Abstract
Background
Elevated pressure, elevated oxygen tension (DOT) and elevated carbon dioxide tension (DCT) are readily encountered at the bottom of large industrial bioreactors and during bioprocesses where pressure is applied for enhancing the oxygen transfer. Yet information about their effect on bacteria and on the gene expression thereof is scarce. To shed light on the cellular functions affected by these specific environmental conditions, the transcriptome of Pseudomonas putida KT2440, a bacterium of great relevance for the production of medium-chain-length polyhydroxyalkanoates, was thoroughly investigated using DNA microarrays.
Results
Very well defined chemostat cultivations were carried out with P. putida to produce high quality RNA samples and ensure that differential gene expression was caused exclusively by changes of pressure, DOT and/or DCT. Cellular stress was detected at 7 bar and elevated DCT in the form of heat shock and oxidative stress-like responses, and indicators of cell envelope perturbations were identified as well.
Globally, gene transcription was not considerably altered when DOT was increased from 40 ± 5 to 235 ± 20% at 7 bar and elevated DCT. Nevertheless, differential transcription was observed for a few genes linked to iron-sulfur cluster assembly, terminal oxidases, glutamate metabolism and arginine deiminase pathway, which shows their particular sensitivity to variations of DOT.
Conclusions
This study provides a comprehensive overview on the changes occurring in the transcriptome of P. putida upon mild variations of pressure, DOT and DCT. Interestingly, whereas the changes of gene transcription were widespread, the cell physiology was hardly affected, which illustrates how efficient reorganization of the gene transcription is for dealing with environmental changes that may otherwise be harmful. Several particularly sensitive cellular functions were identified, which will certainly contribute to the understanding of the mechanisms involved in stress sensing/response and to finding ways of enhancing the stress tolerance of microorganisms.
Keywords: Pseudomonas putida KT2440, Gene transcription, DNA microarrays, Elevated pressure, Dissolved oxygen tension, Environmental stress, mcl-PHA
Background
Among the various methods developed for optimizing bacterial bioprocesses, the application of elevated pressure in the range of 1–10 bar constitutes a simple and effective way to increase the oxygen transfer rate and achieve high cell densities [1-10]. In particular, this method has proved suitable for the production of medium-chain-length polyhydroxyalkanoates (mcl-PHA), biocompatible and biodegradable polymers with a wide range of applications [11-13], by Pseudomonas putida KT2440. Indeed, we demonstrated in a recent work that except for a small decrease of viability, the physiology of P. putida KT2440 (biomass production, nutrient yields, respiratory quotient, mcl-PHA production) was not impaired at 7 bar [2]. Nevertheless, pressure may still induce stress and affect metabolic pathways, cellular machineries and cellular functions when varying from optimal values, in a similar way as deviations of temperature, pH, osmolarity or water availability. Unlike for the latter environmental factors [14-16], the effect of elevated pressure (1–10 bars) on the transcriptome of bacteria, and P. putida KT2440 in particular, has never been investigated in detail.
Bacteria live in nature mostly at a pressure of around 1 bar with the exception of microorganisms found in the sea (pressure increase of about 0.1 bar/m), in the soil (pressure increase of about 0.3 bar/m) or in plant cells (pressures below 1 bar made possible by turgor) [17]. In general, bacteria are able to grow up to at least 300 bar but they cannot survive above 2000 bar [18]. As a result, pressure-based preserving and sterilizing methods have been developed and successfully exploited in the food industry [19] and are now being studied as alternative to heat treatment and gamma ray irradiation for the disinfection of biomaterials [20-25]. Essential processes such as DNA replication, RNA transcription and protein synthesis are functional in Escherichia coli up to at least 500, 200 and 600 bar, respectively [26]. Starting from 1000–2000 bar protein aggregation and nucleoid changes can be observed [27] along with loss of membrane integrity [28]. Filamentous growth [29] and loss of motility [30] are two other phenotypes of cells confronted to high-pressure stress. In addition, the composition of fatty acids in the cell membrane varies at high pressure [31] as well as the production of several membrane proteins such as OmpH and OmpL [32,33], transporters [34] and terminal oxidases [35]. Lastly, induction of heat-shock, cold-shock, and SOS responses was observed following high pressure treatment in E. coli[33,34,36].
Notably, all the effects described above were caused by pressures at least 20 times larger than the ones intended to be applied during bacterial fermentations. Therefore, differences are expected between the transcriptomes of cells subjected to high pressure (> 200 bar) and elevated pressure (< 10 bar). Another important point to consider under elevated pressure is the increase of dissolved oxygen tension (DOT) and dissolved carbon dioxide tension (DCT) resulting from the larger gas solubility. This phenomenon is hardly observed for processes at high pressure since they are normally performed in degassed systems. The occurrence of large DOT during a bioprocess can be avoided by decreasing the aeration or by diluting the aeration gas with nitrogen. In contrast it is much more difficult to maintain DCT values below a certain level, especially in the case of medium- or high-cell-density cultivations that produce significant amounts of CO2. As a result changes in DCT, and in DOT if uncontrolled, can also affect gene transcription at elevated pressure.
In order to unravel the cellular mechanisms used by P. putida KT2440 for sensing and responding to variations of pressure, DOT and DCT, we explored its transcriptome by means of DNA microarrays. Not only was the effect of elevated pressure and DCT studied but also the effect of combined elevated pressure, DCT and DOT. This latter effect was investigated because: (1) such conditions can readily occur if pressure is not applied exactly in parallel with the oxygen demand and (2) the effect of oxidative stress on the gene expression of P. putida KT2440 has not been studied yet.
Here, we report the most relevant cellular mechanisms affected by elevated pressure, DCT and DOT and provide general information for a further, in greater depth investigation of specific genes and cellular functions. In addition to extending the global understanding of stress sensing and stress response mechanisms, this study aims at identifying candidate genes that could be engineered to enhance the resistance and/or productivity of microorganisms used in industrial bioprocesses where such stresses can occur (e. g. mcl-PHA production processes at elevated pressure).
Results
Cultivations with P. putida KT2440 were carried out at 1 bar (scenario “Control”), at 7 bar with a similar DOT as at 1 bar (“Pressure”), and at 7 bar with a higher DOT (“Pressure Oxygen”) (Table 1). Detailed description of these cultivations is reported in a previous work where the effect of pressure on cell physiology was investigated [2]. In the present work we focus on the effect of pressure on the cell transcriptome while using the same biomass samples. These biomass samples were produced in chemostat cultivations where the growth rate, cell density, medium composition, pH, and temperature were precisely controlled in order to avoid any unwanted effect on the gene transcription. The level of dissolved CO2 (CO2,aq and HCO3-) increased from 10 mM at 1 bar to ~20 mM at 7 bar (Table 1). As a result, the changes of gene expression reported for 7 bar may be a consequence of either elevated pressure per se, elevated DCT, or the combination of both factors. The most relevant genes and functions affected in the Pressure and Pressure Oxygen conditions are listed in Table 2.
Table 1.
Characteristics of the chemostat cultivation conditions with P. putida KT2440 (dilution rate = 0.15 h-1)
| |
Control |
Condition 1 |
Condition 2 |
|---|---|---|---|
| Effect investigated | - | Pressure | Pressure Oxygen |
| P [bar] |
1 |
7 |
7 |
| DOT [%] |
40 ± 5 |
45 ± 20 |
235 ± 20 |
| [CO2]tot [mM]* |
10 ± 1 |
21 ± 5 |
19 ± 2 |
| CDW [g L-1]# | 14.0 ± 0.5 | 14.1 ± 0.3 | 13.6 ± 0.8 |
For more details about the cultivation and sampling procedure, please refer to [2].
* Total concentration of dissolved CO2. # Cell dry weight.
Table 2.
Overview of the most relevant cellular functions affected by elevated pressure alone and by elevated pressure and DOT
|
Cellular function |
Differentially expressed genes |
|
|---|---|---|
| Pressure | Pressure oxygen | |
| Stress sensing/stress response |
• aer-1 (-), MCP (-) |
• aer-1 (-), MCP (-) |
| |
• rpoH and heat-shock genes (+) |
• rpoH and heat-shock genes (+) |
| |
• DNA repair (-) |
• DNA repair (-) |
| Control of reactive oxygen species |
• ahpC, glutathione genes (+) |
• ahpC, glutathione genes (+) |
| |
• anr (+) and cyo (+) |
• anr (-) and cyo (+) |
| |
• Cbb3-1 (+), Cbb3-2 (-), Aa3 (-), Cyo (+), azu (+) |
• Cbb3-1 (-), Cbb3-2 (-), Aa3 (-), Cyo (+), azu (-) |
| Cell envelope |
• oprG (+), oprH (+) |
• oprG (-), oprH (+) |
| |
• membrane proteins |
• membrane proteins |
| |
• secretion systems (+) |
• secretion systems (+) |
| Internal pH |
• urease (+) |
• secretion systems (+) |
| |
• secretion systems (+) |
|
| |
• oxidative stress (+) |
• oxidative stress (+) |
| Fe homeostasis/Fe-S clusters |
• siderophore transporters (+), bacterioferritin (-) |
|
| |
|
• siderophore transporters (+), bacterioferritin (-) |
| Biosynthetic metabolism |
|
• ADI (-) |
| • Glu synthesis/metabolism (+) | ||
Induction and repression of gene transcription is indicated by the signs (+) and (-), respectively, and major differences of gene transcription between the two conditions are indicated by the bold font. Abbreviations: ADI, arginine deiminase pathway; Glu, glutamate; MCP, methyl-accepting chemotaxis protein.
Global analyses of P. putida KT2440 transcriptome under elevated pressure and under combined elevated pressure and elevated DOT
We showed previously that the physiology of P. putida KT2440 was hardly affected at 7 bar [2]. No decrease of cell growth rate and PHA-free biomass (total cell dry weight minus PHA weight) production was observed and the respiratory quotient did not vary under these conditions, either. Nevertheless, small changes were observed in the production of mcl-PHA (increase) and in cell viability (decrease). The physiology of cells cultivated in the Pressure Oxygen condition was very similar, with the exception of a small decrease of respiratory quotient and slightly larger changes of polymer production and cell viability.
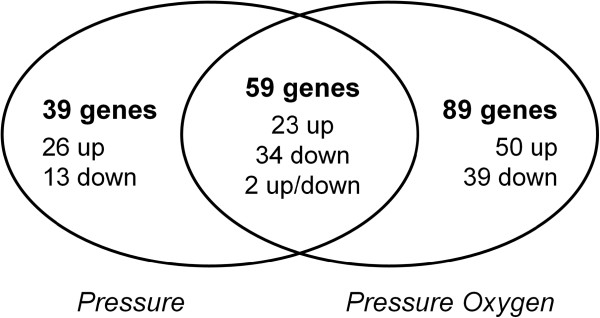
In contrast to these few changes of cell physiology, increasing the pressure from 1 to 7 bar caused extensive changes of gene transcription. The DNA microarray experiments revealed significant differential expression (adjusted p-value ≤ 0.01) for about 100 and 150 genes in the Pressure condition and the Pressure Oxygen condition, respectively (Figure 1, Additional file 1: Table S1-S2). Almost 60 genes were significantly differentially expressed regardless of the DOT level but interestingly, two of them exhibited opposite differential expression (Figure 1, Additional file 1: Table S1). These two genes were coding for the outer membrane porin OprG (PP_0504) and for a hypothetical protein (PP_5390).
Figure 1.
Number of significantly differentially expressed genes. “Pressure” and “Pressure Oxygen” refer to the genes differentially expressed at elevated pressure and at combined elevated pressure and elevated DOT, respectively. The number of genes overexpressed compared to the Control condition is described as “up”, the number of genes underexpressed as “down”, and the number of genes overexpressed in one condition but underexpressed in the other one as “up/down”.
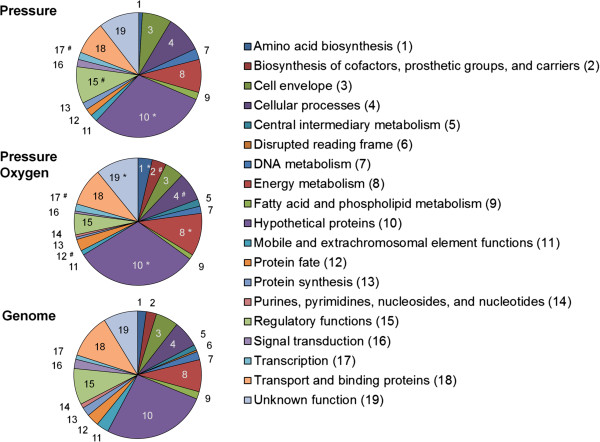
The significantly differentially expressed genes were classified into categories and sub-categories according to the Comprehensive Microbial Resource (CMR) [37]. The gene repartition for the Pressure condition was very similar to the repartition considering the entire genome (Figure 2, Additional file 1: Table S2). In contrast, several categories were over-represented in the Pressure Oxygen condition compared to the genome: genes involved in the biosynthesis of amino acids such as glutamate; in the biosynthesis of cofactors, prosthetic groups, and carriers such as glutathione; in adaptation to atypical conditions; in energy metabolism; in protein folding and stabilization; and, like for Pressure, genes coding for hypothetical proteins and involved in transcription (Additional file 1: Table S3).
Figure 2.
Classification of the significantly differentially expressed genes into role categories. The genes were sorted into the 19 role categories established by the Comprehensive Microbial Resource (CMR). “Pressure” and “Pressure Oxygen” refer to the genes differentially expressed at elevated pressure and at combined elevated pressure and elevated DOT, respectively. For comparison, the gene distribution of the complete P. putida KT2440 genome is also depicted (Genome). For clarity reasons, a number was assigned to each category and is shown in the corresponding sector of the charts. Categories that were significantly over-represented as well as categories containing sub-categories that were significantly over-represented compared to the genome distribution are marked by the asterisk (*) and the hash (#), respectively (for more details, see the Additional file 1: Table S2-S3).
Genes coding for sensing proteins (e. g. methyl-accepting chemotaxis transducer PP_2643) and for proteins located in the cell envelope (e. g. outer membrane proteins OprG and OprH, putative multidrug efflux transporter PP_0906, members of a type VI secretion system PP_3094 and PP_3100) exhibited the highest significant differential expression in both the Pressure and the Pressure Oxygen scenarios (Additional file 1: Table S4). In addition, differential expression of genes coding for a bacterioferritin (PP_1082), for the alkyl hydroperoxide reductase AhpC (PP_2439) involved in detoxification, and for the iron-cluster assembly proteins IscS and IscA (PP_0842 and PP_0844) had a particularly high significance in the Pressure Oxygen condition.
In general, the transcriptomic profiles (Table 3) were supported by qRT-PCR data since out of nine genes tested, eight exhibited similar fold changes with the two methods (Table 4). For the one gene showing discrepant results (aer-1, PP_2257) a highly specific hydrolysis probe base real time PCR had to be performed to establish the down-regulation of the gene in the Pressure and Pressure Oxygen conditions (see the section Methods). At this stage, it is inherently impossible to produce DNA microarray data with neither false positives nor false negatives because of the huge amount of genes (> 5’000) tested simultaneously. As a result, follow-up studies focusing on particular genes should include confirmation experiments such as RT-PCR or production of deletion mutants.
Table 3.
Most relevant genes differentially expressed under elevated pressure ( Pressure ) and under elevated pressure and DOT ( Pressure oxygen )
|
ID |
Name |
Gene |
Pressure |
Pressure oxygen |
||
|---|---|---|---|---|---|---|
| FC | Adj. P-val | FC | Adj. P-val | |||
| Transcription regulators |
||||||
| PP_2088 |
RNA polymerase sigma factor SigX |
sigX |
-1.88 |
1.9 E-03 |
-1.65 |
5.6 E-03 |
| PP_5108 |
RNA polymerase sigma-32 factor |
rpoH |
+1.49* |
3.6 E-03 |
+1.57 |
8.7 E-03 |
| PP_1429 |
sigma E regulatory protein, MucB/RseB |
algN |
+1.64 |
1.5 E-03 |
+1.46* |
1.3 E-02 |
| PP_1863 |
transcriptional regulator LysR family |
|
+2.77 |
3.1 E-03 |
|
|
| PP_2475 |
transcriptional regulator TetR family |
|
+1.59 |
7.8 E-03 |
+1.75 |
7.5 E-03 |
| PP_3439 |
transcriptional regulator AraC family |
|
-1.55 |
7.7 E-03 |
-1.59 |
5.4 E-03 |
| PP_4508 |
transcriptional regulator AraC family |
|
-1.45* |
1.2 E-02 |
-1.55 |
3.6 E-03 |
| Global regulatory proteins | ||||||
| PP_4265 |
transcriptional regulator Anr |
anr |
+1.61 |
1.2 E-02 |
-1.99 |
2.9 E-02 |
| Signal sensing and transduction | ||||||
| PP_2258 |
sensory box protein |
|
-1.51 |
1.6 E-02 |
-1.56 |
3.2 E-02 |
| PP_2643 |
methyl-accepting chemotaxis transducer |
|
-6.82 |
2.9 E-07 |
-15.45 |
8.5 E-08 |
| PP_1761 |
sensory box protein/GGDEF family protein |
|
-1.77 |
1.9 E-03 |
-1.71 |
2.8 E-03 |
| PP_1762 |
conserved hypothetical protein |
|
-1.45* |
2.9 E-02 |
-1.54 |
9.3 E-03 |
| PP_2097 |
sensory box protein |
|
-1.49* |
5.0 E-03 |
-1.52 |
2.0 E-02 |
| PP_4405 |
sensory box protein |
|
+2.04 |
8.0 E-03 |
+1.39* |
8.9 E-02 |
| PP_5324 |
response regulator |
|
-1.53 |
4.3 E-03 |
-1.47* |
9.2 E-03 |
| PP_2356 |
phytochrome family protein (two-component sensor activity) |
|
-1.73 |
4.6 E-03 |
-1.85 |
1.3 E-03 |
| PP_4362 |
Hpt protein (two-component system) |
|
-1.36* |
7.4 E-02 |
-1.52 |
9.0 E-03 |
| PP_4503 |
winged helix family two component transcriptional regulator |
|
+1.56 |
2.3 E-03 |
+1.39* |
8.9 E-02 |
| PP_0997 |
sigma-54 dependent transcriptional regulator/sensory box protein |
|
-1.46 |
2.0 E-02 |
-1.88 |
4.7 E-04 |
| Stress response (Chaperones, DNA repair) | ||||||
| PP_4179 |
heat shock protein HtpG |
htpG |
+1.65 |
1.0 E-02 |
+1.92 |
2.0 E-02 |
| PP_4728 |
heat shock protein GrpE |
grpE |
+1.33* |
6.2 E-02 |
+1.72 |
7.2 E-03 |
| PP_1360 |
co-chaperonin GroES |
groES |
+1.61 |
2.0 E-02 |
+1.77 |
8.2 E-02 |
| PP_1361 |
chaperonin 60 kDa |
groEL |
+1.78 |
1.0 E-02 |
+2.19 |
6.0 E-03 |
| PP_1522 |
cold shock protein CspA |
cspA-1 |
-1.51 |
9.1 E-03 |
-1.75 |
3.3 E-03 |
| PP_1092 |
endonuclease III |
nth |
-1.67 |
1.6 E-03 |
-1.58 |
3.7 E-03 |
| PP_1624 |
group II intron-encoding maturase |
|
-1.68 |
1.3 E-03 |
-1.73 |
1.2 E-03 |
| PP_1630 |
RecX protein |
recX |
+1.54 |
1.5 E-03 |
+1.53 |
1.6 E-02 |
| PP_0483 |
excinuclease ABC A subunit |
uvrA |
|
|
-1.67 |
6.1 E-03 |
| PP_2295 |
antirestriction protein, putative |
|
-1.44* |
9.1 E-03 |
-1.51 |
5.4 E-03 |
| PP_2326 |
universal stress protein family protein |
|
-1.51 |
2.6 E-02 |
-1.51 |
1.3 E-02 |
| Cell envelope | ||||||
| PP_0267 |
outer membrane ferric siderophore receptor putative |
|
+1.54 |
1.2 E-02 |
+1.73 |
6.6 E-03 |
| PP_0504 |
outer membrane protein OprG |
oprG |
+2.55 |
5.8 E-04 |
-3.39 |
3.1 E-04 |
| PP_1185 |
outer membrane protein H1 |
oprH |
+6.63 |
2.9 E-04 |
+4.76 |
1.1 E-04 |
| PP_3293 |
conserved hypothetical protein (predicted ion channel) |
|
-1.47* |
4.5 E-03 |
-1.74 |
6.4 E-03 |
| PP_4454 |
opine ABC transporter permease protein putative |
|
+1.95 |
2.6 E-03 |
+1.36 |
4.6 E-02 |
| PP_4465 |
porin putative |
|
+2.33 |
1.9 E-03 |
+1.60 |
1.1 E-01 |
| PP_2445 |
integral membrane protein TerC |
|
-1.95 |
1.9 E-03 |
-2.25 |
1.3 E-03 |
| PP_1150 |
membrane protein putative |
|
|
|
-4.14 |
3.1 E-04 |
| PP_4592 |
membrane protein putative |
|
-1.74 |
3.1 E-03 |
-2.20 |
1.9 E-03 |
| PP_0916 |
transporter LysE family |
|
|
|
+1.83 |
1.6 E-03 |
| PP_1068 |
amino acid ABC transporter ATP-binding protein |
|
|
|
+1.80 |
1.3 E-03 |
| PP_3023 |
amino acid efflux protein putative |
|
|
|
+1.51 |
6.4 E-03 |
| PP_5073 |
conserved hypothetical protein (periplasmic protein) |
|
+5.70 |
1.9 E-03 |
+5.98 |
8.1 E-06 |
| PP_4841 |
branched-chain amino acid ABC transporter, periplasmic amino acid-binding protein, putative |
|
+1.95 |
8.5 E-03 |
|
|
| PP_4842 |
branched-chain amino acid ABC transporter, permease protein, putative |
|
+3.07 |
4.9 E-03 |
|
|
| PP_5233 |
ammonium transporter |
amtB |
+1.87 |
8.0 E-03 |
|
|
| PP_4867 |
extracellular ligand-binding receptor |
|
+1.79 |
3.3 E-03 |
|
|
| PP_1076 |
glycerol uptake facilitator protein |
glpF |
+2.23 |
3.0 E-03 |
+2.03 |
4.7 E-04 |
| PP_2454 |
ribose ABC transporter |
rbsB |
+1.88 |
1.4 E-02 |
+3.81 |
5.4 E-05 |
| PP_2119 |
ABC efflux tranporter/ATP-binding protein |
|
-2.19 |
4.1 E-03 |
-2.10 |
1.2 E-03 |
| PP_0508 |
conserved hypothetical protein (same operon as efflux ABC transporter ATP-binding prot.) |
|
+2.55 |
8.8 E-03 |
+3.36 |
1.8 E-02 |
| PP_5307 |
biopolymer transport protein ExbD |
exbD |
|
|
+2.30 |
1.8 E-03 |
| PP_0906 |
multidrug efflux RND transporter putative |
|
-2.11 |
1.4 E-04 |
-2.19 |
2.1 E-03 |
| PP_0033 |
sugar transferase putative |
|
-1.74 |
1.9 E-03 |
-2.20 |
9.6 E-03 |
| PP_3422 |
lytic transglycosylase |
ltg |
-1.74 |
1.9 E-03 |
-1.97 |
9.9 E-04 |
| PP_4897 |
N-acetylmuramoyl-L-alanine amidase |
|
+1.67 |
3.6 E-03 |
|
|
| PP_4384 |
flagellar basal body L-ring protein |
|
-1.48* |
5.6 E-03 |
|
|
| Secretion systems | ||||||
| PP_0806 |
surface adhesion protein putative (same operon as type I secretion system) |
|
-1.58 |
6.2 E-03 |
-1.39 |
5.2 E-02 |
| PP_1055 |
type II secretion pathway protein GspN |
gspN |
+1.52 |
3.1 E-03 |
|
|
| PP_3087 |
excinuclease ABC A subunit putative (type VI) |
|
+1.64 |
3.1 E-03 |
+2.27 |
1.7 E-03 |
| PP_3093 |
conserved hypothetical protein (type VI) |
|
+1.65 |
6.1 E-03 |
+2.39 |
3.6 E-03 |
| PP_3094 |
hypothetical protein (type VI) |
|
+1.61 |
1.0 E-02 |
+2.60 |
3.5 E-05 |
| PP_3095 |
chaperone-associated ATPase putative (type VI) |
|
+1.60 |
2.0 E-02 |
+1.79 |
5.6 E-03 |
| PP_3096 |
hypothetical protein (type VI) |
|
+1.60 |
1.1 E-02 |
+1.97 |
2.4 E-02 |
| PP_3098 |
conserved hypothetical protein (type VI) |
|
|
|
+2.27 |
2.3 E-04 |
| PP_3099 |
hypothetical protein (type VI) |
|
+1.65 |
1.5 E-02 |
+2.00 |
1.1 E-02 |
| PP_3100 |
conserved hypothetical protein (type VI) |
|
+2.25 |
2.9 E-04 |
+2.69 |
9.6 E-04 |
| PP_3782 |
hypothetical protein |
|
+1.55 |
1.3 E-02 |
+1.91 |
7.6 E-04 |
| PP_3783 |
conserved hypothetical protein |
|
+1.60 |
2.7 E-03 |
+1.95 |
2.8 E-04 |
| PP_3784 |
conserved domain protein |
|
|
|
+1.78 |
7.7 E-03 |
| PP_3785 |
hypothetical protein |
|
+1.65 |
1.0 E-02 |
+1.87 |
3.2 E-03 |
| PP_3786 |
aminotransferase |
|
+1.45* |
6.2 E-02 |
+1.78 |
7.3 E-03 |
| PP_3787 |
hypothetical protein |
|
+2.10 |
5.3 E-03 |
+2.35 |
9.2 E-04 |
| PP_3788 |
non-ribosomal peptide synthetase putative |
|
+2.35 |
3.7 E-03 |
+2.79 |
4.2 E-05 |
| PP_3790 |
diaminopimelate epimerase |
dapF |
|
|
+1.99 |
1.4 E-03 |
| Biosynthetic metabolism (amino acids - CO2 production/consumption - glutamate metabolism) | ||||||
| PP_1143 |
3-hydroxyisobutyrate |
|
-1.69 |
1.0 E-02 |
-1.95 |
1.8 E-03 |
| PP_4617 |
leucine dehydrogenase |
|
-1.77 |
1.9 E-03 |
-1.62 |
3.0 E-03 |
| PP_4794 |
leucyl-tRNA synthetase |
leuS |
-1.92 |
3.4 E-04 |
-1.73 |
7.5 E-03 |
| PP_0432 |
N-acetyl-gamma-glutamyl-phosphate reductase |
argC |
|
|
+2.03 |
1.7 E-03 |
| PP_0997 |
sigma-54 dependent transcriptional regulator/sensory box protein |
|
-1.46* |
2.0 E-02 |
-1.88 |
4.7 E-04 |
| PP_0999 |
carbamate kinase |
arcC |
|
|
-1.72 |
1.3 E-03 |
| PP_1000 |
ornithine carbamoyltransferase |
argI |
|
|
-1.53 |
9.6 E-03 |
| PP_1001 |
arginine deiminase |
arcA |
|
|
-1.73 |
1.9 E-03 |
| PP_1002 |
arginine/ornithine antiporter |
arcD |
|
|
-1.66 |
1.2 E-02 |
| PP_1079 |
arginine deiminase |
argF |
|
|
+1.69 |
3.9 E-03 |
| PP_3775* |
sarcosine oxidase, putative |
|
|
|
+1.42 |
6.8 E-03 |
| PP_3776 |
rarD protein |
rarD-3 |
|
|
+1.71 |
3.6 E-03 |
| PP_3777 |
hypothetical protein |
|
|
|
+1.52 |
2.4 E-02 |
| PP_3780 |
hypothetical protein |
|
+1.44* |
8.0 E-03 |
+1.72 |
1.4 E-03 |
| PP_4594 |
cystathionine gamma-lyase |
|
-1.69 |
1.9 E-03 |
-1.78 |
1.9 E-02 |
| PP_1631 |
hypothetical protein (possible lysine decarboxylase) |
|
-1.91 |
3.0 E-03 |
-1.62 |
1.3 E-02 |
| PP_3662 |
decarboxylase family protein (possible lysine decarboxylase) |
|
-1.85 |
4.9 E-03 |
-1.69 |
3.9 E-03 |
| PP_1389 |
carboxyphosphonoenolpyruvate phosphonomutase, putative |
|
-1.54 |
3.7 E-03 |
|
|
| PP_5346 |
pyruvate carboxylase subunit B |
oadA |
-1.58 |
1.7 E-02 |
-1.51 |
1.1 E-01 |
| PP_5347 |
pyruvate carboxylase subunit A |
accC-2 |
-2.23 |
1.9 E-03 |
-2.41 |
5.6 E-03 |
| PP_5075 |
glutamate synthase subunit beta |
gltD |
|
|
+1.69 |
1.3 E-02 |
| PP_5409 |
glucosamine--fructose-6-phosphate aminotransferase |
glmS |
|
|
+1.54 |
1.3 E-02 |
| Electron transport | ||||||
| PP_4265 |
transcriptional regulator Anr |
anr |
+1.61 |
1.2 E-02 |
-1.99 |
2.9 E-02 |
| PP_0103 |
cytochrome c oxidase subunit II |
|
-1.41* |
1.2 E-02 |
-1.67 |
2.3 E-03 |
| PP_0104 |
cytochrome c oxidase subunit I |
|
|
|
-1.68 |
5.6 E-03 |
| PP_0111 |
electron transport protein SCO1/SenC |
|
-1.46* |
1.3 E-02 |
-1.44* |
8.7 E-03 |
| PP_0811 |
cyoups2 protein |
cyoups2 |
+1.79 |
1.9 E-03 |
+1.95 |
7.2 E-04 |
| PP_0812 |
cytochrome o ubiquinol oxidase subunit II |
cyoA |
+1.43* |
1.3 E-02 |
+1.59 |
3.0 E-03 |
| PP_0813 |
cytochrome o ubiquinol oxidase, subunit I |
cyoB |
|
|
+1.41* |
9.6 E-02 |
| PP_0816 |
protoheme IX farnesyltransferase |
cyoE-2 |
|
|
+1.41* |
2.1 E-02 |
| PP_4119 |
NADH dehydrogenase I A subunit |
nuoA |
-1.51 |
2.0 E-02 |
-1.66 |
3.6 E-03 |
| PP_4202 |
electron transfer flavoprotein beta subunit |
|
+1.42* |
6.2 E-03 |
+1.82 |
1.4 E-03 |
| PP_4250 |
cytochrome c oxidase cbb3-type, subunit II |
ccoN-1 |
+1.51 |
7.2 E-02 |
-1.56 |
1.0 E-02 |
| PP_4251 |
cytochrome c oxidase cbb3-type, subunit II |
ccoO-1 |
|
|
-2.22 |
1.7 E-03 |
| PP_4252 |
cytochrome c oxidase, cbb3-type, CcoQ subunit |
ccoQ-1 |
|
|
-1.65 |
9.2 E-02 |
| PP_4253 |
cytochrome c oxidase, cbb3-type, subunit III |
ccoP-1 |
|
|
-1.69 |
2.6 E-02 |
| PP_4255 |
cytochrome c oxidase cbb3-type, subunit I |
ccoN-2 |
-1.39* |
3.0 E-02 |
-1.55 |
7.4 E-04 |
| PP_4870 |
azurin |
azu |
+2.22 |
1.9 E-03 |
-1.19 |
2.0 E-01 |
| Fe-S cluster assembly |
|
|
|
|
|
|
| PP_0841 |
iron-sulfur cluster assembly transcription factor IscR |
iscR |
|
|
+1.67 |
2.3 E-03 |
| PP_0842 |
cysteine desulfurase |
iscS-1 |
|
|
+2.50 |
5.7 E-05 |
| PP_0843 |
iron-binding protein IscU |
iscU |
|
|
+2.48 |
1.5 E-04 |
| PP_0844 |
iron-binding protein IscA |
iscA |
|
|
+2.19 |
1.1 E-04 |
| PP_0845 |
co-chaperone HscB |
hscB |
|
|
+2.46 |
7.6 E-04 |
| PP_0846 |
chaperone protein HscA |
hscA |
|
|
+1.88 |
1.7 E-02 |
| PP_0847 |
ferredoxin, 2Fe-2S |
|
|
|
+1.72 |
2.2 E-03 |
| PP_0848 |
conserved hypothetical protein |
|
-1.38 |
4.6 E-02 |
+1.52 |
5.2 E-03 |
| PP_2378 |
yghI protein |
yghI |
|
|
+2.14 |
1.5 E-04 |
| PP_1082 |
bacterioferritin |
bfr |
-1.85 |
1.3 E-02 |
-4.20 |
1.1 E-05 |
| PP_1083 |
BFD domain protein (2Fe-2S)-binding domain protein |
|
|
|
+2.35 |
1.4 E-04 |
| PP_4900 |
iron-sulfur cluster-binding, putative |
|
-1.43* |
4.5 E-03 |
-1.49* |
7.3 E-02 |
| PP_5306 |
ferric siderophore transport system protein ExbB |
exbB |
|
|
+1.87 |
3.0 E-02 |
| PP_5307 |
ferric siderophore transport system inner membrane protein |
exbD |
|
|
+2.30 |
1.8 E-03 |
| PP_5308 |
TonB family protein |
tonB |
|
|
+1.54 |
2.4 E-02 |
| PP_0267 |
outer membrane ferric siderophore receptor putative |
|
+1.54 |
1.2 E-02 |
+1.73 |
6.6 E-03 |
| Detoxification | ||||||
| PP_2439 |
alkyl hydroxide reductase C subunit |
ahpC |
+1.87 |
3.8 E-03 |
+3.89 |
2.5 E-05 |
| PP_3639 |
alkylhydroperoxidase |
|
-1.68 |
6.7 E-03 |
-1.59 |
2.1 E-03 |
| PP_1684 |
major facilitator transporter |
|
|
|
+1.51 |
7.5 E-03 |
| PP_1686 |
glutathione peroxidase |
|
|
|
+2.11 |
3.1 E-04 |
| PP_3444 |
glyoxalase/bleomycin resistance protein/dioxygenase |
|
-1.57 |
4.5 E-03 |
-1.49* |
2.9 E-02 |
| PP_2474 |
glutathione S-transferase family protein |
|
+1.69 |
7.2 E-02 |
+1.67 |
6.7 E-02 |
| PP_3742 | glutathione S-transferase family protein | +1.53 | 5.1 E-03 | |||
The genes were sorted according to their function. Differential expression is expressed in fold change (FC) where positive and negative values indicate how many times the gene transcription is increased and decreased, respectively, compared to the Control. The significance of differential expression is given by the adjusted P-value (Adj. P-val).
* Genes with a fold change slightly below 1.5 but whose differential expression is supported by a low adjusted P-value or by the differential expression of genes belonging to the same operon.
Table 4.
Comparison between the differential expressions measured using DNA microarrays and qRT-PCR
| |
|
|
Pressure |
Pressure oxygen |
||||
|---|---|---|---|---|---|---|---|---|
| DNA microarrays | qRT-PCR | DNA microarrays | qRT-PCR | |||||
| ID |
Name |
Gene |
FC |
Adj. P-val |
FC |
FC |
Adj. P-val |
FC |
| PP_4265 |
transcriptional regulator Anr |
anr |
+ 1.6 |
1.2 E-02 |
+ 1.3 |
- 2.0 |
2.9 E-02 |
- 2.6 |
| PP_2257 |
aerotaxis receptor Aer-1 |
aer-1 |
+ 2.8 |
4.8 E-05 |
- 3.5 |
+ 2.6 |
1.1 E-05 |
- 1.7 |
| PP_2258 |
sensory box protein |
|
- 1.5 |
1.6 E-02 |
- 1.6 |
- 1.6 |
3.2 E-02 |
- 2.2 |
| PP_0504 |
outer membrane OprG |
oprG |
+ 2.5 |
5.8 E-04 |
+ 3.9 |
- 3.4 |
3.1 E-04 |
- 9.4 |
| PP_3087 |
excinuclease ABC A subunit putative (type VI) |
|
+ 1.6 |
3.1 E-03 |
+ 1.4 |
+ 2.3 |
1.7 E-03 |
+ 1.9 |
| PP_0104 |
cytochrome c oxidase subunit I |
|
n. s. |
- 2.3 |
- 1.7 |
5.6 E-03 |
- 4.8 |
|
| PP_4255 |
cytochrome c oxidase cbb3-type, subunit I |
ccoN-2 |
- 1.4 |
3.0 E-02 |
- 1.7 |
- 1.5 |
7.4 E-04 |
- 1.9 |
| PP_4870 |
azurin |
azu |
+ 2.2 |
1.9 E-03 |
+ 2.0 |
- 1.2 |
2.0 E-01 |
- 1.9 |
| PP_2439 | alkyl hydroxide reductase C subunit | ahpC | + 1.9 | 3.8 E-03 | + 2.1 | + 3.9 | 2.5 E-05 | + 3.7 |
Nine of the most relevant genes affected by elevated pressure (Pressure) and elevated pressure and dissolved oxygen (Pressure Oxygen) were tested. Differential expression is given in fold change (FC) where positive and negative values indicate how many times the gene transcription is increased and decreased, respectively, compared to the Control. The significance of differential expression is given by the adjusted P-value (Adj. P-value).
n. s. = no significant differential expression measured.
Role of methyl-accepting chemotaxis sensory transducers PP_2643 and PP_2257 (aer-1) at 7 bar
The gene with the highest significance level of differential expression for both the Pressure and the Pressure Oxygen conditions was the methyl-accepting sensory transducer gene PP_2643 (down-regulated in both conditions, Additional file 1: Table S4). In addition, the aerotaxis receptor gene aer-1 (PP_2257) and its co-transcript PP_2258 were both shown to be repressed under these two conditions by qRT-PCR (Table 4).
Bacterial aerotaxis receptors detect changes in the redox state of the electron transport system via an FAD-containing PAS (Per-Arnt-Sim) system and enable the cells to move towards more favorable environments in a similar way as for photo- or redox-taxis [38]. Aer-1 is one of the three Aer-like receptors of P. putida KT2440, along with Aer-2 (PP_2111) and Aer-3 (PP_4521). These three receptors are all localized at one pole on the cell and have a sequence similarity: 1. predicted PAS homology domain with signature residues for the binding of FAD; 2. hydrophobic membrane-spanning region; 3. methyl-accepting chemotaxis protein (MCP) domain [39]. Whereas a lack of Aer-2 mediates defects in metabolism-dependent taxis and aerotaxis, mutations of aer-1 and aer-3 genes have no reported phenotypes [39]. The gene aer-1 forms a bicistronic operon with PP_2258 which codes for a sensory box protein and whose mutation was shown to cause motility defects in P. putida KT2440 [39].
Since both the DOT level and the medium composition were the same between the Control and the Pressure conditions (Table 1) they cannot be responsible for the repression of aer-1 and PP_2258 transcription. This suggests that this aerotaxis operon may have an alternative role in sensing pressure and/or DCT, possibly via detection of changes in the redox state of the cell.
Regarding PP_2643, no information about its function or regulation is available to date. Nevertheless, since PP_2643 is a member of the MCP family one could speculate that it may work as a partner of aer-1 and PP_2258 for sensing and/or responding to environmental changes. In agreement with it, PP_2643 transcription was significantly repressed in both the Pressure and Pressure Oxygen conditions.
Activation of heat-shock response and repression of the expression of the cold-shock protein CspA and DNA repair proteins at 7 bar
The DNA microarray data revealed the induction of the sigma factor responsible for heat-shock response (RpoH) and of the heat-shock proteins HtpG, GrpE, GroES and GroEL at elevated pressure, i. e. both in the Pressure and Pressure Oxygen conditions (Table 3). RpoH induction is not restricted to dealing with temperature increase but plays a more general role in stress response: it can occur following DNA damage, oxidative stress, exposure to antibiotics and heavy metals, phage infection, and carbon source or amino acid starvation [40]. In addition, heat-shock induction was observed at high pressure (300 bar and above) [41-43] and heat-shock proteins were shown to provide protection against high-pressure induced damage [41].
Quite surprisingly, high pressure does not only induce heat-shock in bacteria but also cold-shock [34,43,44]. The heat-shock and cold-shock responses have been proposed to perform complementary functions, the former coping with protein denaturation/destabilization and the latter helping to maintain membrane fluidity and efficient protein synthesis [27,45-48]. In addition to these two stress responses, Aertsen et al. observed induction of the SOS response (uvrA, recA, and sulA) in E. coli from 750 bar, possibly caused by the disassembly of multisubunit proteins involved in DNA replication [36].
In contrast to these studies performed at high pressure, we observed a repression of the major cold-shock protein CspA (PP_1522) at moderate pressure (Table 3). Also repressed were the DNA repair genes uvrA and nth as well as the group II-intron maturase PP_1624 and the putative antirestriction protein PP_2295. However, RecX, which inhibits the DNA repair protein RecA but is co-transcribed with it [49,50], was induced in both conditions. Therefore, it seems that the DNA repair system and possibly the cold-shock response were to some extent affected for P. putida KT2440 at 7 bar but in the opposite way as reported for E. coli at much higher pressure.
Induction of detoxification agents at elevated pressure
P. putida possesses several detoxification agents dealing with reactive oxygen species (ROS): superoxide dismutases (PP_0915, PP_0946), catalases (PP_0115, PP_0481, PP_2887, PP_2668), the alkylhydroperoxide reductase Ahp (PP_2439-2440, PP_3639), and other various peroxidases (PP_0777, PP_1686, PP_1859, PP_1874, PP_2943, PP_3248, PP_3587). Further antioxidant agents are thioredoxins (PP_0786, PP_5069), glutaredoxins (PP_2958, PP_5054), and glutathione reductase (PP_3819). Exclusively ahpC and genes related with glutathione were differentially expressed in the Pressure and the Pressure Oxygen conditions, with higher extent and significance level for the Pressure Oxygen condition. Induction of ahpC transcription in both conditions was confirmed by qRT-PCR (Table 4). Surprisingly, none of the differentially expressed genes coded for catalases or superoxide dismutases (Table 3). This suggests, first, that the increase in DOT between the Pressure and Pressure Oxygen cultivations did not generate a large production of reactive oxygen species and second, that a mild oxidative stress-like response was induced at elevated pressure. The latter statement is also supported by the differential expression of genes involved in electron transport and in iron homeostasis observed at 7 bar (see below).
Effect of elevated pressure on the cell envelope
One of the most striking outcomes of the DNA microarray analyses was the impressive number of cell envelope proteins that were differentially regulated at elevated pressure. Amongst these were found porins, transporters, secretion system proteins, and various other membrane proteins with known or unknown functions (Table 3). In particular, the outer membrane porins OprG and OprH were induced with a very high significance level in the Pressure condition (Additional file 1: Table S4). OprH was strongly induced in the Pressure Oxygen condition as well but not OprG which was repressed, very likely because of a direct control by the oxygen sensor Anr [51]. The repression and induction of oprG transcription for Pressure and Pressure Oxygen, respectively, were validated by qRT-PCR (Table 4). The role of OprG porin has not been clearly established yet and contradictory results were reported about a possible function in iron and antibiotic uptake [51-54]. In contrast, OprH was shown to be regulated by the PhoP/PhoQ two-component system and overproduced under Mg2+-, Ca2+-, Mn2+-, and Sr2+-low conditions, possibly to stabilize the outer membrane by replacing the divalent cations [55,56]. The induction of these two outer membrane porins at elevated pressure might therefore be the consequence of a destabilized membrane.
A further interesting finding was the clear overexpression of two secretion systems located in the cell membrane at 7 bar: the PP_3084-3101 type VI secretion system and the PP_3775-3790 secretion system possibly involved in phytotoxicity (Table 3). Type VI secretion systems have been described for the first time in 2006 in Vibrio cholerae and Pseudomonas aeruginosa[57,58]. They are involved in the transport of proteins across the cell envelope of Gram-negative bacteria and are key virulence factors of several pathogenic bacteria [59]. Since they are also present in non-pathogenic organisms, alternative functions in host/symbiont communication, biofilm formation, quorum sensing modulation and general stress response have been proposed [60-62]. P. putida KT2440 possesses three such secretion systems: PP_2610-2632, PP_3084-3101, and PP_4066-4085 [61]. The second of these secretion systems was found to be induced both in the Pressure and the Pressure Oxygen conditions (results confirmed by qRT-PCR, see Table 4). Reva et al. showed that one gene of this secretion system (PP_3091) was repressed (and not induced) upon exposure to low temperature, urea, and benzoate stress [16]. This suggests that this type VI secretion system is sensitive to changes of environmental conditions in general. Regarding the PP_3775-3790 secretion system, it is located in a genomic island with atypical sequence features that was probably acquired from exogenous sources [63,64]. This gene cluster contains two distinct parts: ORFs PP_3781-3790 that are involved in the biosynthesis of lipodepsinonapeptide phytotoxins [65,66] and ORFs PP_3775-3780 that have been linked to amino acid biosynthesis and modification [64]. Two members of the H-NS-like MvaT family of transcriptional regulators have been reported to regulate these two gene clusters: TurA mediates the transcription of both PP_3781-3790 and PP_3775-3780 putative operons and its paralogue TurB regulates the transcription of PP_3781-3790 only [64]. In our experiments, we observed an induction of PP_3781-3790 for the Pressure condition and of both PP_3781-3790 and PP_3775-3780 for the Pressure Oxygen, which suggests that TurB and TurA, respectively, may play a role in the gene regulation under these two growth conditions.
Effect of elevated DCT on P. putida transcriptome
Although not very large, the increase in the concentration of dissolved CO2 species [CO2]tot between 1 bar and at 7 bar might be responsible for some of the gene expression changes observed (Table 1). Indeed, we noticed in a previous work that P. putida KT2440 cultivated at 1 bar with DCT levels similar as those found in the Pressure and Pressure Oxygen conditions underwent a few physiological changes (e. g. growth rate, cell yields) compared to cells cultivated at DCT levels similar to those found in the Control condition [2].
High DCT can (i) affect the function of biological membranes thereby interfering with cell division, substrate uptake and transport, (ii) acidify the internal pH, (iii) affect carboxylation/decarboxylation reactions, (iv) alter the physico-chemical properties of enzymes and their function, and (v) regulate virulence and toxin production in several pathogens [67-69].
Perturbations of the cell membrane and induction of virulence-like responses at elevated pressure were strongly suggested by the transcriptome analyses (see above) but these effects may not be restricted to the action of CO2. Only a small number of genes encoding enzymes involved in carboxylation/decarboxylation reactions (PP_1631, PP_1389, PP_3662, PP_5346/oadA, PP_5347/accC-2) were differentially regulated in the Pressure and Pressure Oxygen conditions (Table 3). For some genes, the differential expression was in agreement with an increase of DCT but for others it was not, possibly because of an additional role of these the corresponding enzymes in pH control (e. g. pyruvate carboxylases, see below).
The effect of low pH on E. coli transcriptome was studied by Maurer et al. who observed an acceleration of acid consumption and proton export as well as the coinduction of oxidative stress and heat shock regulons [70]. In addition, low pH was shown to induce the uptake of membrane permeable acids that dissipate the proton potential [71] and to repress sugar transporters and the maltose regulon in order to reduce sugar fermentation and the production of small acids [72]. In contrast, the tricarboxylic acid cycle (TCA) consumes acids and decarboxylation of amino acids (lysine, arginine) produces alkaline amines, which makes them suitable for counterbalancing pH acidification [73-75]. Also, urease, which produces two molecules of ammonia and one of CO2, is used by some microorganisms to increase the pH [76,77]. However, it should be noted that decarboxylases produce CO2 molecules which is in general unfavorable at large DCT. Lastly, acidic pH has been linked to virulence in different pathogens [78-80].
In contrast to most studies investigating the effect of low pH stress, the external pH was constantly controlled to ~ 7 during the experiments presented here. This means that if acidification of the cytoplasm occurred at elevated pressure, it was caused by an increase in the intracellular DCT and not by a flux of protons arising from the extracellular medium. As a result, differences can be expected compared with the studies mentioned above.
The DNA microarray data did not indicate a strong acidification of the cytoplasm. Therefore, the changes observed in the cell envelope and the induction of the virulence-like secretion systems at 7 bar - both of which have been linked to pH acidification and elevated DCT as mentioned above - are more likely to be explained by an elevated DCT than by internal acidification.
Role of the oxygen sensor Anr and effect on the electron transport machinery at elevated pressure and at combined elevated pressure and DOT
Anr is the global transcriptional regulator responsible for oxygen sensing in P. putida and is the homologue of E. coli Fnr [81,82]. Its level has been reported to increase following oxygen limitation, leading to the induction of the arginine deiminase (ADI) pathway, nitrate respiration, hydrogen cyanide biosynthesis and to the overexpression of azurin and oprG in P. aeruginosa[51,82,83]. Anr also coordinates the regulation of three terminal oxidases (see below) and seems to be involved in the regulation of PHB biosynthesis genes in Pseudomonas extremaustralis[84]. The DNA microarray analyses we performed revealed a decrease in the transcription of anr for the Pressure Oxygen condition, which was confirmed by qRT-PCR (Table 4) and is in line with its role as oxygen sensor. Furthermore, Anr repression was supported by the differential expression of three ADI genes arc, argI, and arcA (PP_0999, PP_1000, and PP_1001), of azurin (azu, PP_4870; confirmed by qRT-PC, see Table 4), of oprG (PP_0504), of the Cyo genes cyoA, cyoB and cyoE-2 (PP_0812, PP_0813 and PP0816), and of the cbb3-1 type genes ccoO-1, ccoQ-1, ccoN-1 and ccoP-1 (PP_4250-4253) (Table 3). Unexpectedly, the transcription of anr was found to be induced in the Pressure condition from both DNA microarray and qRT-PCR experiments (Table 4). This suggests that Anr may play an alternative role in the sensing of pressure, carbon dioxide or stress in general.
P. putida KT2440 contains five different terminal oxidases (Figure 3A) with presumably different redox properties, affinity for oxygen and ability to pump protons: the cyanide-insensitive oxidase (CIO), the cytochrome o ubiquinol oxidase (Cyo), the cytochrome aa3 oxidase (Aa3), the cytochrome cbb3-1 oxidase (Cbb3-1) and the cytochrome cbb3-2 oxidase (Cbb3-2). Regulation of the terminal oxidases is quite complex and has not been completely unraveled to date. It involves several regulators amongst which the oxygen sensor Anr [85], the Cyo terminal oxidase [86], and the global regulator Crc (computational prediction) [87] have been identified.
Figure 3.
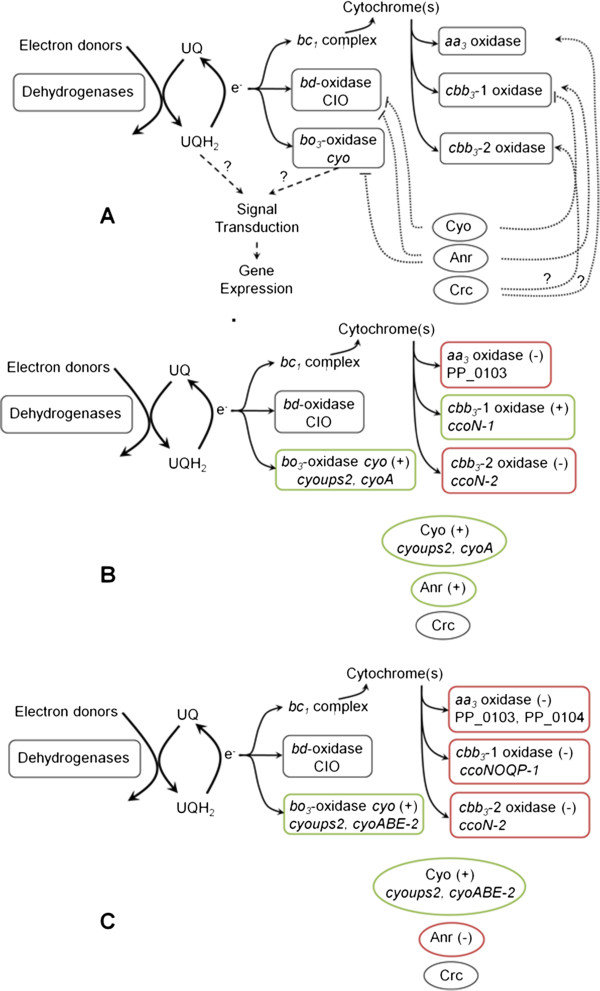
Changes of gene transcription related to the electron transport machinery. A. Model of P. putida KT2440 electron transport chain, adapted from [116]. Electrons are delivered from different electron donors to the ubiquinones located in the cell membrane (UQ). They are then transferred to the terminal ubiquinol oxidases CIO or Cyo, or are directed to the bc1 complex. In the latter case, electrons are fed to the terminal oxidases Aa3, Cbb3-1, or Cbb3-2 via cytochromes. The Cyo oxidase as well as the global regulators Anr and Crc are believed to modulate the expression of some of the terminal oxidase genes (dotted lines, see the text for the details). B. Genes coding for the five terminal oxidases and for their proposed regulators whose expression was up-regulated (+) or down-regulated (-) in the Pressure condition. C. Same as panel B but for the Pressure Oxygen condition.
Cytochrome cbb3-type oxidases have a very high affinity for oxygen and are therefore mostly important under oxygen-limiting conditions [88]. Expression of Cbb3-1 is activated by Anr [85], repressed by Cyo [86] and, together with Cbb3-2, possibly regulated by Crc [87]. In contrast to Cbb3, Cyo is predominant under high oxygen levels. It is repressed by Anr under low-oxygen conditions and during stationary phase and also acts as a global regulator [85,86,89]. CIO expression is repressed by Anr when the cells enter the stationary phase or during growth under oxygen-limiting conditions [85] and is in addition negatively regulated by Cyo [86]. Since Cyo is scarce whereas Anr is in its active form when oxygen is limiting, their effects on CIO regulation are antagonistic. As a matter of fact, the expression of CIO is induced under these conditions which means that the action of CIO is predominant [86] and it was proposed that Anr regulation is used by the cells to prevent an excessive expression of CIO compared to the other terminal oxidases [85]. Lastly, Cbb3-2 and Aa3 are neither regulated by Anr nor by Cyo [85,86] but their expression may be controlled by Crc [87].
The composition of terminal oxidases has been reported to vary with pressure in the piezophilic bacteria Shewanella sp. strain DB-172 F: this strain expressed cytochrome c as terminal oxidase at 1 bar but quinol oxidase at 600 bar [90]. The transcriptome analyses performed in this work revealed changes in the expression of terminal oxidases even at much more moderate pressure: the cytochrome c oxidases Aa3 and Cbb3-2 were repressed at 7 bar while the cytochrome o ubiquinol oxidase Cyo was induced (Table 3, Figure 3B and C). These differential expressions probably arose from the elevated pressure but an effect of the higher DCT or of a change in the cellular redox state cannot be excluded, either.
As expected from the higher DOT, Cbb3-1 (ccoNOQP-1) and Cbb3-2 (ccoN-2) were repressed in the Pressure Oxygen condition (Table 3, Figure 3C) and the repression of the latter was validated by qRT-PCR (Table 4). The down-regulation of Cbb3-1 is in agreement with the repression of Anr (PP_4265) and the activation of Cyo. Cyo, however, was induced both for Pressure and Pressure Oxygen (cyoups-2, cyoA, and cyoups-2, cyoA, cyoB, cyoE-2, respectively) and we observed as well a repression of Cbb3-2 and Aa3 in both cultivation conditions (confirmed by qRT-PCR, Table 4). This suggests that elevated pressure and/or DCT affected the transcription of these three terminal oxidases whereas the expression CIO remained unchanged.
Influence of elevated DOT on iron-sulfur cluster assemblies and on iron homeostasis
Iron-sulfur clusters are ubiquitous cofactors of proteins that are involved in various cellular functions such as catalysis, electron transport, and environment sensing due to their redox properties [91]. These clusters are sensitive to oxygen and prone to decomposition if not deeply buried in the polypeptides [92]. Two different systems are responsible for the assembly and delivery of Fe-S clusters in E. coli: the ISC (iron-sulfur cluster) system, which functions under normal growth conditions and is inactivated under oxidative stress, and the SUF (sulfur assimilation) system, which is induced under oxidative stress and iron scarcity [91]. Only the ISC system is present in P. putida KT2440, which suggests that it is more resistant than the E. coli equivalent or that the strain possesses additional systems, not described so far, able to take over under oxidative stress. Expression of the ISC genes was clearly up-regulated in the Pressure Oxygen condition (PP_0841-0848; Table 3). It should be noted that this does not necessarily imply that the ISC machinery was more effective since Jang and Imlay showed that although the expression of ISC genes was induced by H2O2 stress in E. coli the system was inactive [93].
In addition to the ISC genes, the transcription of yhgI (PP_2378, also known as gntY or nfuA) was induced in the Pressure Oxygen condition. NfuA is involved in the biogenesis of Fe-S clusters in E. coli and Azobacter vinelandii[94,95] and may play a general role in the repair of damaged Fe-S proteins under stress conditions [94]. The nfuA gene was shown to belong to the iscR and rpoH regulons in E. coli[96,97]. Nevertheless, rpoH regulation seems to be less strict in P. putida since its overexpression did not result in the induction of NfuA/YhgI in the Pressure condition.
Not only the Fe-S clusters are sensitive to oxygen but iron itself. Indeed, Fe2+, which is soluble at physiological pH, can be oxidized into Fe3+ which readily precipitates as ferric hydroxide or forms insoluble complexes with anionic salts. Bacteria are endowed with ferritin and bacterioferritin proteins (without and with heme, respectively) that control the intracellular level of iron between ~10-3-10-5 M: they store excess iron in order to prevent the formation of ROS formation via Fenton reactions and release it according to the cellular needs [98,99]. We observed a significant repression of transcription for the bacterioferritin gene bfr (PP_1082) in the Pressure Oxygen condition but induction of the putative bacterioferritin-associated ferredoxin (PP_1083) that is located next to it in P. putida genome. Interestingly, Tuanyok and coworkers detected a similar regulation in Burkholderia pseudomallei and Burkholderia mallei when cultivated under low iron conditions [100]. This suggests that the putative bacterioferritin-associated ferredoxin may be involved in the release of iron from bacterioferritin and that the level of free iron was lower in the Pressure Oxygen condition than in the Control. Lastly, it must be noted that the expression of bfr and of two siderophore transporter genes was differentially regulated in the Pressure condition (Table 3). We propose that this apparent perturbation of iron homeostasis at 7 bar was the consequence of a mild oxidative stress.
Induction of glutamate metabolism and repression of the arginine deiminase pathway at elevated DOT
The global analyses of the transcriptome data suggested an effect of elevated DOT on glutamate biosynthesis (see above). Examination of the genes involved in glutamate metabolism [101] revealed that a glutamate synthase (PP_5075) and a gene involved in the metabolization of glutamine (PP_5409) were induced in the Pressure Oxygen condition (Table 3). The enhancement of glutamate synthesis and metabolism at elevated DOT may be explained by an acceleration of the tricarboxylic acid cycle (TCA) triggered by larger NAD+/NADH and FAD/FADH2 ratios. This acceleration of the TCA could possibly enhance the conversion of glutamate to α-ketoglutarate as well as the subsequent metabolization of the latter compound in order to reduce the α-ketoglutarate pool. Two different systems are available in P. putida KT2440 to produce glutamate from α-ketoglutarate: a system based on glutamate dehydrogenase (Gdh) that works under nitrogen non-limiting conditions and a system based on glutamate synthase (GOGAT) and glutamine synthetase (GS) that works under nitrogen limiting conditions [102,103]. All cultivations including the Control were performed under dual (carbon, nitrogen) limited conditions. Therefore, it is not surprising that the induction of glutamate metabolism we observed involved the GOGAT/GS system and not the Gdh system.
A whole operon encoding proteins from the arginine deiminase (ADI) pathway was found to be repressed in the Pressure Oxygen condition (PP_0997-PP_1002; Table 3). This pathway consists of three enzymes: the arginine deiminase which degrades arginine into citrulline and ammonia (PP_1001), the catabolic ornithine carbamoyltransferase which converts citrulline and phosphate into ornithine and carbamoyl phosphate (PP_1000) and the carbamate kinase which produces ATP, ammonia and CO2 from carbamoyl phosphate (PP_0999). In addition, the genes encoding these three enzymes are flanked with an arginine/ornithine antiporter (PP_1002) and with a sigma-54 dependent transcriptional regulator/sensory box protein (PP_0997) that is likely to regulate the transcription of the operon. Activation of the ADI pathway was reported to occur via Anr regulation in P. aeruginosa and other microorganisms under anaerobic conditions as a mean to generate ATP when terminal electron acceptors such as oxygen and nitrate are scarce [104-107]. The present transcriptome analyses are in agreement with a similar dependency between the transcription of ADI genes, the transcription of anr and oxygen availability in P. putida KT2440.
Conclusions
The DNA microarrays and qRT-PCR experiments performed within this work revealed widespread effects of elevated pressure on the transcriptome of P. putida KT2440 (Table 2). Growth at 7 bar and elevated DCT was apparently a factor of stress for P. putida KT2440, as indicated by the activation of a heat-shock response. In addition, signs of a mild oxidative stress-like response were observed, such as up-regulation of ROS detoxification genes (ahpC, glutathione metabolism genes), changes in the composition of terminal oxidases and differential expression of bacterioferritin and siderophore transporters indicating an alteration of iron homeostasis. It should be noted that these stresses could be efficiently fought by the cells, for which no major physiological defects had been detected [2]. The global regulator Anr known for modulating the cellular response to DOT changes was repressed in the Pressure Oxygen condition but, unexpectedly, induced in the Pressure condition. Thus, it can be concluded that Anr may play an alternative role in the cellular response to elevated pressure and/or DCT. Lastly, the cell envelope appeared to be strongly affected by elevated pressure and/or DCT.
Taken together, the results from the physiological and transcriptomic studies revealed that the variations in pressure, DCT and DOT were important enough to be sensed by the cells and to induce a reorganization of the gene expression pattern while being small enough not to alter significantly the cell physiology.
Furthermore, the data presented here suggest that stimulating the heat-shock and the oxidative-stress responses would be a sensible approach for enhancing the tolerance of P. putida against elevated pressure and/or DCT.
Methods
Culture conditions and sampling
P. putida KT2440 was cultivated in dual (carbon, nitrogen) limited chemostat cultivations in a 16 L high-pressure bioreactor (Bioengineering, Wald, Switzerland) as described previously [2]. The cultivation at 1 bar and DOT = 40% was used as control (Control), the one at 7 bar and DOT = 45% to study the effect of elevated pressure (Pressure), and the one at 7 bar and DOT = 235% to study the effect of combined elevated pressure and elevated DOT (Pressure Oxygen) (Table 1). Three to four culture samples were taken for each cultivation process after steady state was established and at 12 h-intervals. The samples were immediately cooled on ice for 15 min to stop cell growth and RNA degradation. They were then pelleted by centrifugation, incubated 1 h in RNAlater solution (Ambion, Austin, USA), pelleted again, flash frozen in liquid nitrogen, and stored at -80°C until RNA extraction. Aliquots of about 80 OD-unit (OD600 x volume in mL) were prepared. The samples at different time points were not pooled but treated separately and used individually in the four replicate DNA microarrays.
RNA isolation and purification
RNA isolation and purification were performed according to Yuste et al.[108] with minor variations. It involved cell lysis using TRI Reagent® LS (Molecular Research Center Inc., Cincinnati, USA), DNAse treatment with DNase I recombinant (Roche Diagnostics Corporation, Indianapolis, USA), verification of the absence of DNA contamination by polymerase chain reaction (PCR), final RNA purification with RNeasy mini kit (Qiagen, Valencia, USA), and verification of the integrity of the RNA samples by capillary electrophoresis (Agilent 2100 Bioanalyzer, California, USA).
Hybridization and processing of the microarrays
The hybridization experiments were performed with 2-color DNA microarrays designed for P. putida KT2440 [108] and printed by Progenika Biopharma (Vizcaya, Spain). Samples were fluorescently labeled with either Cy3 or Hy5, mixed, and used to hybridize the DNA microarray as reported [108]. Four replicate microarrays were used for each of the two tested conditions (elevated pressure = Pressure, and combined elevated pressure and elevated DOT = Pressure Oxygen). Microarray data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-3632.
The results were statistically analyzed using Limma software package [109] in the R environment (http://www.r-project.org). Normalization of the data was performed within arrays using the method Print-tip loess and between arrays using the method Scale[110]. A linear fit was applied to the four replicate microarrays taking into account the fact that spots were printed in duplicates [111], i. e. differential expression was calculated from eight values. The genes were tested for differential expression using the empirical Bayes-moderated t-statistics. P-values were adjusted for multiple testing with the Benjamini-Hochberg method to control the false discovery rate [112].
Data analyses
The large amount of data generated by these experiments (> 5´000 ORFs tested) was analyzed with the following strategy. In a first step, only the genes exhibiting differential expression with a high significance level were considered (adj. P-value < 0.01; Additional file 1: Table S1-S2) in order to identify the most relevant genes and cell functions associated with the response to pressure and DOT increase. Two genes (ttga and aer-1/PP_2257) were removed from this list and considered as “false positives”: the former belongs to the few extra genes added on the DNA microarray that are absent from P. putida KT2440 genome and the latter showed discrepant expression results in qRT-PCR experiments, possibly because of a lack of specificity of its DNA microarray probe. The significantly differentially expressed genes were sorted into role categories and sub-categories according to the Comprehensive Microbial Resource (CMR) [37]. Fisher’s exact tests were performed in the R environment to determine whether (sub-) categories were over-represented in the Pressure condition and in the Pressure Oxygen condition compared to the genome distribution. The tests were performed with the alternative “greater”, and the significance threshold was set to a P-value of 0.15 in order to obtain a sensible number of relevant functions. In a second step, the expression of specific genes with an adjusted P-value slightly below the significance threshold of 0.01 but with a fold change higher than 1.5 was considered as well (Table 3). Information about the genes was collected from literature search, from the Pseudomonas Genome Database publication [113], and from the Kyoto Encyclopedia of Genes and Genomes (KEGG) [101].
Real Time quantitative Reverse Transcription PCR (qRT-PCR) assay
Reverse transcription reactions for the synthesis of total cDNA were carried out with 4 μg of RNA, 0.5 mM dNTPs, 200 U of SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, USA) and 2.5 μM of random hexamers as primers in the buffer recommended by the manufacturer. Samples were initially heated at 65°C for 5 min, then incubated at 42°C for 2 h, and the amplification was terminated by incubation at 70°C for 15 min. The cDNA obtained was purified using Geneclean Turbo kit (MP Biomedicals, Santa Ana, USA) and its concentration was measured using a NanoPhotometer™ Pearl (Implen, Munich, Germany). Real time PCR was performed using iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, USA) with 0.2 μM of each primer in an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, USA). The primers used for each target gene are listed in the Additional file 1. The samples were initially denatured at 95°C for 5 min, followed by 40 cycles of amplification (95°C, 30 s; 60°C, 30 s and 72°C, 30 s). Three biological and two technical replicates were used for each sample. Data were analyzed with the 2-ΔΔCt method [114] using rpoN as internal control as its expression is known to be constant throughout the growth curve [108,115]. Moreover, we have observed that the levels of rpoN mRNA were similar under the conditions tested (data not shown).
For the analysis of the expression levels of the target gene aer-1 hydrolysis probe-based real time PCR was performed using probe n° 70 from the Universal ProbeLibrary (Roche, Basel, Switzerland), with 5 ng of purified cDNA as template, iQ™ Supermix (Bio-Rad Laboratories, Hercules, USA) and 1.25 μM of each primer (see Additional file for the sequences) in an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, USA). Samples were initially denatured at 95°C for 10 min, followed by 50 cycles of amplification (95°C, 10 s; 60°C, 30 s). Three biological and two technical replicates were used for each sample. Data was analyzed using the 2-ΔCt method [114].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SF carried out the chemostat cultivations for the production of biomass samples together with BH, performed the RNA extraction and purification together with IFE, did the data treatment and interpretation of the DNA microarray experiments with support from IFE and MAP, and drafted the manuscript. PF performed the qRT-PCR experiments. MZ and MAP participated in the design of the study and, together with SP, IFE and PF, helped with the manuscript preparation. All authors read and approved the final manuscript.
Supplementary Material
Significantly differentially expressed genes under elevated pressure (Pressure) and elevated pressure and DOT (Pressure Oxygen). Table S2. Categories and sub-categories significantly over-represented in the lists of significantly differentially expressed genes under elevated pressure. Table S3. Categories and sub-categories significantly over-represented in the lists of significantly differentially expressed genes under combined elevated pressure and elevated DOT. Table S4. Top genes with the most significant differential expression at elevated pressure (Pressure) and at combined elevated pressure and DOT (Pressure Oxygen).
Contributor Information
Stéphanie Follonier, Email: stephanie.follonier@hevs.ch.
Isabel F Escapa, Email: ifescapa@forsyth.org.
Pilar M Fonseca, Email: pfongar@gmail.com.
Bernhard Henes, Email: bernhard.henes@gmail.com.
Sven Panke, Email: sven.panke@bsse.ethz.ch.
Manfred Zinn, Email: manfred.zinn@hevs.ch.
María Auxiliadora Prieto, Email: auxi@cib.csic.es.
Acknowledgements
We are greatly indebted to Prof. José Luis García for helpful discussions. We thank warmly E. Gil and J. M. Gonzalez from the Genomic Unit of Centro Nacional de Biotechnología (CNB) in Madrid for performing the microarray hybridization experiments. We also thank A. Valencia for technical assistance. This work was partially supported by the Swiss National Science Foundation grant 315200-116812/1 and by the Spanish grant BIO2010-21049 from the Comisión Interministerial de Ciencia y Tecnología.
References
- Belo I, Pinheiro R, Mota M. Fed-batch cultivation of Saccharomyces cerevisiae in a hyperbaric bioreactor. Biotechnol Prog. 2003;19:665–671. doi: 10.1021/bp0257067. [DOI] [PubMed] [Google Scholar]
- Follonier S, Henes B, Panke S, Zinn M. Putting cells under pressure: A simple and efficient way to enhance the productivity of medium-chain-length polyhydroxyalkanoate in processes with Pseudomonas putida KT2440. Biotechnol Bioeng. 2012;109:451–461. doi: 10.1002/bit.23312. [DOI] [PubMed] [Google Scholar]
- Knabben I, Regestein L, Grumbach C, Steinbusch S, Kunze G, Büchs J. Online determination of viable biomass up to very high cell densities in Arxula adeninivorans fermentations using an impedance signal. J Biotechnol. 2010;149:60–66. doi: 10.1016/j.jbiotec.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Knabben I, Regestein L, Marquering F, Steinbusch S, Lara AR, Büchs J. High cell-density processes in batch mode of a genetically engineered Escherichia coli strain with minimized overflow metabolism using a pressurized bioreactor. J Biotechnol. 2010;150:73–79. doi: 10.1016/j.jbiotec.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Knoll A, Bartsch S, Husemann B, Engel P, Schroer K, Ribeiro B. High cell density cultivation of recombinant yeasts and bacteria under non-pressurized and pressurized conditions in stirred tank bioreactors. J Biotechnol. 2007;132:167–179. doi: 10.1016/j.jbiotec.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Lopes M, Gomes N, Gonçalves C, Coelho MAZ, Mota M, Belo I. Yarrowia lipolytica lipase production enhanced by increased air pressure. Lett Appl Microbiol. 2008;46:255–260. doi: 10.1111/j.1472-765X.2007.02299.x. [DOI] [PubMed] [Google Scholar]
- Lopes M, Gomes N, Mota M, Belo I. Yarrowia lipolytica growth under increased air pressure: Influence on enzyme production. Appl Biochem Biotechnol. 2009;159:46–53. doi: 10.1007/s12010-008-8359-0. [DOI] [PubMed] [Google Scholar]
- Ma X, Fan D, Shang L, Cai Q, Chi L, Zhu C. Oxygen transfer rate control in the production of human-like collagen by recombinant Escherichia coli. Biotechnol Appl Biochem. 2010;55:169–174. doi: 10.1042/BA20090314. [DOI] [PubMed] [Google Scholar]
- Pinheiro R, Belo I, Mota M. Air pressure effects on biomass yield of two different Kluyveromyces strains. Enzyme Microb Technol. 2000;26:756–762. doi: 10.1016/S0141-0229(00)00168-X. [DOI] [PubMed] [Google Scholar]
- Yang JD, Wang NS. Oxygen mass transfer enhancement via fermentor headspace pressurization. Biotechnol Prog. 1992;8:244–251. doi: 10.1021/bp00015a010. [DOI] [PubMed] [Google Scholar]
- Chen GQ. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev. 2009;38:2434–2446. doi: 10.1039/b812677c. [DOI] [PubMed] [Google Scholar]
- Rai R, Keshavarz T, Roether JA, Boccaccini AR, Roy I. Medium chain length polyhydroxyalkanoates, promising new biomedical materials for the future. Mater Sci Eng R. 2011;72:29–47. doi: 10.1016/j.mser.2010.11.002. [DOI] [Google Scholar]
- Zinn M, Witholt B, Egli T. Occurrence, synthesis and medical application of bacterial polyhydroxyalkanoate. Adv Drug Deliv Rev. 2001;53:5–21. doi: 10.1016/S0169-409X(01)00218-6. [DOI] [PubMed] [Google Scholar]
- Fonseca P, Moreno R, Rojo F. Growth of Pseudomonas putida at low temperature: global transcriptomic and proteomic analyses. Environ Microbiol Rep. 2011;3:329–339. doi: 10.1111/j.1758-2229.2010.00229.x. [DOI] [PubMed] [Google Scholar]
- Gülez G, Dechesne A, Workman CT, Smets BF. Transcriptome dynamics of Pseudomonas putida KT2440 under water stress. Appl Environ Microb. 2012;78:676–683. doi: 10.1128/AEM.06150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva ON, Weinel C, Weinel M, Bohm K, Stjepandic D, Hoheisel JD. Functional genomics of stress response in Pseudomonas putida KT2440. J Bacteriol. 2006;188:4079–4092. doi: 10.1128/JB.00101-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett DH. Microbial life at high-pressures. Sci Prog. 1992;76:479–496. [PubMed] [Google Scholar]
- ZoBell CE, Johnson FH. The influence of hydrostatic pressure on the growth and viability of terrestrial and marine bacteria. J Bacteriol. 1949;57:179–189. doi: 10.1128/jb.57.2.179-189.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheftel JC. Review: High-pressure, microbial inactivation and food preservation. Food Sci Technol Int. 1995;1:75–90. doi: 10.1177/108201329500100203. [DOI] [Google Scholar]
- Brouillet M, Gautier H, Miègeville AF, Bouler JM, Merle C, Caillon J. Inactivation of Staphylococcus aureus in calcium phosphate biomaterials via isostatic compression. J Biomed Mater Res B Appl Biomater. 2009;91B:348–353. doi: 10.1002/jbm.b.31408. [DOI] [PubMed] [Google Scholar]
- Diehl P, Schmitt M, Schauwecker J, Eichelberg K, Gollwitzer H, Gradinger R. Effect of high hydrostatic pressure on biological properties of extracellular bone matrix proteins. Int J Mol Med. 2005;16:285–289. [PubMed] [Google Scholar]
- Diehl P, Steinhauser E, Gollwitzer H, Heister C, Schauwecker J, Milz S. Biomechanical and immunohistochemical analysis of high hydrostatic pressure-treated Achilles tendons. J Orthop Sci. 2006;11:380–385. doi: 10.1007/s00776-006-1031-z. [DOI] [PubMed] [Google Scholar]
- Diehl P, Schauwecker J, Mittelmeier W, Schmitt M. High hydrostatic pressure, a novel approach in orthopedic surgical oncology to disinfect bone, tendons and cartilage. Anticancer Res. 2008;28:3877–3883. [PubMed] [Google Scholar]
- Gollwitzer H, Mittelmeier W, Brendle M, Weber P, Miethke T, Hofmann GO. High hydrostatic pressure for disinfection of bone grafts and biomaterials: An experimental study. Open Orthop J. 2009;3:1–7. doi: 10.2174/1874325000903010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naal FD, Schauwecker J, Steinhauser E, Milz S, von Knoch F, Mittelmeier W. Biomechanical and immunohistochemical properties of meniscal cartilage after high hydrostatic pressure treatment. J Biome. 2008;87B:19–25. doi: 10.1002/jbm.b.31059. [DOI] [PubMed] [Google Scholar]
- Michiels C, Bartlett DH, Aertsen A. High-Pressure Microbiology. Washington: ASM Press; 2008. [Google Scholar]
- Mackey B, Mañas P. In: High-pressure microbiology. Michiels C, Bartlett DH, Aertsen A, editor. Washington DC: ASM Press; 2008. Inactivation of Escherichia coli by high pressure; pp. 53–85. [Google Scholar]
- Pagán R, Mackey B. Relationship between membrane damage and cell death in pressure-treated Escherichia coli cells: differences between exponential- and stationary-phase cells and variation among strains. Appl Environ Microb. 2000;66:2829–2834. doi: 10.1128/AEM.66.7.2829-2834.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZoBell CE, Cobet AB. Filament formation by Escherichia coli at increased hydrostatic pressures. J Bacteriol. 1964;87:710–719. doi: 10.1128/jb.87.3.710-719.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meganathan R, Marquis RE. Loss of bacterial motility under pressure. Nature. 1973;246:525–527. doi: 10.1038/246525a0. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Yayanos AA. Adaptation of the membrane lipids of a deep-sea bacterium to changes in hydrostatic pressure. Science. 1985;228:1101–1103. doi: 10.1126/science.3992247. [DOI] [PubMed] [Google Scholar]
- Bartlett D, Wright M, Yayanos AA, Silverman M. Isolation of a gene regulated by hydrostatic pressure in a deep-sea bacterium. Nature. 1989;342:572–574. doi: 10.1038/342572a0. [DOI] [PubMed] [Google Scholar]
- Welch TJ, Bartlett DH. Isolation and characterization of the structural gene for OmpL, a pressure-regulated porin-like protein from the deep-sea bacterium Photobacterium species strain SS9. J Bacteriol. 1996;178:5027–5031. doi: 10.1128/jb.178.16.5027-5031.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A, Oshima T, Sato T, Nakasone K, Mori H, Kato C. Analysis of hydrostatic pressure effects on transcription in Escherichia coli by DNA microarray procedure. Extremophiles. 2005;9:65–73. doi: 10.1007/s00792-004-0414-3. [DOI] [PubMed] [Google Scholar]
- Qureshi MH, Kato C, Horikoshi K. Purification of a ccb-type quinol oxidase specifically induced in a deep-sea barophilic bacterium, Shewanella sp. strain DB-172 F. Extremophiles. 1998;2:93–99. doi: 10.1007/s007920050047. [DOI] [PubMed] [Google Scholar]
- Aertsen A, Van Houdt R, Vanoirbeek K, Michiels CW. An SOS response induced by high pressure in Escherichia coli. J Bacteriol. 2004;186:6133–6141. doi: 10.1128/JB.186.18.6133-6141.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen T, Beck E, Ganapathy A, Montgomery R, Zafar N, Yang Q. The comprehensive microbial resource. Nucl Acids Res. 2010;38:D340–D345. doi: 10.1093/nar/gkp912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL. Aer on the inside looking out: paradigm for a PAS–HAMP role in sensing oxygen, redox and energy. Mol Microbiol. 2007;65:1415–1424. doi: 10.1111/j.1365-2958.2007.05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarand I, Österberg S, Holmqvist S, Holmfeldt P, Skärfstad E, Parales RE. Metabolism-dependent taxis towards (methyl)phenols is coupled through the most abundant of three polar localized Aer-like proteins of Pseudomonas putida. Environ Microbiol. 2008;10:1320–1334. doi: 10.1111/j.1462-2920.2007.01546.x. [DOI] [PubMed] [Google Scholar]
- Foster PL. Stress responses and genetic variation in bacteria. Mutat Res Fund Mol Mech Mutagen. 2005;569:3–11. doi: 10.1016/j.mrfmmm.2004.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aertsen A, Vanoirbeek K, De Spiegeleer P, Sermon J, Hauben K, Farewell A. Heat shock protein-mediated resistance to high hydrostatic pressure in Escherichia coli. Appl Environ Microb. 2004;70:2660–2666. doi: 10.1128/AEM.70.5.2660-2666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß M, Kosmowsky IJ, Lorenz R, Molitoris HP, Jaenicke R. Response of bacteria and fungi to high-pressure stress as investigated by two-dimensional polyacrylamide gel electrophoresis. Electrophoresis. 1994;15:1559–1565. doi: 10.1002/elps.11501501224. [DOI] [PubMed] [Google Scholar]
- Welch TJ, Farewell A, Neidhardt FC, Bartlett DH. Stress-response of Escherichia coli to elevated hydrostatic pressure. J Bacteriol. 1993;175:7170–7177. doi: 10.1128/jb.175.22.7170-7177.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic M, Hörmann S, Vogel R, Ehrmann M. Transcriptional response reveals translation machinery as target for high pressure in Lactobacillus sanfranciscensis. Arch Microbiol. 2005;184:11–17. doi: 10.1007/s00203-005-0021-4. [DOI] [PubMed] [Google Scholar]
- Aertsen A, Michiels C. In: High-pressure microbiology. Michiels C, Bartlett DH, Aertsen A, editor. Washington DC: ASM Press; 2008. Cellular impact of sublethal pressures on Escherichia coli; pp. 87–100. [Google Scholar]
- Bartlett DH, Kato C, Horikoshi K. High-pressure influences on gene and protein expression. Res Microbiol. 1995;146:697–706. doi: 10.1016/0923-2508(96)81066-7. [DOI] [PubMed] [Google Scholar]
- Broeze RJ, Solomon CJ, Pope DH. Effects of low temperature on in vivo and in vitro protein synthesis in Escherichia coli and Pseudomonas fluorescens. J Bacteriol. 1978;134:861–874. doi: 10.1128/jb.134.3.861-874.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieringer HA, Jones PG, Inouye M. Cold shock and adaptation. BioEssays. 1998;20:49–57. doi: 10.1002/(SICI)1521-1878(199801)20:1<49::AID-BIES8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Pagès V, Koffel-Schwartz N. Fuchs RPP: recX, a new SOS gene that is co-transcribed with the recA gene in Escherichia coli. DNA Repair. 2003;2:273–284. doi: 10.1016/S1568-7864(02)00217-3. [DOI] [PubMed] [Google Scholar]
- Stohl EA, Brockman JP, Burkle KL, Morimatsu K, Kowalczykowski SC, Seifert HS. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J Biol Chem. 2003;278:2278–2285. doi: 10.1074/jbc.M210496200. [DOI] [PubMed] [Google Scholar]
- McPhee JB, Tamber S, Bains M, Maier E, Gellatly S, Lo A. The major outer membrane protein OprG of Pseudomonas aeruginosa contributes to cytotoxicity and forms an anaerobically regulated, cation-selective channel. FEMS Microbiol Lett. 2009;296:241–247. doi: 10.1111/j.1574-6968.2009.01651.x. [DOI] [PubMed] [Google Scholar]
- Chamberland S, Bayer AS, Schollaardt T, Wong SA, Bryan LE. Characterization of mechanisms of quinolone resistance in Pseudomonas aeruginosa strains isolated in vitro and in vivo during experimental endocarditis. Antimicrob Agents Chemother. 1989;33:624–634. doi: 10.1128/AAC.33.5.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Xu C, Ren H, Lin X, Wu L, Wang S. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Pseudomonas aeruginosa responding to ampicilin, kanamycin, and tetracycline resistance. J Proteome Res. 2005;4:2257–2265. doi: 10.1021/pr050159g. [DOI] [PubMed] [Google Scholar]
- Yates JM, Morris G, Brown MRW. Effect of iron concentration and growth rate on the expression of protein G in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1989;58:259–262. doi: 10.1111/j.1574-6968.1989.tb03055.x. [DOI] [PubMed] [Google Scholar]
- Hancock REW, Siehnel R, Martin N. Outer membrane proteins of Pseudomonas. Mol Microbiol. 1990;4:1069–1075. doi: 10.1111/j.1365-2958.1990.tb00680.x. [DOI] [PubMed] [Google Scholar]
- McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J Bacteriol. 2006;188:3995–4006. doi: 10.1128/JB.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle LEH, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Aschtgen MS, Bernard CS, De Bentzmann S, Lloubes R, Cascales E. SciN Is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol. 2008;190:7523–7531. doi: 10.1128/JB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genom. 2009;10:104–117. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Hasic M, Chen C, Wai SN, Milton DL. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ Microbiol. 2009;11:3018–3028. doi: 10.1111/j.1462-2920.2009.02005.x. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VAP. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- Renzi F, Rescalli E, Galli E, Bertoni G. Identification of genes regulated by the MvaT-like paralogues TurA and TurB of Pseudomonas putida KT2440. Environ Microbiol. 2010;12:254–263. doi: 10.1111/j.1462-2920.2009.02064.x. [DOI] [PubMed] [Google Scholar]
- Bender CL, Alarcon-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos VAPM, Heim S, Moore ERB, Strätz M, Timmis KN. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ Microbiol. 2004;6:1264–1286. doi: 10.1111/j.1462-2920.2004.00734.x. [DOI] [PubMed] [Google Scholar]
- Dixon NM, Kell DB. The inhibition by CO2 of the growth and metabolism of micro-organisms. J Appl Bacteriol. 1989;67:109–136. doi: 10.1111/j.1365-2672.1989.tb03387.x. [DOI] [PubMed] [Google Scholar]
- Stretton S, Marshall KC, Dawes IW, Goodman AE. Characterisation of carbon dioxide-inducible genes of the marine bacterium, Pseudomonas sp. S91. FEMS Microbiol Lett. 1996;140:37–42. doi: 10.1111/j.1574-6968.1996.tb08311.x. [DOI] [PubMed] [Google Scholar]
- Stretton S, Goodman AE. Carbon dioxide as a regulator of gene expression in microorganisms. Antonie Leeuwenhoek. 1998;73:79–85. doi: 10.1023/A:1000610225458. [DOI] [PubMed] [Google Scholar]
- Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol. 2005;187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JB, Diez-Gonzalez F. In: Advances in Microbial Physiology. Volume 39. Poole RK, editor. Waltham: Academic; 1997. The effects of fermentation acids on bacterial growth; pp. 205–234. [DOI] [PubMed] [Google Scholar]
- Chagneau C, Heyde M, Alonso S, Portalier R, Laloi P. External-pH-dependent expression of the maltose regulon and ompF gene in Escherichia coli is affected by the level of gycerol kinase, encoded by glpK. J Bacteriol. 2001;183:5675–5683. doi: 10.1128/JB.183.19.5675-5683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger EA, Redding KE, Plumb T, Childs LC, Meng SY, Bennett GN. Construction of lac fusions to the inducible arginine-and lysine decarboxylase genes of Escherichia coli K12. Mol Microbiol. 1989;3:609–620. doi: 10.1111/j.1365-2958.1989.tb00208.x. [DOI] [PubMed] [Google Scholar]
- Gong S, Richard H, Foster JW. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J Bacteriol. 2003;185:4402–4409. doi: 10.1128/JB.185.15.4402-4409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J Bacteriol. 2002;184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DR, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterol. 1998;114:58–70. doi: 10.1016/S0016-5085(98)70633-X. [DOI] [PubMed] [Google Scholar]
- Sissons CH, Perinpanayagam HER, Hancock EM, Cutress TW. pH regulation of urease levels in Streptococcus salivarius. J Dent Res. 1990;69:1131–1137. doi: 10.1177/00220345900690050301. [DOI] [PubMed] [Google Scholar]
- Bearson BL, Wilson L, Foster JW. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J Bacteriol. 1998;180:2409–2417. doi: 10.1128/jb.180.9.2409-2417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behari J, Stagon L. Calderwood SB: pepA, a gene mediating pH regulation of virulence genes in Vibrio cholerae. J Bacteriol. 2001;183:178–188. doi: 10.1128/JB.183.1.178-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D, Tatham D, Williamson ED, Titball RW. The pH 6 antigen of Yersinia pestis binds to b1-linked galactosyl residues in glycosphingolipids. Infect Immun. 1998;66:4545–4548. doi: 10.1128/iai.66.9.4545-4548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M, Gamper M, Zimmermann A, Haas D. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol. 1991;173:1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann A, Reimmann C, Galimand M, Haas D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol Microbiol. 1991;5:1483–1490. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]
- Vijgenboom E, Busch JE, Canters GW. In vivo studies disprove an obligatory role of azurin in denitrification in Pseudomonas aeruginosa and show that azu expression is under control of RpoS and ANR. Microbiology. 1997;143:2853–2863. doi: 10.1099/00221287-143-9-2853. [DOI] [PubMed] [Google Scholar]
- Tribelli PM, Méndez BS, López NI. Oxygen-sensitive global regulator, Anr, is involved in the biosynthesis of poly(3-hydroxybutyrate) in Pseudomonas extremaustralis. J Mol Microbiol Biotechnol. 2010;19:180–188. doi: 10.1159/000320261. [DOI] [PubMed] [Google Scholar]
- Ugidos A, Morales G, Rial E, Williams HD, Rojo F. The coordinate regulation of multiple terminal oxidases by the Pseudomonas putida ANR global regulator. Environ Microbiol. 2008;10:1690–1702. doi: 10.1111/j.1462-2920.2008.01586.x. [DOI] [PubMed] [Google Scholar]
- Morales G, Linares JF, Beloso A, Albar JP, Martinez JL, Rojo F. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J Bacteriol. 2004;186:1337–1344. doi: 10.1128/JB.186.5.1337-1344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne P, Barret M, O'Gara F, Morrissey J. Computational prediction of the Crc regulon identifies genus-wide and species-specific targets of catabolite repression control in Pseudomonas bacteria. BMC Microbiol. 2010;10:300. doi: 10.1186/1471-2180-10-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HD, Zlosnik JEA, Ryall B. In: Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Robert KP, editor. Vol. 52. Waltham: Academic; 2006. pp. 1––71. (Advances in Microbial Physiology). [DOI] [PubMed] [Google Scholar]
- Dinamarca MA, Aranda-Olmedo I, Puyet A, Rojo F. Expression of the Pseudomonas putida OCT plasmid alkane degradation pathway is modulated by two different global control signals: evidence from continuous cultures. J Bacteriol. 2003;185:4772–4778. doi: 10.1128/JB.185.16.4772-4778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi MH, Kato C, Horikoshi K. Purification of two pressure-regulated c-type cytochromes from a deep-sea barophilic bacterium, Shewanella sp. strain DB-172 F. FEMS Microbiol Lett. 1998;161:301–309. doi: 10.1111/j.1574-6968.1998.tb12961.x. [DOI] [Google Scholar]
- Lill R. Function and biogenesis of iron–sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol Microbiol. 2010;78:1448–1467. doi: 10.1111/j.1365-2958.2010.07418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini S, Gerez C, Choudens SO, Sanakis Y, Fontecave M, Barras F. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J Biol Chem. 2008;283:14084–14091. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Naik SG, O'Carroll IP, Huynh BH, Dean DR, Johnson MK. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J Biol Chem. 2008;283:14092–14099. doi: 10.1074/jbc.M709161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol. 2006;60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-shock factor, s32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006;20:1776–1789. doi: 10.1101/gad.1428206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrondo MA. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 2003;22:1959–1968. doi: 10.1093/emboj/cdg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil EC. Ferritin. Ltd: John Wiley & Sons; 2006. [Google Scholar]
- Tuanyok A, Kim HS, Nierman WC, Yu Y, Dunbar J, Moore RA. Genome-wide expression analysis of iron regulation in Burkholderia pseudomallei and Burkholderia mallei using DNA microarrays. FEMS Microbiol Lett. 2005;252:327–335. doi: 10.1016/j.femsle.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas AB, Canosa I, Santero E. Transcriptome analysis of Pseudomonas putida in response to nitrogen availability. J Bacteriol. 2008;190:416–420. doi: 10.1128/JB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- Gamper M, Zimmermann A, Haas D. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J Bacteriol. 1991;173:4742–4750. doi: 10.1128/jb.173.15.4742-4750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CD, Winteler H, Abdelal A, Haas D. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J Bacteriol. 1999;181:2459–2464. doi: 10.1128/jb.181.8.2459-2464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercenier A, Simon JP, Vander Wauven C, Haas D, Stalon V. Regulation of enzyme synthesis in the arginine deiminase pathway of Pseudomonas aeruginosa. J Bacteriol. 1980;144:159–163. doi: 10.1128/jb.144.1.159-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga M, Champomier-Verges M, Zagorec M, Perez-Martinez G. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J Bacteriol. 1998;180:4154–4159. doi: 10.1128/jb.180.16.4154-4159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste L, Hervas AB, Canosa I, Tobes R, Jimenez JI, Nogales J. Growth phase-dependent expression of the Pseudomonas putida KT2440 transcriptional machinery analysed with a genome-wide DNA microarray. Environ Microbiol. 2006;8:165–177. doi: 10.1111/j.1462-2920.2005.00890.x. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Molec Biol. 2004;3:3–28. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc. 1995;57:289–300. [Google Scholar]
- Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock REW. Pseudomonas genome database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucl Acids Res. 2009;37:D483–D488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Cases II, de Lorenzo V, Pérez-Martín J. Involvement of s54 in exponential silencing of the Pseudomonas putida TOL plasmid Pu promoter. Mol Microbiol. 1996;19:7–17. doi: 10.1046/j.1365-2958.1996.345873.x. [DOI] [PubMed] [Google Scholar]
- Morales G, Ugidos A, Rojo F. Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ Microbiol. 2006;8:1764–1774. doi: 10.1111/j.1462-2920.2006.01061.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Significantly differentially expressed genes under elevated pressure (Pressure) and elevated pressure and DOT (Pressure Oxygen). Table S2. Categories and sub-categories significantly over-represented in the lists of significantly differentially expressed genes under elevated pressure. Table S3. Categories and sub-categories significantly over-represented in the lists of significantly differentially expressed genes under combined elevated pressure and elevated DOT. Table S4. Top genes with the most significant differential expression at elevated pressure (Pressure) and at combined elevated pressure and DOT (Pressure Oxygen).