Abstract
Human population growth, technological advances, and changing social behaviors lead to the selection of new microbial pathogens. Antimicrobial drugs, vaccines, diagnostics, and treatments for emerging infectious diseases must be developed. The selective forces that drive the emergence of new infectious diseases, and the implications for our survival, are just beginning to be understood.
Emerging infectious diseases are caused by new or previously unrecognized microorganisms (Figure 1 and Table 1). Although the term became part of the journalist’s lexicon in the 1990s, emerging infectious diseases have long been recognized as an important outcome of host-pathogen evolution. Because emerging infections may have severe public health consequences, they are a focus of both the popular press and scientific research. Emerging infections can be tracked by the hour on the Internet (ProMED-mail, www.promedmail.org) and in scientific journals (e.g., Emerging Infectious Diseases). This review series, motivated by the appearance of severe acute respiratory syndrome (SARS), the first emerging infectious disease of the 21st century, uses six different pathogens to illustrate the principles that govern the emergence of infectious diseases.
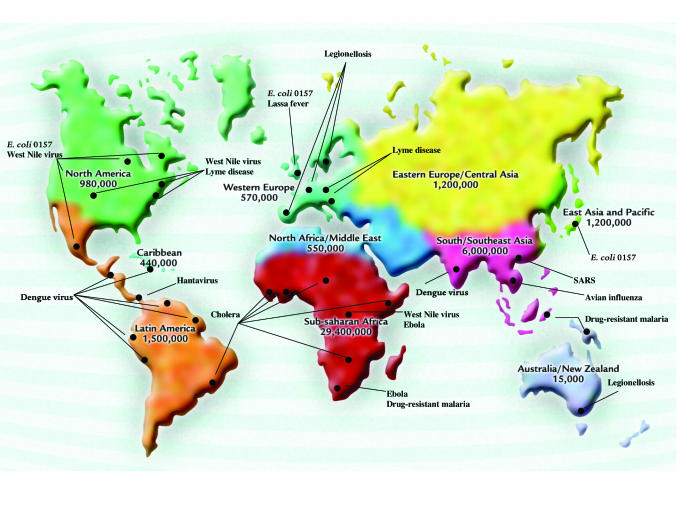
Figure 1.
Emerging and re-emerging infectious diseases. Numbers indicate the people estimated to be living with HIV/AIDS at the end of 2002. Locations of recent outbreaks of emerging infectious diseases are indicated. Source: WHO (7).
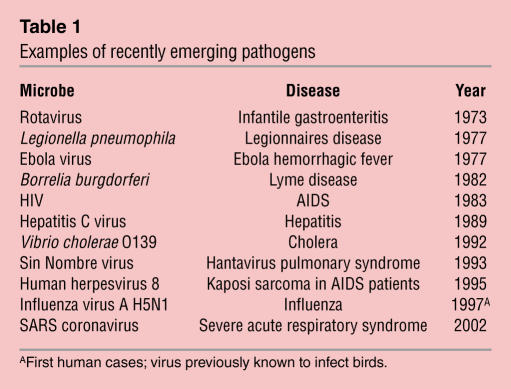
Table 1.
Examples of recently emerging pathogens
Changes in human populations and environment
Many factors contribute to the emergence of new infectious diseases. They include the increasing growth and mobility of the world’s population, overcrowding in cities with poor sanitation, massive food preparation and international distribution, unsanitary food preparation, exposure of humans to disease vectors and reservoirs, and ecological changes that alter the composition and size of insect vectors and animal reservoirs.
Long before “emerging infectious disease” was uttered, a change in the host population and the environment led to the emergence of poliomyelitis. This ancient disease has probably afflicted humans since the Eighteenth Egyptian Dynasty (1580–1350 BCE). The infection was endemic in the human population until the first half of the 20th century, when epidemics appeared. The emergence of poliomyelitis as an epidemic disease has been ascribed to a dramatic change in human life-style. The virus is spread by oral-fecal contamination. With poor sanitation, the virus circulated freely, infecting children at an early age when the development of paralysis is rare. The industrialization and urbanization of the 19th and 20th centuries led to improved sanitation, which interrupted the normal pattern of poliovirus transmission. Poliovirus infections were delayed until later in life, at which time the risk for developing paralytic disease is much greater.
West Nile virus, the topic of the review in this series by Gould and Fikrig (1), is an example of a known human pathogen that later emerged to infect new areas of the globe. The virus was isolated in the West Nile district of Uganda in 1937 but was not encountered in the Western Hemisphere until 1999, when it appeared in New York City. The virus has since spread throughout North America, where it has reached epidemic status. There is yet no answer to the fascinating question of why the infection emerged in New York City in 1999.
Animal origins of emerging infections
Some emerging infections are caused by microbes that originate in nonhuman vertebrates. Hantavirus pulmonary syndrome was first noted in the Four Corners area of New Mexico in 1993. The disease is caused by Sin Nombre virus, which is endemic in the deer mouse (Peromyscus maniculatus). Why humans first became infected with this rodent virus is not known, but an increase in the deer mouse population might have been a factor. In 1992–93, abundant rainfall produced a large crop of piñon nuts, which are food for both humans and the deer mouse. As the mouse population rose, contact with humans increased. The virus is excreted in mouse droppings, and contaminated blankets or dust from floors provided opportunities for human infection. Because humans are not the natural host for Sin Nombre virus, the human disease is rare. The emergence of Lyme disease, discussed in the review by Steere, Coburn, and Glickstein (2), followed a similar course. The infection has probably been present in North America for millennia, but it was only in the 20th century that conditions in the northeastern US changed to favor the propagation of Lyme disease.
Two other viruses that probably originated in nonhuman vertebrate hosts are HIV and the SARS coronavirus. HIV, the cause of AIDS, most likely arose from interspecies transmission between nonhuman primates and humans. The chimpanzee Pan troglodytes troglodytes was probably the origin of this virus, as this primate harbors strains of simian immunodeficiency virus that are closest in sequence to HIV-1. The ability to diagnose HIV infection within the first 3 weeks of exposure may allow new opportunities for treatment and prevention, as discussed in this review series by Pilcher et al. (3). The SARS coronavirus was first noted in China in the fall of 2002. The origin of this virus is unknown, but it may have been transmitted from animals to humans. A SARS-like coronavirus has been isolated from a palm civet and other animals in Guangdong Province, where the disease originated. The campaign to control SARS demonstrated the power of global mechanisms of health governance and revolutionized the response to emerging infectious diseases, as discussed in the review by Fidler (4).
Re-emerging infectious diseases
Some emerging infectious diseases are caused not by new pathogens, but by the re-emergence of microbes that had been successfully controlled. The mosquito-borne dengue virus was shown in 1903 to be the causative agent of dengue fever, a disease accompanied by fever, rash, and arthralgia. Fifty years later, a new disease caused by the virus, dengue hemorrhagic fever, was identified in Southeast Asia. The Pan American Health Organization led efforts to eradicate the mosquito vector of dengue virus, Aedes aegypti, from most of Central and South America. As a consequence, dengue fever and dengue hemorrhagic fever were largely eliminated from all but Southeast Asia. Unfortunately, the mosquito-eradication programs ended in the 1970s, leading to reinfestation of the Americas. Since then, dengue virus has been reintroduced into the Caribbean, the Pacific, Australia, and the Indian subcontinent and has appeared for the first time in China, Venezuela, and Brazil. Intercontinental transport of car tires containing mosquito eggs has been implicated in the spread of the virus. Currently there are an estimated 100 million cases of dengue fever and several hundred thousand cases of dengue hemorrhagic fever each year, and 2.5 billion people are at risk for infection. The series article by Rothman presents the challenges involved in developing a dengue vaccine (5).
Emerging resistance to antimicrobial drugs
As problematic as the threat of emerging infectious diseases is the emergence of drug-resistant pathogens, which can turn a conquered microbe into a new threat. Low-cost antibiotics can no longer clear infections of Escherichia coli, Neisseria gonorrhoeae, Pneumococcus, Shigella, and Staphylococcus aureus. The cost and length of treatment of many common diseases are increased, or infections may be refractory to treatment. The changing dynamics of a growing population (e.g., the relatively new phenomenon of children’s day care centers), and the overuse of antibiotics, both in humans and in the food chain, have contributed to the emergence of drug-resistant microbes. The review by White on drug-resistant malaria illustrates the magnitude of this problem (6). The emergence of resistance of Plasmodium falciparum to chloroquine, once a mainstay in the prevention of malaria, has led to a global resurgence of this disease.
Humans are always providing new ways to meet new pathogens. Air travel, dam construction, hot tubs, air conditioning, blood transfusion, deforestation, day care centers, and urbanization are some of the technological and social changes that influence the spread of microbes. Most importantly, growth of the human population continues, providing interactions with other humans and the environment on an unprecedented scale, and ensuring the propagation of new diseases.
Footnotes
Nonstandard abbreviations used: severe acute respiratory syndrome (SARS).
Conflict of interest: The author has declared that no conflict of interest exists.
References
- 1.Gould, L.H., and Fikrig, E. 2004. West Nile virus: a growing concern? J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 2.Steere, A.C., Coburn, J., and Glickstein, L. 2004. The emergence of Lyme disease. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 3.Pilcher, C.D., Eron, J.J., Jr., Galvin, S., Gay, C., and Cohen, M.S. 2004. Acute HIV revisited: new opportunities for treatment and prevention. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 4.Fidler DP. Germs, governance, and global public health in the wake of SARS. J. Clin. Invest. 2004;113:799–804. doi:10.1172/JCI200421328. doi: 10.1172/JCI21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothman, A.L. 2004. Dengue: defining protective versus pathologic immunity. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 6.White, N.J. 2004. Antimalarial drug resistance. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 7.WHO. 2002. Archives by disease. http://www.who.int/csr/don/archive/disease/en/.