Abstract
The dwarf tomato variety Micro-Tom is regarded as a model system for functional genomics studies in tomato. Various tomato genomic tools in the genetic background of Micro-Tom have been established, such as mutant collections, genome information and a metabolomic database. Recent advances in tomato genome sequencing have brought about a significant need for reverse genetics tools that are accessible to the larger community, because a great number of gene sequences have become available from public databases. To meet the requests from the tomato research community, we have developed the Micro-Tom Targeting-Induced Local Lesions IN Genomes (TILLING) platform, which is comprised of more than 5000 EMS-mutagenized lines. The platform serves as a reverse genetics tool for efficiently identifying mutant alleles in parallel with the development of Micro-Tom mutant collections. The combination of Micro-Tom mutant libraries and the TILLING approach enables researchers to accelerate the isolation of desirable mutants for unraveling gene function or breeding. To upgrade the genomic tool of Micro-Tom, the development of a new mutagenized population is underway. In this paper, the current status of the Micro-Tom TILLING platform and its future prospects are described.
Keywords: Micro-Tom, TILLING, tomato, reverse genetics
Introduction
Tomato (Solanum lycopersicum) is an important vegetable crop that is commercially valuable worldwide. It is also a model plant of Solanaceae species because of its relatively small genomic size (950 Mb) and well-conserved genome organization (e.g., chromosome number) with other solanaceous plants (Hille et al. 1989, Van der Hoeven et al. 2002). Additionally, as tomato bears berry-type fruits, it is widely used for studying many aspects of fruit biology including development, ripening and metabolism (Carrari and Fernie 2006). A genome project has been advanced for tomato and its genome was entirely sequenced by the International Solanaceae Genomics Project (SOL). A comprehensive database including the majority of the tomato genome sequence and a large number of expressed sequence tags (ESTs) is presented through the Solanaceae Genomics Network (SGN) website (http://solgenomics.net/) (Bombarely et al. 2011). The genome information for mining target gene sequences is now available from the public database. It is also important to characterize candidate genes harboring roles in attractive tomato-specific traits. Combining the genome information and mutant resources is an effective and valuable strategy for accelerating functional genomics studies.
A dwarf tomato variety Micro-Tom has been extensively used for functional genomic studies because of its attractive features such as small plant size, a short life cycle and the existence of functional genomic tools. Micro-Tom also has an efficient transformation system, a metabolite database, a cDNA database, a genetic map with DNA markers and a mutant database (Aoki et al. 2010, Iijima et al. 2008, Saito et al. 2011, Shirasawa et al. 2010, Sun et al. 2006). The features of Micro-Tom make it possible to carry out large-scale mutant screening, even in limited spaces including greenhouses or growth chambers. It has been reported that MicroTom can be grown at high density, i.e., up to 1357 plants/m2 (Meissner et al. 1997). The Micro-Tom mutants are applicable to breeding programs for an efficient exploration of favorable agronomic traits as germplasm lines.
In recent years, the Micro-Tom TILLING (Targeting-Induced Local Lesions IN Genomes) platform has been developed as a reverse genetics tool of tomato that provides an opportunity to isolate novel mutant alleles for functional genomic studies and breeding (Okabe et al. 2011, 2012). TILLING technology was initially reported as a high-throughput reverse genetics tool for screening mutants. This technique was reported by McCallum et al. (2000a, 2000b). TILLING enables researchers to efficiently identify induced point mutations that are caused by chemical mutagens, such as ethyl methanesulfonate (EMS) in a large-scale mutant population of Arabidopsis. Although RNA interference and insertional mutagenesis by T-DNA or transposon tagging are effective reverse genetic tools for investigating gene function, their availability is often restricted to plant species harboring an efficient transformation system (Baulcombe 2004, Hirschi et al. 2003). The TILLING approach has several advantages compared to other reverse genetic tools. (1) EMS has been known to induce point mutations randomly in the whole genome, which makes it possible to produce a broad spectrum of mutant alleles, including loss-of function mutations such as truncations, splice junction mutations and leaky alleles, without performing plant transformation. (2) The TILLING approach could be applied to any plant species, and several mutants can be obtained from a relatively small-scale mutant population that contains on the order of 103 individuals (Emmanuel and Levy 2002). (3) Mutant alleles isolated by TILLING and the identified mutations can be directly used in breeding programs as parental germplasm materials or markers, because they are non-transgenic and the induced mutation is stable.
A number of agronomically important mutants in crop species have been identified by TILLING, such as the wheat waxy mutant alleles and the SBE IIa mutant alleles that display altered amylose content (Botticella et al. 2011, Slade et al. 2005, 2012), the potato granule-bound starch synthase I (waxy) mutant allele that produces high-amylopectin starch in the tuber (Muth et al. 2008), the Brasica napus FAE1 mutant allele that results in the altered content of erucic acid in the seed oil (Wang et al. 2008) and the melon CmACO1 mutant that produces fruit with enhanced shelf life (Dahmani-Mardas et al. 2010). By using a combined biochemical screen and TILLING approach, acyanogenic lines suitable for forage production have been successfully identified in Sorghum bicolor (Blomstedt et al. 2012). The TILLING technology was initially developed for use in functional genomic studies; it has now become a valuable tool in crop breeding for improving desirable traits in various species.
This review on TILLING technology discusses techniques for discovering induced point mutations from mutant populations and the current status of the platform in tomato. The review then focuses on the potential for the Micro-Tom TILLING platform in molecular breeding programs in tomato.
TILLING techniques for discovery of induced mutations
Several types of systems and methods have been reported for identifying point mutations. The denaturing high performance liquid chromatograpy (dHPLC) method was first reported as an original TILLING method that involves the generation of an Arabidopsis EMS mutant population, DNA isolation and pooling, PCR amplification of the target region, heteroduplex formation and identification of the heteroduplex (McCallum et al. 2000b). Although it was an automatic system, it was not feasible to scale up the method for high-throughput screening (Henikoff et al. 2004). Subsequently, the dHPLC method was replaced by the LI-COR DNA analyzer system, including PCR amplification with infra-red dye-labeled primers, digestion of heteroduplex DNAs using single strand-specific endonuclease CEL I (Oleykowski et al. 1998), followed by polyacrylamide electrophoresis and visualization of mutations (Colbert et al. 2001). The TILLING procedure with the use of single strand-specific endonuclease has been widely used in detecting mutations because of its cost-effectiveness and detection sensitivity. In addition, an alternative enzyme of CEL I, Arabidopsis endonuclease 1 (ENDO1), has been used in several TILLING platforms (Dahmani-Mardas et al. 2010, Minoia et al. 2010, Piron et al. 2010, Triques et al. 2007). The advantages of ENDO1 include the higher sensitivity for recognizing mismatches in DNA strands and the simple expression and purification method of the recombinant ENDO1 protein (Okabe et al. 2011, Triques et al. 2007, 2008). Alternative techniques without the enzymatic procedure and use of expensive reagents (i.e., labeled primers) were reported, such as high resolution DNA melting analysis (HRM) and conformation-sensitive capillary electrophoresis (CSCE) (Botticella et al. 2011, Gady et al. 2009). Although LI-COR/ENDO1 (CEL I) is regarded as a very efficient system for detecting mutations, time-consuming steps such as endonuclease digestion, sample purification and polyacrylamide gel electrophoresis are required. By contrast, when using the HRM and CSCE technologies, the only procedure required is the PCR amplification of the region of interest before analysis of the sample. The HRM method performs melting-curve analysis of PCR products intercalated with a fluorescent dye (LCGreen Plus+) by using LightScanner to discriminate the melting temperature difference between heteroduplex DNA and homoduplex DNA (Botticella et al. 2011, Gady et al. 2009, Ishikawa et al. 2010). The CSCE method detects the difference in the migration speed of heteroduplex DNAs compared to homoduplexes. A typical read length of HRM and CSCE is PCR fragments of 200–500 bp, which is shorter than the LI-COR/ENDO1 technology, which can screen a region of up to 1.5 kbp, or even up to 1.8 kb (Okabe Y, personal communication). When screening specific regions in genes of interest, such as functional domains, motifs and segmented short exons, HRM is efficient and cost-effective. HRM has been successfully employed as a high-throughput method for mutation discovery in tomato, wheat and Medaka (Botticella et al. 2011, Gady et al. 2009, Ishikawa et al. 2010). An alternative TILLING system that combines agarose gel electrophoresis and CEL I nuclease or CEL I-like nuclease from Brassica petiole has been constructed in soybean and rice, as a low-cost system without expensive equipment (Hoshino et al. 2010, Sato et al. 2006). The system in soybean enables a single person to screen approximately 6000 independent mutant lines in 1 day (Anai 2012).
In recent years, next-generation sequencing (NGS) technology has been applied to identify induced mutations in model plant and crop species including Arabidopsis, rice, wheat, tobacco and tomato (Reddy et al. 2012, Rigola et al. 2009, Tsai et al. 2011, Uchida et al. 2011). Rigola et al. (2009) first reported a TILLING approach using the Keypoint technology. They employed the Roche 454 GS FLX system and sequenced 28 pooled DNA samples isolated from 15,000 M2 plants representing 3008 M2 families in tomato. This was achieved by using a three-dimensional (3D) pooling strategy with 12 or eight DNA samples on each axis (e.g., x, y and z axis with 12, 8 and 8 samples, respectively) for screening EMS-induced mutations in a targeted 287 bp region of the SleIF4E gene. They obtained a total of 667,864 sequencing reads with an average read length of 254 bases per single run and identified two mutations through the use of a bioinformatics pipeline. The Illumina GA II platform has also been applied to TILLING in rice and wheat (Tsai et al. 2011). These authors took advantage of a multidimensional pooling strategy for screening induced mutations in the same manner as the GS FLX system. A DNA sample of 768 individuals was used for searching new mutations in rice (Till et al. 2007) and wheat (Uauy et al. 2009) mutant populations composed of 768 individuals, which were previously characterized by using CEL I-based TILLING screening. A number of new mutations, several previously identified mutations, and some false-positives were successfully identified by the NGS-based approach. This platform could sequence 40 million reads of 40 bp lengths (i.e., 1,600,000 kb of sequence) with a desired coverage of 2500 per base for the number of 1.5 kb targets amplified from a pool of 96 individuals in a single lane. In current applications of NGS-based TILLING, targeted PCR products amplified from mutant DNA pools have been sequenced, because the cost of sequencing the whole genome of mutant populations is still prohibitive. These studies demonstrate that the TILLING approach is constantly evolving with the advent of new technology that engenders greater efficiency in high-throughput screening.
TILLING platform in tomato
To date, TILLING platforms have been developed in several crop species including rice (Suzuki et al. 2008, Till et al. 2007), soybean (Cooper et al. 2008), maize (Till et al. 2004), tomato (Gady et al. 2009, Minoia et al. 2010, Piron et al. 2010), melon (Dahmani-Mardas et al. 2010, González et al. 2011), wheat (Slade et al. 2005) and barley (Caldwell et al. 2004, Talamè et al. 2008). These studies report that TILLING is an efficient tool for mutation discovery in crop species, similarly as in Arabidopsis.
In tomato, four independent TILLING platforms that consist of a variety of genetic backgrounds (i.e., M82, Micro-Tom, Red Setter and TPAADASU) have been developed by different groups (Table 1). M82 has frequently been used for tomato genetic analysis during the past decade, because the genetic tools, including high-quality mutant population, linkage map, and DNA marker and introgression lines, are now available (Eshed and Zamir 1994, Menda et al. 2004). To efficiently generate a saturated tomato mutant population, EMS has been widely used as an effective mutagen. Mutagenesis using 0.5–1.0% EMS has been frequently reported in tomato and the resu1ts suggest that 1.0% EMS is regarded as an appropriate condition for mutagenizing tomato seeds in several varieties. The mutation frequency of the TILLING platform ranges from one mutation per 322–737 kb. Several novel mutant alleles have been identified from a tomato EMS mutant population using the TILLING approach. Piron et al. (2010) successfully isolated a novel potyvirus-resistant mutant, namely the SleIF4E1 allele. Jones et al. (2012) isolated SlDET1 mutant alleles, which are allelic to high pigment 2 (hp2), for improving the nutritional quality of fruit. They were screened from a M82 EMS mutant population developed by Menda et al. (2004). Gady et al. (2012) isolated phytoene synthase (Psy1) mutant alleles from a TPAADASU EMS-mutagenized population, namely SlPsy1 P192L and W180stop lines, which are leaky and knockout alleles, respectively. We have previously isolated novel mutant alleles of an ethylene receptor Solanum lycopersicum ETHYLENE RESPONSE 1 (SlETR1) gene, Sletr1-1 and Sletr1-2, by using the Micro-Tom TILLING platform. This study provided novel parental germplasm for improving fruit shelf life in tomato (Okabe et al. 2011). These reports demonstrated that the tomato TILLING platform is a valuable tool for molecular breeding of desirable traits. In a recent advance of our project, we upgraded the Micro-Tom TILLING platform by preparing additional DNA pools. Approximately 2000 mutant lines were mutagenized by treatment with 0.7–1.0% EMS. This DNA pool has approximately one mutation per 600 kb and is newly available for screening mutants by TILLING. Thus, the current Micro-Tom TILLING platform consists of a total of approximately 5000 EMS-mutagenized lines (Table 2). Mutation frequency was calculated as follows: total screened length per total number of mutations that represents the length of nucleotide which contains one mutation. And average number of alleles was calculated as follows: total number of mutant lines per mutation frequency. Preliminary tests estimate that an average of 5.8 alleles (2180 lines/ 1710 kb + 872 lines/737 kb + 2033 lines/600 kb = 5.8 alleles) per kb is obtained when screening 5000 EMS-mutagenized lines. The average number of mutant alleles per screening of 1 kb in other tomato platforms is 1.5–2.0 times higher in M82 (4759 lines/574 kb = 8.3 alleles), Red Setter (3885 lines/574 kb + 1284 lines/322 kb = 10.8 alleles) and TPPADASU (8225 lines/737 kb = 11.2 alleles). However, the performance of the Micro-Tom TILLING platform will reach similar levels to other tomato platforms by continuous efforts to improve the quality of the platform.
Table 1.
Overview of TILLING platform in tomato
| Cultivar | Population size | EMS dose | Mutation frequency | Screening Method | Screened gene | Reference |
|---|---|---|---|---|---|---|
| M82 | 4759 | 0.7% | 1/574 kb | LI-COR/ENDO1 | eIF4E, DET1, COP1-like, DDB1a, COP10, NAM, ACO1, E8, DHS, Rab11a, PG, MET1, Exp1 CRTISO, CUL4 | Piron et al. (2010) |
| Micro-Tom | 2180, 872 | 0.5%, 1.0% | 1/1710 kb, 1/737 kb | LI-COR/ENDO1 | ETR1-ETR6, GABA-T1, GABA-T3 SSADH, PL | Okabe et al. (2011) |
| Red Setter | 3885, 1284 | 0.7%, 1.0% | 1/574 kb, 1/322 kb | LI-COR/ENDO1 | Rab11a, PG, Exp1, RIN, GR, Lcy-a, Lcy-b | Minoia et al. (2010) |
| TPPADASU | 8225 | 1.0% | 1/737 kb | HRM and CSCE | ARF7, ProDH, PSY1, Sus2 | Gady et al. (2009) |
Table 2.
Summary of the current Micro-Tom TILLING platform
| Category | Year | EMS condition | Population size | Mutation frequency |
|---|---|---|---|---|
| Population 1 | 2005–2006 | 0.5% | 2180 | 1/1710 kb |
| Population 2 | 2008–2009 | 1.0% | 872 | 1/737 kb |
| Population 3 (New) | 2010–2011 | 1.0%, 0.7% | 2033 | ~1/600 kb |
| Population 4 (New) | 2012–2013 | 1.0% + 1.0% | ~1900 | ND* |
ND, not determined
Potential of the Micro-Tom TILLING platform in tomato breeding
During the past decade, Micro-Tom has been recognized as an attractive material for functional genomics studies as described in a large number of articles (Aoki et al. 2010, Emmanuel and Levy 2002, Matsukura et al. 2008, Meissner et al. 1997, Saito et al. 2011). The availability of Micro-Tom for tomato breeding has been recognized because of its clear advantages and potentials. In our routine experiments, Micro-Tom has been routinely managed at approximately 100–200 plants/m2 and can also be managed at a maximum density of 1357 plants/m2 (Meissner et al. 1997). By contrast, common tomato cultivars may be grown at a maximum density of approximately 4 plants/m2. With respect to the cultivation management in a greenhouse, the working efficiency could increase 20-fold (i.e., up to 300-fold) per unit area by the use of Micro-Tom. This advantage would accelerate breeding programs for an efficient screening of desired agronomic traits. In a case study, we reported that the combination of a Micro-Tom EMS-mutant library and the TILLING technology was useful for efficiently obtaining genetic variation in fruit shelf life by manipulating an ethylene receptor gene SlETR1 (Okabe et al. 2011, 2012). Several new allelic mutants of indole-3-acetic acid 9 (iaa9) that produce parthenocarpic fruits at a high rate were identified from EMS-induced and γ-ray irradiated Micro-Tom mutant populations (Saito et al. 2011). Additionally, some agronomically important traits in tomato (e.g., fruit set, fruit ripening, fruit development and carbohydrate metabolism) have been studied using Micro-Tom as a model system (Itkin et al. 2009, Obiadalla-Ali et al. 2004, Serrani et al. 2007, 2008, Vrebalov et al. 2009, Wang et al. 2009, Yin et al. 2010). These studies revealed that agronomically important genetic traits are not greatly altered in Micro-Tom, as compared with other common cultivars. In support of this, Carvalho et al. (2011) reported the development of MicroTom near isogenic lines (NILs) that introgressed a suite of hormonal and photomorphogenetic mutant alleles relevant to auxin sensitivity or endogenous levels of auxin, ethylene, abscisic acid, gibberellin, brassinosteroids and light responses, which were originally identified in wild relatives or spontaneous mutants of common cultivars. They observed expected phenotypes including altered hormonal and light responses in the NILs, indicating that the genetic background of Micro-Tom did not affect the expression of mutant phenotypes. These results demonstrate that Micro-Tom is applicable to conduct breeding of common tomato cultivars. Because mutant screening and evaluation of the phenotype will be synergistically advanced by combining a MicroTom mutant library and the TILLING platform, the MicroTom TILLING platform will become an alternative tool for improving agronomically important traits in tomato in mutant-based breeding.
Future prospects
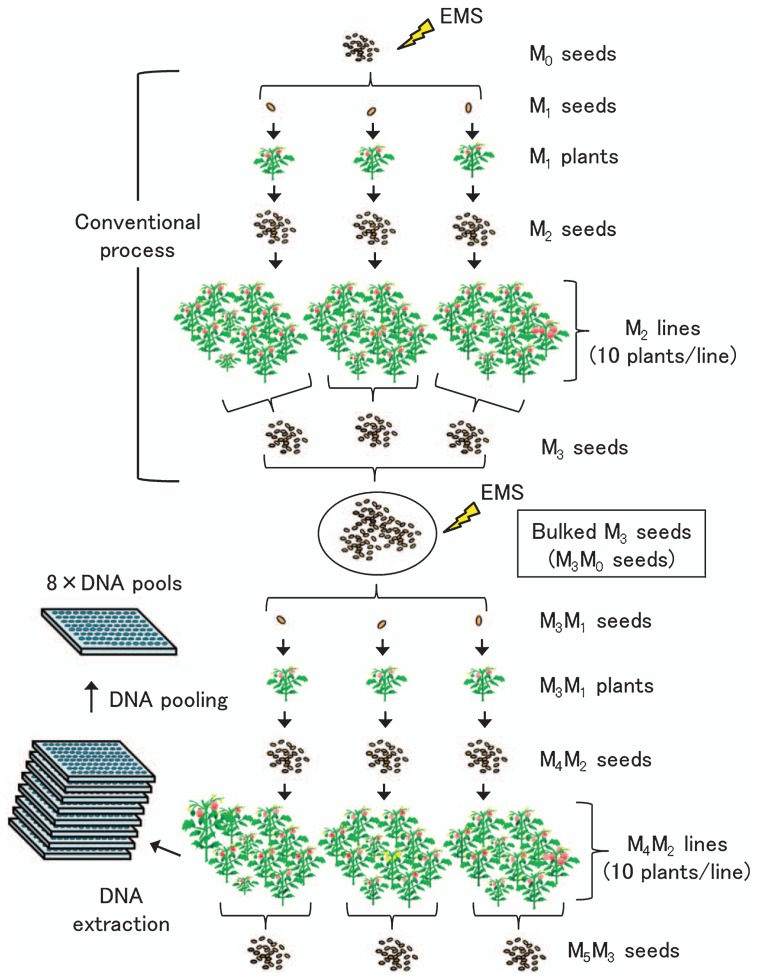
Several TILLING methods for identifying induced mutations have been developed; they have advantages and disadvantages depending on each situation and the experimental goals. The low-cost TILLING methods, such as HRM and agarose-gel-based system, are suitable for setting the initial TILLING platform because there is no requirement to prepare special equipment for the detection of mutations. The NGS platform is regarded as one of the most high-throughput screening systems. This tool could be applied in addition to conventional TILLING methods with decreasing sequencing costs. A next approach may be resequencing mutant lines using NGS and in silico procedures will gradually be employed in TILLING. Emerging technologies will enable rapid recovery of allelic series of target genes from mutant populations without time-consuming steps. Thus, high-quality Micro-Tom mutant resources would be increasingly valuable. Currently, to further improve the availability of the Micro-Tom TILLING platform, we intend to increase the scale of mutant populations by developing double EMS-mutagenized lines that might have high-frequency mutations, which would reduce the cost and labor for screening (Fig. 1). Approximately 2200 M3M1 plants have been generated; the progeny consist of at least 1900 M4M2 lines that could be available for screening mutants by TILLING in 2013 (Table 2). The goal in our study is to upgrade the Micro-Tom TILLING platform to a nearly-saturated level composed of 7000–9000 lines, which will enable us to obtain more than 10 alleles per kb. Because the TILLING platform has been closely linked to the Micro-Tom mutant database TOMATOMA (http://tomato.nbrp.jp/indexEn.html), additional mutant populations will become beneficial resources for both forward- and reverse genetic studies and provide opportunities to obtain more novel mutants and allelic variations of target genes. Also, the mutant screening of several genes involved in important traits is currently underway. A large number of important new alleles could be identified from the Micro-Tom mutant population by the TILLING approach. Because there is a growing demand for public access to the tomato TILLING platform to recover desirable mutants, we will continuously upgrade the MicroTom TILLING platform.
Fig. 1.
Flow chart for constructing mutant populations with double-EMS mutagenesis. The conventional process of EMS mutagenesis (upper panel) was described by Saito et al. (2011). In the following steps, bulked M3 (M3M0) seeds were mutagenized with 1.0% EMS and M3M1 seeds were sown in the greenhouse to harvest M4M2 seeds from single M3M1 plants. Ten M4M2 seeds were sown and grown as an M4M2 line and M5M3 seeds were harvested from the same M4M2 lines as a bulked seed. Mutant DNA was extracted from each M4M2 line (10 plants per line) and pooled as 8-fold DNA for TILLING screening.
Acknowledgments
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan [grant-in-aid (to H.E.)], National BioResource Project (No. ADD20035) and Bio-Oriented Technology Research Advancement Institution (BRAIN) [the ‘Research and Development program for New Bio-industry Initiatives’ (to H.E.)].
Literature Cited
- Anai, T. (2012) Potential of a mutant-based reverse genetic approach for functional genomics and molecular breeding in soybean. Breed. Sci. 61: 462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, K., Yano, K., Suzuki, A., Kawamura, S., Sakurai, N., Suda, K., Kurabayashi, A., Suzuki, T., Tsugane, T., and Watanabe, M.et al. (2010) Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics 11: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Blomstedt, C. K., Gleadow, R.M., O’Donnell, N., Naur, P., Jensen, K., Laursen, T., Olsen, C.E., Stuart, P., Hamill, J.D., and Møller, B.L.et al. (2012) A combined biochemical screen and TILLING approach identifies mutations in Sorghum bicolor L. Moench resulting in acyanogenic forage production. Plant Biotechnol. J. 10: 54–66 [DOI] [PubMed] [Google Scholar]
- Bombarely, A., Menda, N., Tecle, I.Y., Buels, R.M., Strickler, S., Fischer-York, T., Pujar, A., Leto, J., Gosselin, J., and Mueller, L.A. (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 39: 1149–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botticella, E., Sestili, F., Hernandez-Lopez, A., Phillips, A., and Lafiandra, D. (2011) High resolution melting analysis for the detection of EMS induced mutations in wheat SBEIIa genes. BMC Plant Biol. 11: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell, D.G., McCallum, N., Shaw, P., Muehlbauer, G.J., Marshall, D.F., and Waugh, R. (2004) A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare L.). Plant J. 40: 143–150 [DOI] [PubMed] [Google Scholar]
- Carrari, F., and Fernie, A.R. (2006) Metabolic regulation underlying tomato fruit development. J. Exp. Bot. 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Carvalho, R.F., Campos, M.L., Pino, L.E., Crestana, S.L., Zsögön, A., Lima, J.E., Benedito, V.A., and Peres, L.E. (2011) Convergence of developmental mutants into a single tomato model system: ‘Micro-Tom’ as an effective toolkit for plant development research. Plant Methods 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert, T., Till, B.J., Tompa, R., Reynolds, S., Steine, M.N., Yeung, A.T., McCallum, C.M., Comai, L., and Henikoff, S. (2001) High-throughput screening for induced point mutations. Plant Physiol. 126: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, J.L., Till, B.J., Laport, R.G., Darlow, M.C., Kleffner, J.M., Jamai, A., El-Mellouki, T., Liu, S., Ritchie, R., and Nielsen, N.et al. (2008) TILLING to detect induced mutations in soybean. BMC Plant Biol. 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmani-Mardas, F., Troadec, C., Boualem, A., Lévêque, S., Alsadon, A.A., Aldoss, A.A., Dogimont, C., and Bendahmane, A. (2010) Engineering melon plants with improved fruit shelf life using the TILLING approach. PloS One 5: e15776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanuel, E., and Levy, A.A. (2002) Tomato mutants as tools for functional genomics. Curr. Opin. Plant Biol. 5: 112–117 [DOI] [PubMed] [Google Scholar]
- Eshed, Y., and Zamir, D. (1994) A genomic library of Lycopersicon pennellii in L. esculentum: A tool for fine mapping of genes. Euphytica 79: 175–179 [Google Scholar]
- Gady, A.L., Hermans, F.W., VandeWal, M.H., vanLoo, E.N., Visser, R.G., and Bachem, C.W. (2009) Implementation of two high through-put techniques in a novel application: detecting point mutations in large EMS mutated plant populations. Plant Methods 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gady, A.L., Vriezen, W.H., Van de Wal, M.H., Huang, P., Bovy, A.G., Visser, R.G., and Bachem, C.W. (2012) Induced point mutations in the phytoene synthase 1 gene cause differences in carotenoid content during tomato fruit ripening. Mol. Breed. 29: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, M., Xu, M., Esteras, C., Roig, C., Monforte, A.J., Troadec, C., Pujol, M., Nuez, F., Bendahmane, A., and Garcia-Mas, J.et al. (2011) Towards a TILLING platform for functional genomics in Piel de Sapo melons. BMC Res. Notes 4: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., Till, B.J., and Comai, L. (2004) TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135: 630–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, J., Koornneef, M., Ramanna, M.S., and Zabel, P. (1989) Tomato: a crop species amenable to improvement by cellular and molecular methods. Euphytica 42: 1–23 [Google Scholar]
- Hirschi, K.D. (2003) Insertional mutants: a foundation for assessing gene function. Trends in Plant Sci. 8: 205–207 [DOI] [PubMed] [Google Scholar]
- Hoshino, T., Takagi, Y., and Anai, T. (2010) Novel GmFAD2-1b mutant alleles created by reverse genetics induce marked elevation of oleic acid content in soybean seeds in combination with GmFAD2-1a mutant alleles. Breed. Sci. 60: 419–425 [Google Scholar]
- Iijima, Y., Nakamura, Y., Ogata, Y., Tanaka, K., Sakurai, N., Suda, K., Suzuki, T., Suzuki, H., Okazaki, K., and Kitayama, M.et al. (2008) Metabolite annotations based on the integration of mass spectral information. Plant J. 54: 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, T., Kamei, Y., Otozai, S., Kim, J., Sato, A., Kuwahara, Y., Tanaka, M., Deguchi, T., Inohara, H., and Tsujimura, T.et al. (2010) High-resolution melting curve analysis for rapid detection of mutations in a Medaka TILLING library. BMC Mol. Biol. 11: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin, M., Seybold, H., Breitel, D., Rogachev, I., Meir, S., and Aharoni, A. (2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Jones, M.O., Piron-Prunier, F., Marcel, F., Piednoir-Barbeau, E., Alsadon, A.A., Wahb-Allah, M.A., Al-Doss, A.A., Bowler, C., Bramley, P.M., and Fraser, P.D.et al. (2012) Characterisation of alleles of tomato light signalling genes generated by TILLING. Phytochemistry 79: 78–86 [DOI] [PubMed] [Google Scholar]
- Matsukura, C., Aoki, K., Fukuda, N., Mizoguchi, T., Asamizu, E., Saito, T., Shibata, D., and Ezura, H. (2008) Comprehensive resources for tomato functional genomics based on the miniature model tomato Micro-Tom. Curr. Genomics 9: 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A., and Henikoff, S. (2000a) Targeted screening for induced mutations. Nat. Biotechnol. 18: 455–457 [DOI] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A., and Henikoff, S. (2000b) Targeting induced local lesions in genomes (TILLING) for plant functional genomes. Plant Physiol. 123: 439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda, N., Semel, Y., Peled, D., Eshed, Y., and Zamir, D. (2004) In silico screening of a saturated mutation library of tomato. Plant J. 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Meissner, R., Jacobson, Y., Melamed, S., Levyatuv, S., Shalev, G., Ashri, A., Elkind, Y., and Levy, A.A. (1997) A new model system for tomato genetics. Plant J. 12: 1465–1472 [Google Scholar]
- Minoia, S., Petrozza, A., D’Onofrio, O., Piron, F., Mosca, G., Sozio, G., Cellini, F., Bendahmane, A., and Carriero, F. (2010) A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 3: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth, J., Hartje, S., Twyman, R.M., Hofferbert, H.R., Tacke, E., and Prüfer, D. (2008) Precision breeding for novel starch variants in potato. Plant Biotechnol. J. 6: 576–584 [DOI] [PubMed] [Google Scholar]
- Obiadalla-Ali, H., Fernie, A.R., Kossmann, J., and Lloyd, J.R. (2004) Developmental analysis of carbohydrate metabolism in tomato (Lycopersicon esculentum cv. Micro-Tom) fruits. Physiol. Plant. 120: 196–204 [DOI] [PubMed] [Google Scholar]
- Okabe, Y., Asamizu, E., Saito, T., Matsukura, C., Ariizumi, T., Brès, C., Rothan, C., Mizoguchi, T., and Ezura, H. (2011) Tomato TILLING technology: development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol. 52: 1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe, Y., Asamizu, E., Ariizumi, T., Shirasawa, K., Tabata, S., and Ezura, H. (2012) Availability of Micro-Tom mutant library combined with TILLING in molecular breeding of tomato fruit shelf-life. Breed. Sci. 62: 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleykowski, C.A., BronsonMullins, C.R., Godwin, A.K., and Yeung, A.T. (1998) Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 26: 4597–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron, F., Nicolaï, M., Minoïa, S., Piednoir, E., Moretti, A., Salgues, A., Zamir, D., Caranta, C., and Bendahmane, A. (2010) An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PloS One 5: e11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, T.V., Dwivedi, S., and Sharma, N.V. (2012) Development of TILLING by sequencing platform towards enhanced leaf yield in tobacco. Industrial Crops and Products 40: 324–335 [Google Scholar]
- Rigola, D., vanOeveren, J., Janssen, A., Bonné, A., Schneiders, H., van der Poel, H.J., van Orsouw, N.J., Hogers, R.C., de Both, M.T., and van Eijk, M.J. (2009) High-throughput detection of induced mutations and natural variation using KeyPoint technology. PLoS One 4: e4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, T., Ariizumi, T., Okabe, Y., Asamizu, E., Hiwasa-Tanase, K., Fukuda, N., Mizoguchi, T., Yamazaki, Y., Aoki, K., and Ezura, H. (2011) TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 52: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y., Shirasawa, K., Takahashi, Y., Nishimura, M., and Nishio, T. (2006) Mutant Selection from Progeny of Gamma-ray-irradiated Rice by DNA Heteroduplex Cleavage using Brassica Petiole Extract. Breed. Sci. 56: 179–183 [Google Scholar]
- Serrani, J.C., Sanjuán, R., Ruiz-Rivero, O., Fos, M., and García-Martínez, J.L. (2007) Gibberellin regulation of fruit set and growth in tomato. Plant Physiol. 145: 246–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrani, J.C., Ruiz-Rivero, O., Fos, M., and García-Martínez, J.L. (2008) Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J. 56: 922–934 [DOI] [PubMed] [Google Scholar]
- Shirasawa, K., Isobe, S., Hirakawa, H., Asamizu, E., Fukuoka, H., Just, D., Rothan, C., Sasamoto, S., Fujishiro, T., and Kishida, Y.et al. (2010) SNP discovery and linkage map construction in cultivated tomato. DNA Res. 17: 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade, A.J., Fuerstenberg, S.I., Loeffler, D., Steine, M.N., and Facciotti, D. (2005) A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 23: 75–81 [DOI] [PubMed] [Google Scholar]
- Slade, A.J., McGuire, C., Loeffler, D., Mullenberg, J., Skinner, W., Fazio, G., Holm, A., Brandt, K.M., Steine, M.N., and Goodstal, J.F.et al. (2012) Development of high amylose wheat through TILLING. BMC Plant Biol. 12: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., Eiguchi, M., Kumamaru, T., Satoh, H., Matsusaka, H., Moriguchi, K., Nagato, Y., and Kurata, N. (2008) MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics 279: 213–223 [DOI] [PubMed] [Google Scholar]
- Sun, H.J., Uchii, S., Watanabe, S., and Ezura, H. (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 47: 426–431 [DOI] [PubMed] [Google Scholar]
- Talamè, V., Bovina, R., Sanguineti, M.C., Tuberosa, R., Lundqvist, U., and Salvi, S. (2008) TILLMore, a resource for the discovery of chemically induced mutants in barley. Plant Biotechnol. J. 6: 477–485 [DOI] [PubMed] [Google Scholar]
- Triques, K., Sturbois, B., Gallais, S., Dalmais, M., Chauvin, S., Clepet, C., Aubourg, S., Rameau, C., Caboche, M., and Bendahmane, A. (2007) Characterization of Arabidopsis thaliana mismatch specific endonucleases: application to mutation discovery by TILLING in pea. Plant J. 51: 1116–1125 [DOI] [PubMed] [Google Scholar]
- Triques, K., Piednoir, E., Dalmais, M., Schmidt, J., Le Signor, C., Sharkey, M., Caboche, M., Sturbois, B., and Bendahmane, A. (2008) Mutation detection using ENDO1: application to disease diagnostics in humans and TILLING and Eco-TILLING in plants. BMC Mol. Biol. 9: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till, B.J., Reynolds, S.H., Weil, C., Springer, N., Burtner, C., Young, K., Bowers, E., Codomo, C.A., Enns, L.C., and Odden, A.R.et al. (2004) Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till, B.J., Cooper, J., Tai, T.H., Colowit, P., Greene, E.A., Henikoff, S., and Comai, L. (2007) Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, H., Howell, T., Nitcher, R., Missirian, V., Watson, B., Ngo, K.J., Lieberman, M., Fass, J., Uauy, C., and Tran, R.K.et al. (2011) Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol. 156: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy, C., Paraiso, F., Colasuonno, P., Tran, R.K., Tsai, H., Berardi, S., Comai, L., and Dubcovsky, J. (2009) A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 9: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, N., Sakamoto, T., Kurata, T., and Tasaka, M. (2011) Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 52: 716–722 [DOI] [PubMed] [Google Scholar]
- Van der Hoeven, R., Ronning, C., Giovannoni, J., Martin, G., and Tanksley, S. (2002) Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov, J., Pan, I.L., Arroyo, A.J., McQuinn, R., Chung, M., Poole, M., Rose, J., Seymour, G., Grandillo, S., and Giovannoni, J.et al. (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N., Wang, Y., Tian, F., King, G.J., Zhang, C., Long, Y., Shi, L., and Meng, J. (2008) A functional genomics resource for Brassica napus: development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytol. 180: 751–765 [DOI] [PubMed] [Google Scholar]
- Wang, H., Schauer, N., Usadel, B., Frasse, P., Zouine, M., Hernould, M., Latché, A., Pech, J.C., Fernie, A.R., and Bouzayen, M. (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21: 1428–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y.G., Kobayashi, Y., Sanuki, A., Kondo, S., Fukuda, N., Ezura, H., Sugaya, S., and Matsukura, C. (2010) Salinity induces carbohydrate accumulation and sugar-regulated starch biosynthetic genes in tomato (Solanum lycopersicum L. cv. ‘Micro-Tom’) fruits in an ABA- and osmotic stress-independent manner. J. Exp. Bot. 61: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]