fluid is insensitive, with reported sensitivity ranging from 0% to 6% and the conventional microbiological diagnostic methods are slow and may not be sensitive enough for establishing a diagnosis in a timely manner.[26,29] Moreover, the validity of molecular diagnostic techniques in peritoneal dialysis patients has not been established.[30] Treatment delay is the most significant factor contributing to high morbidity and mortality due to TB peritonitis. Early diagnosis and treatment of disease are extremely important for improving the outcome. Standard antituberculous chemotherapy is highly effective. Therefore, it is prudent to start empirical treatment with anti-tubercular chemotherapy in CAPD patients with peritonitits that is refractory to broad-spectrum antibiotics while awaiting the results of the mycobacterial cultures to improve the outcome and preserve peritoneal membrane integrity.
Conclusion
In our CAPD patients, the main causes of peritonitis were gram-positive and gram-negative microorganisms and a lesser percentage of other agents. The rate of gram-positive peritonitis was higher than that of gram-negative peritonitis. The higher incidence of gram-positive peritonitis in our CAPD patients was due to their break in the sterile technique. Mycobacterial infections should be suspected in all episodes of culture-negative peritonitis especially those which do not respond to usual antimicrobial therapy.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Prasad N, Gupta A, Sharma RK, Prasad KN, Gulati S, Sharma AP. Outcome of gram-positive and gram-negative peritonitis in patients on continuous ambulatory peritoneal dialysis: A single-center experience. Perit Dial Int. 2003;23:S144–7. [PubMed] [Google Scholar]
- 2.Fried LF, Bernardini J, Johnston JR, Piraino B. Peritonitis influences mortality in peritoneal dialysis patients. J Am Soc Nephrol. 1996;7:2176–82. doi: 10.1681/ASN.V7102176. [DOI] [PubMed] [Google Scholar]
- 3.Bunke CM, Brier ME, Golper TA. Outcomes of single organism peritonitis in peritoneal dialysis: Gram negatives versus gram positives in the Network 9 Peritonitis Study. Kidney Int. 1997;52:524–9. doi: 10.1038/ki.1997.363. [DOI] [PubMed] [Google Scholar]
- 4.Troidle L, Gorban-Brennan N, Kliger A, Finkelstein F. Differing outcomes of gram-positive and gram-negative peritonitis. Am J Kidney Dis. 1998;32:623–8. doi: 10.1016/s0272-6386(98)70026-5. [DOI] [PubMed] [Google Scholar]
- 5.Chow KM, Szeto CC, Leung CB, Kwan BC, Law MC, Li PK. A risk analysis of continuous ambulatory peritoneal dialysis-related peritonitis. Perit Dial Int. 2005;25:374–9. [PubMed] [Google Scholar]
- 6.Vikrant S. Continuous ambulatory peritoneal dialysis: A viable modality of renal replacement therapy in a hilly state of India. Indian J Nephrol. 2007;17:165–9. [Google Scholar]
- 7.Piraino B, Bailie GR, Bernardini J, Boeschoten E, Gupta A, Holmes C, et al. Peritoneal dialysis-related infections recommendations: 2005 update. Perit Dial Int. 2005;25:107–31. [PubMed] [Google Scholar]
- 8.Health Protection Agency. Investigation of Continuous Ambulatory Peritoneal Dialysis Fluid. UK Standards for Microbiology Investigations. B 25 Issue 5.2. 2012. [Last cited on 2012]. http://ww.hpa.org.uk/SMI/pdf .
- 9.Spencer RC, Ahmad WK. Laboratory diagnosis of peritonitis in continuous ambulatory peritoneal dialysis by lysis and centrifugation. J Clin Pathol. 1986;39:925–6. doi: 10.1136/jcp.39.8.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall RJ. A simple lysis method for the culture of dialysis fluid from patients on continuous ambulatory peritoneal dialysis. J Hosp Infect. 1992;20:59–60. doi: 10.1016/0195-6701(92)90066-u. [DOI] [PubMed] [Google Scholar]
- 11.Performance Standards for Antimicrobial Susceptibility Testing. CLSI Approved Standard M100-S15. Wayne, PA: Clinical and Laboratory Standards Institute; 2005. Clinical and Laboratory Standards Institute. [Google Scholar]
- 12.Parsad KN. Diagnosis of peritonitis and exit site infection in patients on continuous ambulatory peritoneal dialysis. Peritoneal Dial Soc India. 2007;1:55–62. [Google Scholar]
- 13.Gokal R. Peritoneal dialysis. Prevention and control of infection. Drugs Aging. 2000;17:269–82. doi: 10.2165/00002512-200017040-00003. [DOI] [PubMed] [Google Scholar]
- 14.Tranaeus A, Heimbürger O, Granqvist S. Diverticular disease of the colon: A risk factor for peritonitis in continuous peritoneal dialysis. Nephrol Dial Transplant. 1990;5:141–7. doi: 10.1093/ndt/5.2.141. [DOI] [PubMed] [Google Scholar]
- 15.Zelenitsky S, Barns L, Findlay I, Alfa M, Ariano R, Fine A, et al. Analysis of microbiological trends in peritoneal dialysis-related peritonitis from 1991 to 1998. Am J Kidney Dis. 2000;36:1009–13. doi: 10.1053/ajkd.2000.19103. [DOI] [PubMed] [Google Scholar]
- 16.Davies SJ, Ogg CS, Cameron JS, Poston S, Noble WC. Staphylococcus aureus nasal carriage, exit-site infection and catheter loss in patients treated with continuous ambulatory peritoneal dialysis (CAPD) Perit Dial Int. 1989;9:61–4. [PubMed] [Google Scholar]
- 17.Dasgupta MK, Ward K, Noble PA, Larabie M, Costerton JW. Development of bacterial biofilms on silastic catheter materials in peritoneal dialysis fluid. Am J Kidney Dis. 1994;23:709–16. doi: 10.1016/s0272-6386(12)70281-0. [DOI] [PubMed] [Google Scholar]
- 18.Rosman JB, Johnson DW. Enterococcal peritonitis in peritoneal dialysis: The danger from within? Perit Dial Int. 2011;31:518–21. doi: 10.3747/pdi.2011.00041. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad M, Krishnan A, Kelman E, Allen V, Bargman JM. Listeria monocytogenes peritonitis in a patient on peritoneal dialysis: A case report and review of the literature. Int Urol Nephrol. 2008;40:815–9. doi: 10.1007/s11255-008-9411-2. [DOI] [PubMed] [Google Scholar]
- 20.Borràs M. Antibiotic resistance in gram-negative peritonitis. Perit Dial Int. 2009;29:274–6. [PubMed] [Google Scholar]
- 21.Ashline V, Stevens A, Carter MJ. Nosocomial peritonitis related to contaminated dialysate warming water. Am J Infect Control. 1981;9:50–2. doi: 10.1016/s0196-6553(81)80032-6. [DOI] [PubMed] [Google Scholar]
- 22.Keithi-Reddy SR, Gupta KL, Jha V, Sud K, Singh SK, Kohli HS, et al. Spectrum and sensitivity pattern of gram-negative organisms causing CAPD peritonitis in India. Perit Dial Int. 2007;27:205–7. [PubMed] [Google Scholar]
- 23.Prasad KN, Prasad N, Gupta A, Sharma RK, Verma AK, Ayyagari A. Fungal peritonitis in patients on continuous ambulatory peritoneal dialysis: A single centre Indian experience. J Infect. 2004;48:96–101. doi: 10.1016/s0163-4453(03)00119-1. [DOI] [PubMed] [Google Scholar]
- 24.Eisenberg ES, Leviton I, Soeiro R. Fungal peritonitis in patients receiving peritoneal dialysis: Experience with 11 patients and review of the literature. Rev Infect Dis. 1986;8:309–21. doi: 10.1093/clinids/8.3.309. [DOI] [PubMed] [Google Scholar]
- 25.Lui SL, Tang S, Li FK, Choy BY, Chan TM, Lo WK, et al. Tuberculosis infection in Chinese patients undergoing continuous ambulatory peritoneal dialysis. Am J Kidney Dis. 2001;38:1055–60. doi: 10.1053/ajkd.2001.28599. [DOI] [PubMed] [Google Scholar]
- 26.Chau TN, Leung VK, Wong S, Law ST, Chan WH, Luk IS, et al. Diagnostic challenges of tuberculosis peritonitis in patients with and without end-stage renal failure. Clin Infect Dis. 2007;45:e141–6. doi: 10.1086/523727. [DOI] [PubMed] [Google Scholar]
- 27.Manohar A, Simjee AE, Haffejee AA, Pettengell KE. Symptoms and investigative findings in 145 patients with tuberculous peritonitis diagnosed by peritoneoscopy and biopsy over a five year period. Gut. 1990;31:1130–2. doi: 10.1136/gut.31.10.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapoor VK. Abdominal tuberculosis. Postgrad Med J. 1998;74:459–67. doi: 10.1136/pgmj.74.874.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abraham G, Mathews M, Sekar L, Srikanth A, Sekar U, Soundarajan P. Tuberculous peritonitis in a cohort of continuous ambulatory peritoneal dialysis patients. Perit Dial Int. 2001;21:S202–4. [PubMed] [Google Scholar]
- 30.Vadivel N, Tucker JK, Trikudanathan S, Heher E, Singh AK. Tuberculous peritonitis: A race against time. Kidney Int. 2006;70:969–72. doi: 10.1038/sj.ki.5001610. [DOI] [PubMed] [Google Scholar]


