Abstract
As tomatoes are one of the most important vegetables in the world, improvements in the quality and yield of tomato are strongly required. For this purpose, omics approaches such as metabolomics and transcriptomics are used not only for basic research to understand relationships between important traits and metabolism but also for the development of next generation breeding strategies of tomato plants, because an increase in the knowledge improves the taste and quality, stress resistance and/or potentially health-beneficial metabolites and is connected to improvements in the biochemical composition of tomatoes. Such omics data can be applied to network analyses to potentially reveal unknown cellular regulatory networks in tomato plants. The high-quality tomato genome that was sequenced in 2012 will likely accelerate the application of omics strategies, including next generation sequencing for tomato breeding. In this review, we highlight the current studies of omics network analyses of tomatoes and other plant species, in particular, a gene coexpression network. Key applications of omics approaches are also presented as case examples to improve economically important traits for tomato breeding.
Keywords: metabolomics, transcriptomics, biochemical trait, Solanum lycopersicum, tomato, multinetwork, coexpression analysis
Introduction
The tomato (Solanum lycopersicum) is one of the most important vegetables in the world, both as a fresh fruit and as the main ingredient of staple products like tomato puree and ketchup. As consumer demands for more variation, improvement of quality and taste, year-round availability and human health benefits have increased, scientists have had to obtain new insights into underlying genetic factors and regulation of metabolic pathways related to biochemical traits to accommodate such demands.
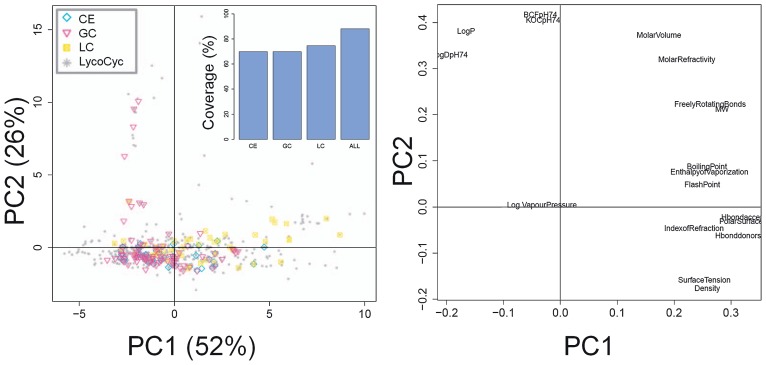
The omics approaches such as transcriptomics, proteomics and metabolomics constitute a trilogy in the post-genomics era to elucidate key steps in cellular events. Metabolomics, transcriptomics, or an integration of these 2 omics strategies have been used to investigate the metabolic networks of tomatoes to improve the quality and yield, because an increased knowledge that improves taste and quality, stress resistance, and/or potentially health-beneficial metabolites (e.g. antioxidants) is connected to the improvement of the biochemical composition of tomato plants. The tomato genome was sequenced in 2012 (Sato et al. 2012); therefore, annotations of tomato gene expression arrays provided by manufacturers can now be more precise using the information. Furthermore, metabolomic approaches based on chromatographic separation techniques connected with mass spectrometry (MS) as well as nuclear magnetic resonance spectroscopy (NMR) have been widely used for tomato metabolomics research because metabolites have beneficial traits such as taste, fragrance, softness and colour and are the ultimate phenotypic representatives of homeostasis in highly complex biochemical networks (Bovy et al. 2007, Deshmukh et al. 2003, Kusano et al. 2011a, Le Gall et al. 2003, Moco et al. 2008, Stark et al. 2008, Tikunov et al. 2005). Fig. 1 presents the current coverage of the tomato metabolome using our MS-based metabolomics platforms that consist of gas chromatography–electron ionization–time-of-flight–MS (GC-EI-TOF-MS), ultraperformance liquid chromatography–electron splay ionization–quadrupole–TOF–MS (UPLC-ESI-Q-TOF-MS) and capillary electrophoresis–ESI–TOF–MS (CE-ESI-TOF-MS) (Kusano et al. 2011a). We have covered more than 80% of the tomato metabolome when we evaluated the coverage by comparing physicochemical properties of the detected metabolites and those in the LycoCyc database (http://solgenomics.net/tools/solcyc/) (Mazourek et al. 2009).
Fig. 1.
To-date coverage of the tomato metabolome using the MS-based metabolomics platform in PRIMe (Platform for RIKEN Metabolomics, http://prime.psc.riken.jp/). Principal component analysis was performed using the physicochemical properties of the metabolomic dataset obtained from Kusano et al. (2011a) and those from the LycoCyc database (http://solgenomics.net/tools/solcyc/) (Mazourek et al. 2009). We used the latest version of the metabolite information in our custom database and ChemSpider (http://www.chemspider.com/). PC, principal component.
In this review, we will first highlight omics network studies to identify and infer cellular regulatory networks that have crucial roles in metabolic regulations of tomato plants and other plant species using gene-to-gene correlation analysis, which were generated by microarray and next-generation sequencer (NGS)-based technologies. Second, key applications of omics approaches aimed to improve economically important traits for tomato breeding are presented.
Revealing coordinated gene networks controlling tomato metabolism
A high-quality genome sequence of the tomato (Sato et al. 2012) facilitates a better understanding of molecular mechanisms regulating important traits such as yield and fruit quality characteristics. In this section, we focus on the role of the ‘omics’ network analysis using microarray- and NGS-based technologies in plants, including tomatoes.
Genomic- and post-genomic resources in tomatoes
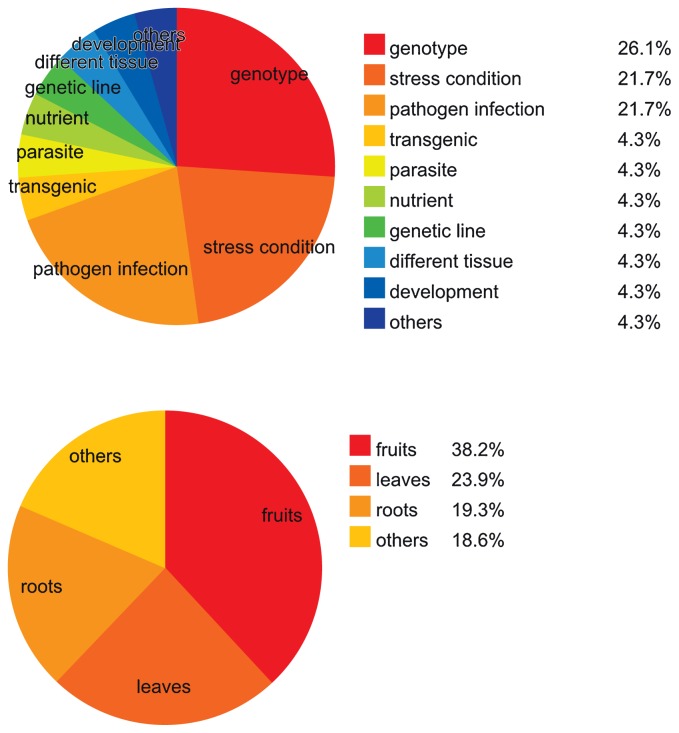
The Expressed Sequence Tag (EST) database for the tomato has many sequences corresponding to 40,000 Uni-Genes (http://www.sgn.cornell.edu) (Mueller et al. 2005). A large-scale collection of >13,000 full-length cDNAs generated from the tomato cultivar Micro-Tom has been previously reported (Aoki et al. 2010). TOMATOMICS, which is the integrated omics database for tomato plants, is constructed from the latest UniGene set (KTU4) that is made up of 125,883 ESTs from 9 cDNA libraries and other publicly available 196,912 ESTs from Sol Genomics Network (SGN), resulting in 58,083 UniGenes (http://bioinf.mind.meiji.ac.jp/tomatomics/). These genomic resources contain fundamental information reflecting complex gene expression in a plant cell. Developments in microarray technology have had a striking impact on the ability of researchers to monitor the expression of thousands of gene simultaneously. In the tomato, many types of microarray platforms, including TOM1, TOM2, Affymetrix GeneChip, Agilent custom array and TomatoArray (COMBIMATRIX), have enabled the investigation of responses to several stress conditions (Cantu et al. 2009, Khodakovskaya et al. 2011, Sun et al. 2010), the comparison of the expression profiles of wild-type and transgenic or mutant plants (Kumar et al. 2012, Martinelli et al. 2009, Nashilevitz et al. 2010, Povero et al. 2011) and the study of host-pathogen interactions (Alkan et al. 2012, Balaji et al. 2008, Owens et al. 2012). Archives of these comprehensive databases are in public repositories like NCBI GEO (Barrett et al. 2009) and ArrayExpress (Parkinson et al. 2009). Fig. 2 shows information about the collection of 393 Affymetrix tomato GeneChip data from NCBI GEO, ArrayExpress and TFGD (Fei et al. 2011). Recently, there have been tomato expression datasets generated by NGS-based RNA-sequencing (RNA-seq) and digital expression analysis; 7 RNA-seq datasets have been stored in the Sequence Read Archive (SRA) at NCBI (http://www.ncbi.nlm.nih.gov/sra). Other public databases for tomato research are summarized by Suzuki and the colleagues (Suzuki et al. 2009).
Fig. 2.
The current status of (A) experimental information and (B) tissues in 23 tomato microarray datasets. We used a collection of 393 Affymetrix tomato GeneChip data from the NCBI GEO, ArrayExpress and TFGD (Fei et al. 2011) public databases for this calculation (collection date, July 2012).
Meta-analysis of plant transcript profiles: gene coexpression
The publicly available datasets mentioned above have facilitated the development of in silico tools and databases to predict the functions of unknown genes. These are useful for model plants such as Arabidopsis and rice (see (Aoki et al. 2007, Usadel et al. 2009) and also for research on crops like tomatoes. Most tools and databases use a ‘gene coexpression’ approach. Gene coexpression is based on expression similarities between gene pairs across many experimental samples. It is predicted that the coexpressed gene pairs share a similar function in biological processes and have related regulatory mechanisms. In coexpression approaches, Pearson’s product-moment correlation coefficient is the most widely used for similarity measures. The Pearson’s correlation coefficient, r, can range from −1 to 1. An r = 1 indicates a perfect positive linear relationship between gene expressions and r = −1 indicates a perfect negative relationship. An r = 0 implies no linear relationship between gene expressions. The calculation of Pearson’s correlation coefficient is not robust for outliers and assumes that the data are from a standard normal distribution. On the other hand, Spearman’s rank correlation coefficient does not depend on a linear relationship and is more resistant to outliers than Pearson’s correlation coefficient. In many cases, such associations can be described as a ‘coexpression networks’, where nodes represent genes and links between nodes represent significant correlations that are higher than a threshold, resulting in a large-scale undirected graph. Accordingly, for the interpretation of coexpressed gene pairs, properties concerning their similarity measures should be noted.
Detecting meaningful associations in transcriptomic data
As several groups have demonstrated, coexpression network-based approaches are useful to characterise gene functions involved mainly in secondary metabolites such as flavonoids and glucosinolates in Arabidopsis (Saito et al. 2008, Usadel et al. 2009). However, a correlation does not always reflect a linear relationship and does not necessarily reflect causal relationships. Markowetz and Spang indicated that coexpression networks visualised by undirected graphs cannot easily explain the difference between direct and indirect dependencies in gene networks, although they should contain causal regulatory relationships (Markowetz and Spang 2007). A partial correlation coefficient may be used for constructing coexpression networks. It measures the correlation between 2 gene expressions x and y conditioning on 1 variable z. Although partial correlation still does not indicate causal relationships, it can be useful to exclude many indirect relationships (de la Fuente et al. 2004). In this framework, Ma et al. (2007) have inferred Arabidopsis coregulation patterns between genes using the graphical Gaussian model (GGM), which is a robust estimation of a direct relationship between variables (Ma et al. 2007). By measuring general dependencies between variables, mutual information based on information theory can identify coexpressed genes in large-scale high-throughput data (Steuer et al. 2002). In contrast to Pearson’s correlation, Galili and colleagues presented a novel bioinformatics tool called ‘Gene Coordination’ (Less and Galili 2009, Less et al. 2011). This method is based on the number of biological perturbations (e.g. drought stress), in which both genes of a given gene pair are significantly upregulated or significantly downregulated together, compared to non-treated controls. The authors demonstrated that the approach identified highly coordinated genes involved in the aspartate-family pathway in Arabidopsis. Reshef et al. (2011) proposed the maximal information coefficient (MIC), which is a novel measure of the association between variables (Reshef et al. 2011). This method can identify both linear and nonlinear relationships (i.e. is not limited to linear relationships), allowing us to explore variable relationships in a given data set in a non-biased manner. Construction of coexpression networks using RNA-seq data of tomatoes will be increased in the near future, as described for mice RNA-seq data (Lancu et al. 2012).
Expanding network concepts from single- to multinetwork
Integrating other omics data is another promising strategy to predict unknown gene functions. The first integrative study to construct a comprehensive multinetwork model of molecular interactions in Arabidopsis was performed using multiple microarray datasets treated by different combinations of carbon and nitrogen sources (Gutierrez et al. 2007, 2008). Gutierrez et al. integrated multiple information, including protein-DNA interactions, protein-protein interactions, metabolic pathways and molecular interactions from literature mining. Coruzzi and coworkers experimentally assessed those subnetworks inferred from the multinetwork (Gifford et al. 2008, Katari et al. 2010, Vidal et al. 2010). From a probabilistic point of view, functional networks integrated from multiple genomic-scale data have recently emerged for several species, including Arabidopsis and rice (Lee et al. 2010, 2011). Of these, AraNet (Lee et al. 2010) includes a million functional links among 19,647 corresponding to ~73% of the total Arabidopsis genes and the functional map demonstrates the usefulness of the network by characterising the predicted function of several genes based on the reverse genetics approach (Table 1).
Table 1.
Omics databases introduced in the article
| species | database | URL | reference |
|---|---|---|---|
| Arabidopsis | AraNet | http://www.functionalnet.org/aranet/ | (Lee et al. 2010) |
| CYPedia | http://www-ibmp.u-strasbg.fr/~CYPedia/ | (Ehlting et al. 2008) | |
| VirtualPlant | http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/ | (Katari et al. 2010) | |
| rice | OryzaExpress | http://riceball.lab.nig.ac.jp/oryzaexpress/ | (Hamada et al. 2011) |
| soybean | Soybean Proteome Database | http://proteome.dc.affrc.go.jp/Soybean/ | (Sakata et al. 2009) |
| tomato | TOMATOMICS | http://bioinf.mind.meiji.ac.jp/tomatomics/ | |
| Arabidopsis and six crop species | PlaNet | http://aranet.mpimp-golm.mpg.de | (Mutwil et al. 2011) |
Comparative coexpression networks: from model to crop plants
Attempts to compare multiple molecular networks are growing rapidly (Fukushima et al. 2009a, Kourmpetis et al. 2011, Lysenko et al. 2011). Investigations of coexpression between adjacent genes along genomes in Arabidopsis and rice demonstrated that the physically neighbouring genes were relatively highly coexpressed (Ren et al. 2005, 2007, Zhan et al. 2006). In bacterial genomes, multiple gene clusters of coregulated genes called operons were found, which have similar functions or belong to the same pathways. For a long time, eukaryotic genomes were not considered to have operons. However, several lines of evidence indicating the presence of operon-like gene clusters in plants have recently been reported (Field and Osbourn 2008, Field et al. 2011), including genes related with triterpene biosynthetic pathways in Arabidopsis and oat, genes in the cyclic hydroxamic acid pathways in maize and genes associated with diterpenoid momilactone production in rice (see the reviews by Mizutani and Ohta (2010), Takos and Rook (2012)). CYPedia is an expression database of Arabidopsis cytochrome P450 monooxygenases (CYPs) (Ehlting et al. 2008). It may be useful to have knowledge about coexpressed gene pairs between each CYP and a gene in the database; the orthologous gene pairs can be searched in tomato transcript datasets. Furthermore, novel statistical methods have recently emerged to extract operon-like gene clusters, including CYPs (Wada et al. 2012).
Using a combination of sequence similarity and gene co-expression, we can examine similar expression patterns and differences among multiple species, including humans and mice (Piasecka et al. 2012) and important cereal crops (Davidson et al. 2012, Van Bel et al. 2012). PlaNet (Mutwil et al. 2011) provides multiple coexpression networks from 7 plants based on an algorithm of network comparisons. A tool in PlaNet can extract conserved network modules across plant species, allowing us to identify reliable homologs. There exists a method available that evaluates the number and significance of shared orthologs between coexpressed modules (Chikina and Troyanskaya 2011, Movahedi et al. 2011, Zarrineh et al. 2011). Interestingly, many groups have reported conserved modules associated with photosynthesis, translation, cell division and DNA metabolism in dicot and monocot plants (Ficklin and Feltus 2011, Fukushima et al. 2008, 2009b, 2012, Mao et al. 2009, Mentzen and Wurtele 2008, Movahedi et al. 2011, Mutwil et al. 2011).
Differential network analyses in plant science
Generally, graph clustering such as Markov clustering (Van Dongen 2000) and DPClus (Altaf-Ul-Amin et al. 2006) can be used for detecting coexpressed modules or clusters in a non-biased manner. Graph clustering is an algorithm for efficiently extracting densely connected genes in coexpression networks. Using this type of network-based approach and >60 GeneChips, Ozaki and colleagues characterised transcription factor regulating flavonoid pathways in the tomato (Ozaki et al. 2010). This approach has also provided insights into transcriptional organization in Arabidopsis and rice as well as the tomato (Fukushima et al. 2009b, 2012, Ma et al. 2007, Mao et al. 2009, Mentzen and Wurtele 2008). Together with conserved coexpressions, a differential network strategy (Ideker and Krogan 2012) has been applied to animals and plants (Choi et al. 2005, de la Fuente 2010, Fukushima et al. 2012, Gillis and Pavlidis 2009). Differential metabolomic correlation analysis has been used for dissecting complex metabolism (Fukushima et al. 2011, Morgenthal et al. 2006, Weckwerth et al. 2004). As we introduced in the review (Table 1), we believe that omics network approaches are useful to understand underlying molecular mechanisms regulating important agronomic traits such as yield and fruit development in the tomato.
Omics-related research approaches for improvement of tomato fruit quality and yield
In this section, we will introduce application examples of omics approaches to improve agronomically and economically important traits as well as metabolite composition in the tomato.
Metabolite quantitative trait loci analyses
Plant breeding uses genetic variation to identify interesting traits and characteristics for growers and consumers. Uncovering the genetic basis of quantitative variation in the tomato is important for breeding purposes. Expression quantitative trait loci (eQTL) analysis, as a genome-wide association study, is becoming a powerful tool to identify the ‘hot spot(s)’ of genetic locations(s) of DNA sequence variation at the mRNA transcript level across a segregating population. Association analysis between genome-wide genetic variation and the levels of metabolites is also a promising approach in this field. In recent studies, eQTL analysis has been applied to Arabidopsis, maize, wheat and tree species (see the review by Druka et al. (2010)), whereas metabolite QTL (mQTL) analysis has been applied to Arabidopsis, rice, maize, potato, populus and tomatoes (Carreno-Quintero et al. 2012, Lisec et al. 2008, Matsuda et al. 2012, Morreel et al. 2006, Riedelsheimer et al. 2012, Toubiana et al. 2012, Wentzell et al. 2007). Of these, we will summarise the works of mQTL analysis based on metabolite profiling approaches to improve interesting traits in the tomato.
Tomato fruit quality is one of the most important traits. The utility of introgression lines (ILs), which were generated by crossing the cultivar Solanum lycopersicum ver. M82 with its wild relative S. pennellii, enables mQTL analysis of each ripe pericarp of the ILs using GC-EI-MS (Schauer et al. 2006). This was the first publication of mQTL analysis using metabolomics and correlation networks in the world. The authors quantified 74 metabolites in the metabolite profiles that were obtained from 2 independent experiments across 2 years (each data matrix: 76 ILs vs. 74 metabolites). Using these data matrices, 889 mQTLs were identified by analysis of variance (ANOVA) tests. Next, correlation network analysis was conducted to integrate metabolic and phenotypic traits of the ILs, then the network was visualised by cartographic representation to display the patterns of intra- and intermodule connections (Guimera and Nunes Amaral 2005). The results showed that 50% of the metabolites were considered to be morphologically associated. Additional mQTLs were identified by examining changes in the metabolite levels in the fruit profiles of the additional year’s harvest and then the extent of heritability of metabolites was investigated (Schauer et al. 2008). Metabolite profiling of the fruit pericarp of the heterozygous hybrids between ILs and M82 (ILHs) as well as that of the 76 homozygous ILs was performed. Then, the authors identified 332 putative mQTLs. Furthermore, the use of additional ILHs provided the chance to classify each putative QTL derived from S. pennellii into 4 types of mode-of-inheritance categories using the method described by Semel et al. (2006). The authors found that most of the putative wild species QTLs showed an increasing effect on the metabolite content when compared to that in the S. lycopersicum line and that these effects were inherited in a dominant or additive manner.
Toubiana and colleagues demonstrated mQTL analysis using tomato seeds of 76 ILs; metabolite profiles of each fruit pericarp were harvested in parallel (Schauer et al. 2006, 2008) to determine the association between seed quality traits and mQTLs (Toubiana et al. 2012). They compared the extent of the broad-sense heritability of each metabolite content in seed and fruit tissues of the ILs and the control line M82 using ANOVA and coefficient of variation tests. The observed patterns suggested that the broad-sense heritability of metabolite content in the seed was greater than that in the fruit. By using the seed and fruit datasets, network analysis based on metabolite-to-metabolite associations was applied to investigate the degree of correlation between metabolites in the seed and the fruit. Metabolite-to-metabolite correlation analyses have been widely applied to visualise genotype-dependent relationships in Arabidopsis (Allen et al. 2010, Kusano et al. 2007), potato (Weckwerth et al. 2004), melon (Biais et al. 2009) and the tomato (Ursem et al. 2008). This approach revealed more intensified correlation relationships in seed metabolism than those found in the fruit. Furthermore, the correlation analysis emphasized the centrality of the amino acid module in the seed metabolic network.
Taken together, these approaches introduced here show that mQTL analysis in combination with introgression breeding is likely to be useful not only to enhance biochemical traits but also to find important ‘hubs’, which are highly connected genes and/or metabolites in coexpression modules, in tomato metabolism.
Parthenocarpy
Parthenocarpy is the ability to produce fruits in the absence of pollination. It is an economically valuable trait for many horticultural crops and vegetables, including tomatoes. Parthenocarpy can prevent mechanical vibration of the flowers to endure pollen shedding. For manufacturing, parthenocarpic tomatoes can reduce processing of tomato products. As such, parthenocarpy is an important trait for tomato breeding. However, the molecular mechanism of fruit development, particularly the onset of ovary development, remains unclear. To address the issue, omics approaches have been applied by combining the reverse genetic approach.
It is known that downregulation of indole acetic acid 9 (IAA9) in tomatoes showed pollination-independent fruit production (Wang et al. 2005). IAA9 is a negative auxin response regulator belonging to the Aux/IAA transcription factor gene family (Abel et al. 1995, Nebenfuhr et al. 2000). Integrated analysis of transcript and metabolite profiling of MicroTom tomato lines downregulated in the expression of the IAA9 gene was applied to investigate underlying molecular events during fruit set (Wang et al. 2009). Three independent experiments were carefully designed; these datasets provided information about (i) natural pollination-induced fruit set, (ii) pollination-dependent fruit set and (iii) genotype-dependent comparison of fruit set between the control (wild type) and 2 independent transgenic antisense lines. Such comprehensive analysis generated a model of the molecular events mediated by IAA9 and those underlying the fruit set process in tomatoes. The model showed that 1455 genes were involved in bud-to-flower transition and 1650 genes were involved in flower-to-fruit transition. Particularly, novel pathways such as photosynthesis, auxin and ethylene signalling as well as the requirement of a high number of transcriptional regulators associated with natural pollination fruit set and pollination-independent fruit set were found using the omics approach.
The second example is the transcript profiling analysis of a parthenocarpic line, the pat3/pat4 mutant (Pascual et al. 2009). Three major mutants that show parthenocarpic growth in tomatoes: pat, pat-2 and pat/3pat4 have been identified (Carmi et al. 2003, Fos et al. 2001, Gorguet et al. 2008, Philouze 1983, 1985). Pascual and colleagues performed transcript profiling of the tomato carpel and fruit of the pat3/pat4 mutant and the UC82 line as a representative parthenocarpic and non-parthenocarpic line, respectively. Time-series experiments allowed the extraction of 2842 differentially expressed genes during carpel development and fruit set. Of these, the major differences between pat3/pat4 and UC82 lines were observed at the anthesis stage. Genes involved in cell division and cell cycle events and genes responsible for gibberellins (GAs) and ethylene biosynthetic pathways were highly expressed in the pat3/pat4 lines at the anthesis stage, suggesting that the transition point at the anthesis stage seems to be shorter in the parthenocarpic line pat3/pat4 than in the non-parthenocarpic line, and this event may be regulated by phytohormones such as GAs and ethylene.
The third example of a parthenocarpy study is downregulation of chalcone synthase (CHS), which encodes the first step of the enzyme in the flavonoid pathway (Schijlen et al. 2007). Flavonoids are a type of phenylpropanoid, which are involved in one of the major pathways in the plant kingdom. Although the study did not employ an omics approach, the data were interesting because suppression of CHS caused not only inhibition of pigment accumulation in the flower tissue but also male sterility in petunia (van der Meer et al. 1992). These data imply that flavonoids and/or genes in flavonoid pathways may have crucial roles in fruit development. Indeed, RNAi-mediated suppression of CHS in tomato plants caused parthenocarpy. These findings encourage us to apply omics approaches for the study of parthenocarpy, particularly focusing on metabolomic changes in flavonoids in parthenocarpic tomatoes.
Investigation of other important traits using omics approaches
Omics approaches have been applied to improve other beneficial traits, e.g. fruit architecture and biochemical traits. Aharoni’s group performed transcript and metabolite profiling of tomato peel (outer layers) and flesh (pericarp after removal of the peel) tissues to obtain new insights into gene expression patterns and metabolite composition of the fruit surface (Mintz-Oron et al. 2008). As expression patterns and changes in metabolite levels, including primary and secondary metabolites in peel and flesh tissues, were well captured at the 5 growth stages in fruit, omics approaches have a great potential to obtain insights for improvement of the tomato fruit surface. To date, the GC-EI-MS technique has been used for determination of cutin and wax components in the tomato peel (Adato et al. 2009, Saladie et al. 2007). This technique is one of the gold standards for metabolomics. Thus, it is easy to apply peak pretreatment and data alignment techniques developed in the metabolomics field for cutin and wax analyses. Using our metabolomics pipeline based on GC-EI-TOF-MS, we recently conducted metabolite fingerprinting analysis of cutin and wax fractions of a tomato mutant, which have both sticky peel (pe) and light green (lg) mutations, as an application example of metabolomic techniques for cutin and wax analysis, although complete identification of cutin and wax monomers requires authentic standards (Kimbara et al. 2012).
Tomato fruits contain flavonoids, which are health-protecting components in the human diet. Recently, flavonoids have been shown to have critical roles in plant physiology, e.g. auxin transport, allelopathy, sterility and stress resistance (Buer et al. 2010, Mol et al. 1998, Peer and Murphy 2007, Ulm and Nagy 2005). It is well known that flavonoid accumulation is strongly induced to protect against ultraviolet (UV)-induced damage when plants are exposed to UV-B light (Kootstra 1994, Kusano et al. 2011b). Giuntini and colleagues treated tomato fruits with a normal sunlight spectrum deprived of the UV-B region using a poly-ethylene film (Giuntini et al. 2008). They used 2 tomato genotypes: the hybrid Esperanza F1 with low lycopene levels in the fruit and the hybrid DRW 5981 with high lycopene content in the fruit. Tomato fruits derived from the 2 cultivars were harvested at the 3 growth stages, and then flavonoid content was quantified using LC-ESI-MS. The flavonoid profiles of the 2 cultivars showed distinct patterns in the presence or absence of the UV-B region. A similar approach was employed by the same group to investigate how the high pigment-1 mutant responds under UV-B depleted conditions; this mutant accumulates fruit pigments and has a mutation in the tomato HIGH-PIGMENT1/UV-DAMAGEDDNA-BINDING PROTEIN 1 (HP1/LeDDB1) gene (Calvenzani et al. 2010). They found that (i) flavonoid biosynthetic genes and genes involved in light signal transduction were induced by UV-B at the early growth stage and (ii) the expression level of LeDDB1 was not regulated by UV-B.
Conclusion and remarks
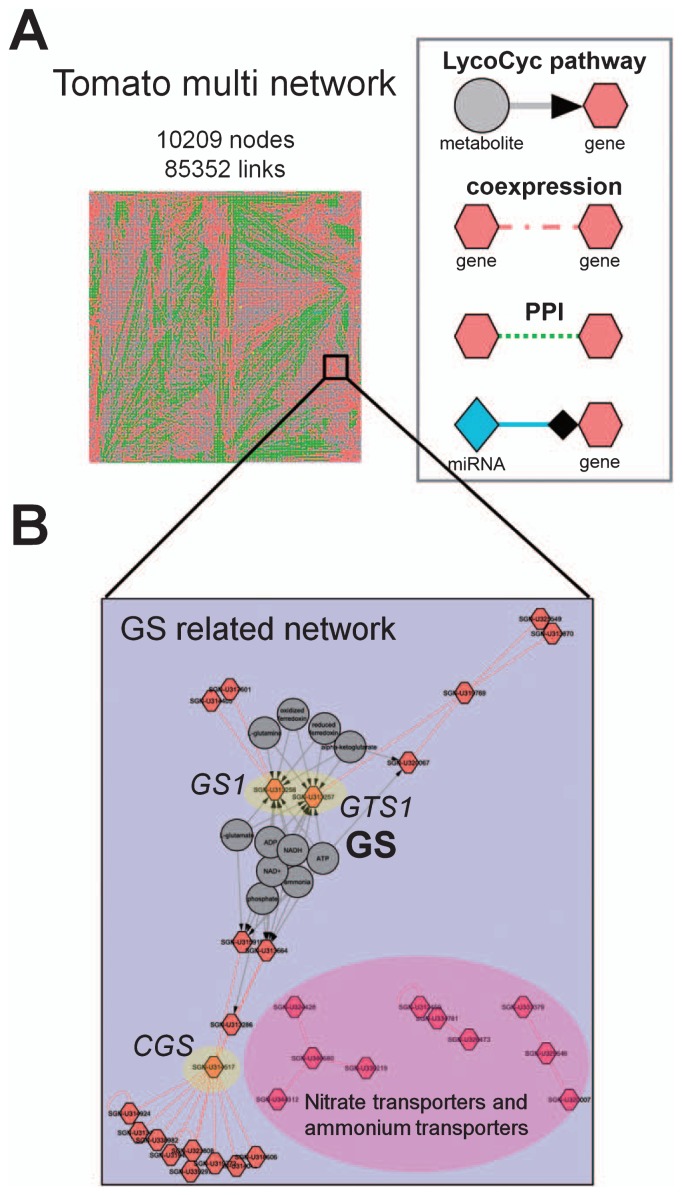
Tomato genome information provides us with more precise gene annotations and probe sets for microarray analysis and thus, we can use more ‘reliable’ microarray chips, such as Arabidopsis and rice, at a lower cost than at present. More datasets, including microarray and RNA-seq data will be stored in public databases like NCBI GEO. Publicly available datasets enable us to generate coexpression networks in tomatoes. Highly connected hub genes in coexpression modules tend to be important for genomic reasons. It should be noted that such ‘hub’ genes are not always good candidate genes for tomato breeding when these hub genes were found to be present in a certain gene cluster (for example, see Fig. 3). Cytosolic glutamine synthetase (GS) and plastidic GS have crucial roles in ammonium assimilation and recycling in various plant species (Bernard and Habash 2009, Diaz et al. 2010, Kusano et al. 2011c). These plants often showed lethal or visual phenotypic changes when these hub genes were knocked out (Martin et al. 2006, Tabuchi et al. 2005). To identify genes and metabolites that contribute to important traits for tomato breeding, multiple datasets obtained from phenotyping and integrated omics analysis as introduced in this review provide great opportunities to conduct a systems biological approach based on multinetwork analysis. This approach will shed light on the points of robustness and weakness in metabolic systems of tomato plants; breeders and breeding companies may use this information for the development of next generation breeding strategies.
Fig. 3.
An example of a tomato multinetwork. (A) We constructed a tomato multinetwork that consisted of 10209 nodes and 85352 links. Information for the tomato metabolic pathway was obtained from the LycoCyc database (http://solgenomics.net/tools/solcyc/) (Mazourek et al. 2009), coexpression information (Fukushima et al. 2012), protein-protein interactions from interolog (Yu et al. 2004) and miRNA-gene relationships from PMRD (Zhang et al. 2010). (B) An expansion of the glutamine synthetase (GS)-related network in the multinetwork. Because the genes annotated as GS1 (SGN-U313258), GTS1 (SGN-U313257) and CGS (SGN-U314517) have many links to other genes, these genes are thought to be ‘hub’ gene candidates. PPI, protein-protein interaction; miRNA, micro RNA; GTS, putative GS; CGS, chloroplastic GS.
Acknowledgements
The work was partly supported by the foundation of Bio-oriented Technology Research Advancement Institution (BRAIN), Japan and a Grant-in-Aid for Young Scientists (B; grant no. 23700355 to A.F.) from the Ministry of Education, Culture, Sports, Science and Technology.
Literature Cited
- Abel, S., Nguyen, M.D., and Theologis, A. (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Adato, A., Mandel, T., Mintz-Oron, S., Venger, I., Levy, D., Yativ, M., Dominguez, E., Wang, Z.H., De Vos, R.C.H., and Jetter, R.et al. (2009) Fruit-surface flavonoid accumulation in tomato is controlled by a SIMYB12-regulated transcriptional network. PLoS Genet. 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan, N., Fluhr, R., and Prusky, D. (2012) Ammonium secretion during Colletotrichum coccodes infection modulates salicylic and jasmonic acid pathways of ripe and unripe tomato fruit. Mol. Plant Microbe Interact. 25: 85–96 [DOI] [PubMed] [Google Scholar]
- Allen, E., Moing, A., Ebbels, T.M., Maucourt, M., Tomos, A.D., Rolin, D., and Hooks, M.A. (2010) Correlation Network Analysis reveals a sequential reorganization of metabolic and transcriptional states during germination and gene-metabolite relationships in developing seedlings of Arabidopsis. BMC Syst. Biol. 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf-Ul-Amin, M., Shinbo, Y., Mihara, K., Kurokawa, K., and Kanaya, S. (2006) Development and implementation of an algorithm for detection of protein complexes in large interaction networks. BMC Bioinformatics 7: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, K., Ogata, Y., and Shibata, D. (2007) Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 48: 381–390 [DOI] [PubMed] [Google Scholar]
- Aoki, K., Yano, K., Suzuki, A., Kawamura, S., Sakurai, N., Suda, K., Kurabayashi, A., Suzuki, T., Tsugane, T., and Watanabe, M.et al. (2010) Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics 11: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji, V., Mayrose, M., Sherf, O., Jacob-Hirsch, J., Eichenlaub, R., Iraki, N., Manulis-Sasson, S., Rechavi, G., Barash, I., and Sessa, G. (2008) Tomato transcriptional changes in response to Clavibacter michiganensis subsp. michiganensis reveal a role for ethylene in disease development. Plant Physiol. 146: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, T., Troup, D.B., Wilhite, S.E., Ledoux, P., Rudnev, D., Evangelista, C., Kim, I.F., Soboleva, A., Tomashevsky, M., and Marshall, K.A.et al. (2009) NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37: D885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, S.M., and Habash, D.Z. (2009) The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 182: 608–620 [DOI] [PubMed] [Google Scholar]
- Biais, B., Allwood, J.W., Deborde, C., Xu, Y., Maucourt, M., Beauvoit, B., Dunn, W.B., Jacob, D., Goodacre, R., and Rolin, D.et al. (2009) 1H NMR, GC-EI-TOFMS, and data set correlation for fruit metabolomics: application to spatial metabolite analysis in melon. Anal. Chem. 81: 2884–2894 [DOI] [PubMed] [Google Scholar]
- Bovy, A., Schijlen, E., and Hall, R.D. (2007) Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): the potential for metabolomics. Metabolomics 3: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer, C.S., Imin, N., and Djordjevic, M.A. (2010) Flavonoids: new roles for old molecules. J. Integr. Plant Biol. 52: 98–111 [DOI] [PubMed] [Google Scholar]
- Calvenzani, V., Martinelli, M., Lazzeri, V., Giuntini, D., Dall’Asta, C., Galaverna, G., Tonelli, C., Ranieri, A., and Petroni, K. (2010) Response of wild-type and high pigment-1 tomato fruit to UV-B depletion: flavonoid profiling and gene expression. Planta 231: 755–765 [DOI] [PubMed] [Google Scholar]
- Cantu, D., Blanco-Ulate, B., Yang, L., Labavitch, J.M., Bennett, A.B., and Powell, A.L. (2009) Ripening-regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol. 150: 1434–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi, N., Salts, Y., Dedicova, B., Shabtai, S., and Barg, R. (2003) Induction of parthenocarpy in tomato via specific expression of the rolB gene in the ovary. Planta 217: 726–735 [DOI] [PubMed] [Google Scholar]
- Carreno-Quintero, N., Acharjee, A., Maliepaard, C., Bachem, C.W., Mumm, R., Bouwmeester, H., Visser, R.G., and Keurentjes, J.J. (2012) Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol. 158: 1306–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikina, M.D., and Troyanskaya, O.G. (2011) Accurate quantification of functional analogy among close homologs. PLoS Comput. Biol. 7: e1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J.K., Yu, U., Yoo, O.J., and Kim, S. (2005) Differential coexpression analysis using microarray data and its application to human cancer. Bioinformatics 21: 4348–4355 [DOI] [PubMed] [Google Scholar]
- Davidson, R.M., Gowda, M., Moghe, G., Lin, H., Vaillancourt, B., Shiu, S.H., Jiang, N., and Robin Buell, C. (2012) Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J. 71: 492–502 [DOI] [PubMed] [Google Scholar]
- de la Fuente, A., Bing, N., Hoeschele, I., and Mendes, P. (2004) Discovery of meaningful associations in genomic data using partial correlation coefficients. Bioinformatics 20: 3565–3574 [DOI] [PubMed] [Google Scholar]
- de la Fuente, A. (2010) From ‘differential expression’ to ‘differential networking’—identification of dysfunctional regulatory networks in diseases. Trends Genet. 26: 326–333 [DOI] [PubMed] [Google Scholar]
- Deshmukh, A.P., Simpson, A.J., and Hatcher, P.G. (2003) Evidence for cross-linking in tomato cutin using HR-MAS NMR spectroscopy. Phytochemistry 64: 1163–1170 [DOI] [PubMed] [Google Scholar]
- Diaz, P., Betti, M., Sanchez, D.H., Udvardi, M.K., Monza, J., and Marquez, A.J. (2010) Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol. 188: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Druka, A., Potokina, E., Luo, Z.W., Jiang, N., Chen, X.W., Kearsey, M., and Waugh, R. (2010) Expression quantitative trait loci analysis in plants. Plant Biotechnol. J. 8: 10–27 [DOI] [PubMed] [Google Scholar]
- Ehlting, J., Sauveplane, V., Olry, A., Ginglinger, J.F., Provart, N.J., and Werck-Reichhart, D. (2008) An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, Z., Joung, J.G., Tang, X., Zheng, Y., Huang, M., Lee, J.M., McQuinn, R., Tieman, D.M., Alba, R., and Klee, H.J.et al. (2011) Tomato Functional Genomics Database: a comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res. 39: D1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficklin, S.P., and Feltus, F.A. (2011) Gene coexpression network alignment and conservation of gene modules between two grass species: maize and rice. Plant Physiol. 156: 1244–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, B., and Osbourn, A.E. (2008) Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Field, B., Fiston-Lavier, A.S., Kemen, A., Geisler, K., Quesneville, H., and Osbourn, A.E. (2011) Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl. Acad. Sci. USA 108: 16116–16121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fos, M., Proano, K., Nuez, F., and Garcia-Martinez, J.L. (2001) Role of gibberellins in parthenocarpic fruit development induced by the genetic system pat-3/pat-4 in tomato. Physiol. Plant. 111: 545–550 [DOI] [PubMed] [Google Scholar]
- Fukushima, A., Wada, M., Kanaya, S., and Arita, M. (2008) SVD-based anatomy of gene expressions for correlation analysis in Arabidopsis thaliana. DNA Res. 15: 367–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima, A., Kusano, M., Redestig, H., Arita, M., and Saito, K. (2009a) Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 13: 532–538 [DOI] [PubMed] [Google Scholar]
- Fukushima, A., Kanaya, S., and Arita, M. (2009b) Characterizing gene coexpression modules in Oryza sativa based on a graph-clustering approach. Plant Biotechnol. 26: 485–493 [Google Scholar]
- Fukushima, A., Kusano, M., Redestig, H., Arita, M., and Saito, K. (2011) Metabolomic correlation-network modules in Arabidopsis based on a graph-clustering approach. BMC Syst. Biol. 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima, A., Nishizawa, T., Hayakumo, M., Hikosaka, S., Saito, K., Goto, E., and Kusano, M. (2012) Exploring tomato gene functions based on coexpression modules using graph clustering and differential coexpression approaches. Plant Physiol. 158: 1487–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford, M.L., Dean, A., Gutierrez, R.A., Coruzzi, G.M., and Birnbaum, K.D. (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis, J., and Pavlidis, P. (2009) A methodology for the analysis of differential coexpression across the human lifespan. BMC Bioinformatics 10: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntini, D., Lazzeri, V., Calvenzani, V., Dall’Asta, C., Galaverna, G., Tonelli, C., Petroni, K., and Ranieri, A. (2008) Flavonoid profiling and biosynthetic gene expression in flesh and peel of two tomato genotypes grown under UV-B-depleted conditions during ripening. J. Agric. Food Chem. 56: 5905–5915 [DOI] [PubMed] [Google Scholar]
- Gorguet, B., Eggink, P.M., Ocana, J., Tiwari, A., Schipper, D., Finkers, R., Visser, R.G., and van Heusden, A.W. (2008) Mapping and characterization of novel parthenocarpy QTLs in tomato. Theor. Appl. Genet. 116: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera, R., and Nunes Amaral, L.A. (2005) Functional cartography of complex metabolic networks. Nature 433: 895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, R.A., Lejay, L.V., Dean, A., Chiaromonte, F., Shasha, D.E., and Coruzzi, G.M. (2007) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 8: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, R.A., Stokes, T.L., Thum, K., Xu, X., Obertello, M., Katari, M.S., Tanurdzic, M., Dean, A., Nero, D.C., and McClung, C.R.et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl. Acad. Sci. USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, K., Hongo, K., Suwabe, K., Shimizu, A., Nagayama, T., Abe, R., Kikuchi, S., Yamamoto, N., Fujii, T., and Yokoyama, K.et al. (2011) OryzaExpress: an integrated database of gene expression networks and omics annotations in rice. Plant Cell Physiol. 52: 220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker, T., and Krogan, N.J. (2012) Differential network biology. Mol. Syst. Biol. 8: 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari, M.S., Nowicki, S.D., Aceituno, F.F., Nero, D., Kelfer, J., Thompson, L.P., Cabello, J.M., Davidson, R.S., Goldberg, A.P., and Shasha, D.E.et al. (2010) VirtualPlant: a software platform to support systems biology research. Plant Physiol. 152: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodakovskaya, M.V., de Silva, K., Nedosekin, D.A., Dervishi, E., Biris, A.S., Shashkov, E.V., Galanzha, E.I., and Zharov, V.P. (2011) Complex genetic, photothermal, and photoacoustic analysis of nanoparticle-plant interactions. Proc. Natl. Acad. Sci. USA 108: 1028–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbara, J., Yoshida, M., Ito, H., Hosoi, K., Kusano, M., Kobayashi, M., Ariizumi, T., Asamizu, E., and Ezura, H. (2012) A novel class of sticky peel and light green mutations causes cuticle deficiency in leaves and fruits of tomato (Solanum lycopersicum). Planta 236: 1559–1570 [DOI] [PubMed] [Google Scholar]
- Kootstra, A. (1994) Protection from UV-B-induced DNA damage by flavonoids. Plant Mol. Biol. 26: 771–774 [DOI] [PubMed] [Google Scholar]
- Kourmpetis, Y.A., van Dijk, A.D., van Ham, R.C., and ter Braak, C.J. (2011) Genome-wide computational function prediction of Arabidopsis proteins by integration of multiple data sources. Plant Physiol. 155: 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R., Sharma, M.K., Kapoor, S., Tyagi, A.K., and Sharma, A.K. (2012) Transcriptome analysis of rin mutant fruit and in silico analysis of promoters of differentially regulated genes provides insight into LeMADS-RIN-regulated ethylene-dependent as well as ethylene-independent aspects of ripening in tomato. Mol. Genet. Genomics 287: 189–203 [DOI] [PubMed] [Google Scholar]
- Kusano, M., Fukushima, A., Arita, M., Jonsson, P., Moritz, T., Kobayashi, M., Hayashi, N., Tohge, T., and Saito, K. (2007) Unbiased characterization of genotype-dependent metabolic regulations by metabolomic approach in Arabidopsis thaliana. BMC Syst. Biol. 1: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano, M., Redestig, H., Hirai, T., Oikawa, A., Matsuda, F., Fukushima, A., Arita, M., Watanabe, S., Yano, M., and Hiwasa-Tanase, K.et al. (2011a) Covering chemical diversity of genetically-modified tomatoes using metabolomics for objective substantial equivalence assessment. PLoS One 6: e16989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano, M., Tohge, T., Fukushima, A., Kobayashi, M., Hayashi, N., Otsuki, H., Kondou, Y., Goto, H., Kawashima, M., and Matsuda, F.et al. (2011b) Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 67: 354–369 [DOI] [PubMed] [Google Scholar]
- Kusano, M., Tabuchi, M., Fukushima, A., Funayama, K., Diaz, C., Kobayashi, M., Hayashi, N., Tsuchiya, Y.N., Takahashi, H., and Kamata, A.et al. (2011c) Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice. Plant J. 66: 456–466 [DOI] [PubMed] [Google Scholar]
- Lancu, O.D., Kawane, S., Bottomly, D., Searles, R., Hitzemann, R., and McWeeney, S. (2012) Utilizing RNA-Seq Data for de-novo coexpression network inference. Bioinformatics 28: 1592–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall, G., Colquhoun, I.J., Davis, A.L., Collins, G.J., and Verhoeyen, M.E. (2003) Metabolite profiling of tomato (Lycopersicon esculentum) using 1H NMR spectroscopy as a tool to detect potential unintended effects following a genetic modification. J. Agric. Food Chem. 51: 2447–2456 [DOI] [PubMed] [Google Scholar]
- Lee, I., Ambaru, B., Thakkar, P., Marcotte, E.M., and Rhee, S.Y. (2010) Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat. Biotechnol. 28: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Seo, Y.S., Coltrane, D., Hwang, S., Oh, T., Marcotte, E.M., and Ronald, P.C. (2011) Genetic dissection of the biotic stress response using a genome-scale gene network for rice. Proc. Natl. Acad. Sci. USA 108: 18548–18553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Less, H., and Galili, G. (2009) Coordinations between gene modules control the operation of plant amino acid metabolic networks. BMC Syst. Biol. 3: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Less, H., Angelovici, R., Tzin, V., and Galili, G. (2011) Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell 23: 1264–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec, J., Meyer, R.C., Steinfath, M., Redestig, H., Becher, M., Witucka-Wall, H., Fiehn, O., Torjek, O., Selbig, J., and Altmann, T.et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J. 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko, A., Defoin-Platel, M., Hassani-Pak, K., Taubert, J., Hodgman, C., Rawlings, C.J., and Saqi, M. (2011) Assessing the functional coherence of modules found in multiple-evidence networks from Arabidopsis. BMC Bioinformatics 12: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, S., Gong, Q., and Bohnert, H.J. (2007) An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 17: 1614–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, L., Van Hemert, J.L., Dash, S., and Dickerson, J.A. (2009) Arabidopsis gene co-expression network and its functional modules. BMC Bioinformatics 10: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowetz, F., and Spang, R. (2007) Inferring cellular networks—a review. Bmc Bioinformatics 8 (Suppl 6): S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, A., Lee, J., Kichey, T., Gerentes, D., Zivy, M., Tatout, C., Dubois, F., Balliau, T., Valot, B., and Davanture, M.et al. (2006) Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18: 3252–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli, F., Uratsu, S.L., Reagan, R.L., Chen, Y., Tricoli, D., Fiehn, O., Rocke, D.M., Gasser, C.S., and Dandekar, A.M. (2009) Gene regulation in parthenocarpic tomato fruit. J. Exp. Bot. 60: 3873–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, F., Okazaki, Y., Oikawa, A., Kusano, M., Nakabayashi, R., Kikuchi, J., Yonemaru, J., Ebana, K., Yano, M., and Saito, K. (2012) Dissection of genotype-phenotype associations in rice grains using metabolome quantitative trait loci analysis. Plant J. 70: 624–636 [DOI] [PubMed] [Google Scholar]
- Mazourek, M., Pujar, A., Borovsky, Y., Paran, I., Mueller, L., and Jahn, M.M. (2009) A dynamic interface for capsaicinoid systems biology. Plant Physiol. 150: 1806–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentzen, W.I., and Wurtele, E.S. (2008) Regulon organization of Arabidopsis. BMC Plant Biol. 8: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron, S., Mandel, T., Rogachev, I., Feldberg, L., Lotan, O., Yativ, M., Wang, Z., Jetter, R., Venger, I., and Adato, A.et al. (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol. 147: 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani, M., and Ohta, D. (2010) Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 61: 291–315 [DOI] [PubMed] [Google Scholar]
- Moco, S., Forshed, J., De Vos, R.C.H., Bino, R.J., and Vervoort, J. (2008) Intra- and inter-metabolite correlation spectroscopy of tomato metabolomics data obtained by liquid chromatography-mass spectrometry and nuclear magnetic resonance. Metabolomics 4: 202–215 [Google Scholar]
- Mol, J., Grotewold, E., and Koes, R. (1998) How genes paint flowers and seeds. Trends Plant Sci. 3: 212–217 [Google Scholar]
- Morgenthal, K., Weckwerth, W., and Steuer, R. (2006) Metabolomic networks in plants: Transitions from pattern recognition to biological interpretation. Biosystems 83: 108–117 [DOI] [PubMed] [Google Scholar]
- Morreel, K., Goeminne, G., Storme, V., Sterck, L., Ralph, J., Coppieters, W., Breyne, P., Steenackers, M., Georges, M., and Messens, E.et al. (2006) Genetical metabolomics of flavonoid biosynthesis in Populus: a case study. Plant J. 47: 224–237 [DOI] [PubMed] [Google Scholar]
- Movahedi, S., Van de Peer, Y., and Vandepoele, K. (2011) Comparative network analysis reveals that tissue specificity and gene function are important factors influencing the mode of expression evolution in Arabidopsis and rice. Plant Physiol. 156: 1316–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, L.A., Solow, T.H., Taylor, N., Skwarecki, B., Buels, R., Binns, J., Lin, C., Wright, M.H., Ahrens, R., and Wang, Y.et al. (2005) The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol. 138: 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil, M., Klie, S., Tohge, T., Giorgi, F.M., Wilkins, O., Campbell, M.M., Fernie, A.R., Usadel, B., Nikoloski, Z., and Persson, S. (2011) PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23: 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashilevitz, S., Melamed-Bessudo, C., Izkovich, Y., Rogachev, I., Osorio, S., Itkin, M., Adato, A., Pankratov, I., Hirschberg, J., and Fernie, A.R.et al. (2010) An orange ripening mutant links plastid NAD(P)H dehydrogenase complex activity to central and specialized metabolism during tomato fruit maturation. Plant Cell 22: 1977–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr, A., White, T.J., and Lomax, T.L. (2000) The diageotropica mutation alters auxin induction of a subset of the Aux/IAA gene family in tomato. Plant Mol. Biol. 44: 73–84 [DOI] [PubMed] [Google Scholar]
- Owens, R.A., Tech, K.B., Shao, J.Y., Sano, T., and Baker, C.J. (2012) Global analysis of tomato gene expression during Potato spindle tuber viroid infection reveals a complex array of changes affecting hormone signaling. Mol. Plant Microbe Interact. 25: 582–598 [DOI] [PubMed] [Google Scholar]
- Ozaki, S., Ogata, Y., Suda, K., Kurabayashi, A., Suzuki, T., Yamamoto, N., Iijima, Y., Tsugane, T., Fujii, T., and Konishi, C.et al. (2010) Coexpression analysis of tomato genes and experimental verification of coordinated expression of genes found in a functionally enriched coexpression module. DNA Res. 17: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson, H., Kapushesky, M., Kolesnikov, N., Rustici, G., Shojatalab, M., Abeygunawardena, N., Berube, H., Dylag, M., Emam, I., and Farne, A.et al. (2009) ArrayExpress update—from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 37: D868–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual, L., Blanca, J.M., Canizares, J., and Nuez, F. (2009) Transcriptomic analysis of tomato carpel development reveals alterations in ethylene and gibberellin synthesis during pat3/pat4 parthenocarpic fruit set. BMC Plant Biol. 9: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A., and Murphy, A.S. (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci. 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Philouze, J. (1983) Natural parthenocarpy in tomato. 1. review of bibliography. Agronomie 3: 611–620 [Google Scholar]
- Philouze, J. (1985) Natural parthenocarpy in tomato. 2. study of a varietal collection. Agronomie 5: 47–54 [Google Scholar]
- Piasecka, B., Robinson-Rechavi, M., and Bergmann, S. (2012) Correcting for the bias due to expression specificity improves the estimation of constrained evolution of expression between mouse and human. Bioinformatics 28: 1865–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povero, G., Gonzali, S., Bassolino, L., Mazzucato, A., and Perata, P. (2011) Transcriptional analysis in high-anthocyanin tomatoes reveals synergistic effect of Aft and atv genes. J. Plant Physiol. 168: 270–279 [DOI] [PubMed] [Google Scholar]
- Ren, X.Y., Fiers, M.W., Stiekema, W.J., and Nap, J.P. (2005) Local coexpression domains of two to four genes in the genome of Arabidopsis. Plant Physiol. 138: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, X.Y., Stiekema, W.J., and Nap, J.P. (2007) Local coexpression domains in the genome of rice show no microsynteny with Arabidopsis domains. Plant Mol. Biol. 65: 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef, D.N., Reshef, Y.A., Finucane, H.K., Grossman, S.R., McVean, G., Turnbaugh, P.J., Lander, E.S., Mitzenmacher, M., and Sabeti, P.C. (2011) Detecting novel associations in large data sets. Science 334: 1518–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer, C., Lisec, J., Czedik-Eysenberg, A., Sulpice, R., Flis, A., Grieder, C., Altmann, T., Stitt, M., Willmitzer, L., and Melchinger, A.E. (2012) Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc. Natl. Acad. Sci. USA 109: 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., Hirai, M.Y., and Yonekura-Sakakibara, K. (2008) Decoding genes with coexpression networks and metabolomics—‘majority report by precogs’. Trends Plant Sci. 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Sakata, K., Ohyanagi, H., Nobori, H., Nakamura, T., Hashiguchi, A., Nanjo, Y., Mikami, Y., Yunokawa, H., and Komatsu, S. (2009) Soybean proteome database: a data resource for plant differential omics. J. Proteome Res. 8: 3539–3548 [DOI] [PubMed] [Google Scholar]
- Saladie, M., Matas, A.J., Isaacson, T., Jenks, M.A., Goodwin, S.M., Niklas, K.J., Xiaolin, R., Labavitch, J.M., Shackel, K.A., and Fernie, A.R.et al. (2007) A reevaluation of the key factors that influence tomato fruit softening and integrity. Plant Physiol. 144: 1012–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S., Tabata, S., Hirakawa, H., Asamizu, E., Shirasawa, K., Isobe, S., Kaneko, T., Nakamura, Y., Shibata, D., and Aoki, K.et al. (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer, N., Semel, Y., Roessner, U., Gur, A., Balbo, I., Carrari, F., Pleban, T., Perez-Melis, A., Bruedigam, C., and Kopka, J.et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Schauer, N., Semel, Y., Balbo, I., Steinfath, M., Repsilber, D., Selbig, J., Pleban, T., Zamir, D., and Fernie, A.R. (2008) Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schijlen, E.G., de Vos, C.H., Martens, S., Jonker, H.H., Rosin, F.M., Molthoff, J.W., Tikunov, Y.M., Angenent, G.C., van Tunen, A.J., and Bovy, A.G. (2007) RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol. 144: 1520–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semel, Y., Nissenbaum, J., Menda, N., Zinder, M., Krieger, U., Issman, N., Pleban, T., Lippman, Z., Gur, A., and Zamir, D. (2006) Overdominant quantitative trait loci for yield and fitness in tomato. Proc. Natl. Acad. Sci. USA 103: 12981–12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, R.E., Yan, B., Stanley-Fernandez, S.M., Chen, Z.J., and Garbow, J.R. (2008) NMR characterization of hydration and thermal stress in tomato fruit cuticles. Phytochemistry 69: 2689–2695 [DOI] [PubMed] [Google Scholar]
- Steuer, R., Kurths, J., Daub, C.O., Weise, J., and Selbig, J. (2002) The mutual information: detecting and evaluating dependencies between variables. Bioinformatics 18 (Suppl 2): S231–240 [DOI] [PubMed] [Google Scholar]
- Sun, W., Xu, X., Zhu, H., Liu, A., Liu, L., Li, J., and Hua, X. (2010) Comparative transcriptomic profiling of a salt-tolerant wild tomato species and a salt-sensitive tomato cultivar. Plant Cell Physiol. 51: 997–1006 [DOI] [PubMed] [Google Scholar]
- Suzuki, A., Suwabe, K., and Yano, K. (2009) Web databases for omics data in tomato. J. Japan. Soc. Hort. Sci. 78: 23–31 [Google Scholar]
- Tabuchi, M., Sugiyama, K., Ishiyama, K., Inoue, E., Sato, T., Takahashi, H., and Yamaya, T. (2005) Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J. 42: 641–651 [DOI] [PubMed] [Google Scholar]
- Takos, A.M., and Rook, F. (2012) Why biosynthetic genes for chemical defense compounds cluster. Trends Plant Sci. 17: 383–388 [DOI] [PubMed] [Google Scholar]
- Tikunov, Y., Lommen, A., de Vos, C.H., Verhoeven, H.A., Bino, R.J., Hall, R.D., and Bovy, A.G. (2005) A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 139: 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana, D., Semel, Y., Tohge, T., Beleggia, R., Cattivelli, L., Rosental, L., Nikoloski, Z., Zamir, D., Fernie, A.R., and Fait, A. (2012) Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PloS Genet. 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R., and Nagy, F. (2005) Signalling and gene regulation in response to ultraviolet light. Curr. Opin. Plant Biol. 8: 477–482 [DOI] [PubMed] [Google Scholar]
- Ursem, R., Tikunov, Y., Bovy, A., van Berloo, R., and van Eeuwijk, F. (2008) A correlation network approach to metabolic data analysis for tomato fruits. Euphytica 161: 181–193 [Google Scholar]
- Usadel, B., Obayashi, T., Mutwil, M., Giorgi, F.M., Bassel, G.W., Tanimoto, M., Chow, A., Steinhauser, D., Persson, S., and Provart, N.J. (2009) Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ. 32: 1633–1651 [DOI] [PubMed] [Google Scholar]
- Van Bel, M., Proost, S., Wischnitzki, E., Movahedi, S., Scheerlinck, C., VandePeer, Y., and Vandepoele, K. (2012) Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer, I.M., Stam, M.E., vanTunen, A.J., Mol, J.N., and Stuitje, A.R. (1992) Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen, S. (2000) Graph Clustering by Flow Simulation. PhD thesis. University of Utrecht [Google Scholar]
- Vidal, E.A., Araus, V., Lu, C., Parry, G., Green, P.J., Coruzzi, G.M., and Gutierrez, R.A. (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, M., Takahashi, H., Altaf-Ul-Amin, M., Nakamura, K., Hirai, M.Y., Ohta, D., and Kanaya, S. (2012) Prediction of operon-like gene clusters in the Arabidopsis thaliana genome based on co-expression analysis of neighboring genes. Gene 503: 56–64 [DOI] [PubMed] [Google Scholar]
- Wang, H., Jones, B., Li, Z.G., Frasse, P., Delalande, C., Regad, F., Chaabouni, S., Latche, A., Pech, J.C., and Bouzayen, M. (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Schauer, N., Usadel, B., Frasse, P., Zouine, M., Hernould, M., Latche, A., Pech, J.C., Fernie, A.R., and Bouzayen, M. (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21: 1428–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth, W., Loureiro, M.E., Wenzel, K., and Fiehn, O. (2004) Differential metabolic networks unravel the effects of silent plant phenotypes. Proc. Natl. Acad. Sci. USA 101: 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell, A.M., Rowe, H.C., Hansen, B.G., Ticconi, C., Halkier, B.A., and Kliebenstein, D.J. (2007) Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 3: 1687–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., Luscombe, N.M., Lu, H.X., Zhu, X., Xia, Y., Han, J.D., Bertin, N., Chung, S., Vidal, M., and Gerstein, M. (2004) Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 14: 1107–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrineh, P., Fierro, A.C., Sanchez-Rodriguez, A., De Moor, B., Engelen, K., and Marchal, K. (2011) COMODO: an adaptive coclustering strategy to identify conserved coexpression modules between organisms. Nucleic Acids Res. 39: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, S., Horrocks, J., and Lukens, L.N. (2006) Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J. 45: 347–357 [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Yu, J., Li, D., Liu, F., Zhou, X., Wang, T., Ling, Y., and Su, Z. (2010) PMRD: plant microRNA database. Nucleic Acids Res. 38: D806–813 [DOI] [PMC free article] [PubMed] [Google Scholar]