Abstract
Leymus mollis (Triticeae; Poaceae) is a useful genetic resource for wheat (Triticum aestivum L.) breeding via wide hybridization to introduce its chromosomes and integrate its useful traits into wheat. Leymus mollis is highly tolerant to abiotic stresses such as drought and salinity and resistant to various diseases, but the genetic mechanisms controlling its physiological tolerance remain largely unexplored. We identified and cloned an allene oxide cyclase (AOC) gene from L. mollis that was strongly expressed under salt stress. AOC is involved in biosynthesis of jasmonic acid, an important signaling compound that mediates a wide range of adaptive responses. LmAOC cDNA consisted of 717 bp, coding for a protein with 238 amino acids that was highly similar to AOCs from barley (Hordeum vulgare) and other monocots. Subcellular localization using Nicotiana benthamiana confirmed it as a chloroplast-localized protein. LmAOC was found to be a multiple-copy gene, and that some copies were conserved and efficiently expressed in wheat–Leymus chromosome addition lines. LmAOC expression was upregulated under drought, heat, cold and wounding stresses, and by jasmonic acid and abscisic acid. Our results suggest that LmAOC plays an important role in L. mollis adaptation to abiotic stresses and it could be useful for wheat improvement.
Keywords: allene oxide cyclase, jasmonic acid, Leymus mollis, LmAOC, drought stress response, salinity, wheat
Introduction
Plants are often exposed to biotic and abiotic stresses in their natural environments. Salinity and drought stresses are the major abiotic stresses that adversely affect plant growth and productivity worldwide. To adapt and survive, plants have developed complex defense mechanisms to perceive various signals and respond optimally to environmental stresses. Phytohormones such as jasmonic acid, abscisic acid (ABA) and ethylene are low-molecular-weight molecules that regulate the protective responses of plants against both biotic and abiotic stresses (Fujita et al. 2006). Jasmonic acid is particularly important signaling molecule that is involved in activating gene expression during a range of plant responses (Wu et al. 2011). The role of jasmonic acid in regulating plant stress responses often requires the elevation of its endogenous levels by de novo synthesis (León et al. 1999). The biosynthesis of jasmonic acid starts with conversion of linolenic acid to cis-12-oxo-phytodienoic acid in the chloroplasts by the sequential action of the enzymes lipoxygenase, allene oxide synthase, and allene oxide cyclase (AOC). Jasmonic acid synthesis proceeds with the action of a cytoplasmic 12-oxo-phytodienoic acid reductase and three rounds of β-oxidation that take place in the peroxisomes (León et al. 1999). Within the process of jasmonic acid biosynthesis, AOC is considered especially important because its specificity determines the correct stereochemical structure of jasmonic acid through its ability to convert the extremely unstable allene oxide derivative 12,13(S)-epoxy-octadecatrienoic acid (12,13-EOT) to cis-12-oxo-phytodienoic acid (Wu et al. 2011, Ziegler et al. 2000).
AOC genes have been cloned from several plant species, including tomato (Ziegler et al. 2000), barley (Maucher et al. 2004) and mangrove (Yamada et al. 2002), and a small gene family (AOC1 to AOC4) has been cloned from Arabidopsis thaliana. Characterization of these genes has provided insights into their biochemical and physiological roles in triggering responses and adaptation to biotic and abiotic stresses. Wu et al. (2011) have studied the expression and functional diversity of the AOC gene family in soybean. Their studies showed that six AOC genes were distributed in different chromosomes and had specific and complex expression patterns in multiple organs and under several stresses.
Common wheat (Triticum aestivum L., 2n = 42, genome AABBDD) is one of the most important staple food crops worldwide. It is grown under irrigated and rain-fed conditions, in areas where both types of agriculture are threatened by salinization (Colmer et al. 2006). In addition, wheat is affected by periodic drought in around 50% of its cultivated area (Rajaram 2001). Genetic improvement of wheat resistance to salinity and drought stresses is therefore a key target for many breeding programs. Introgression of agronomically important genes and QTLs from wild relatives into wheat is considered a sustainable and economically viable solution (Nevo and Chen 2010). Engineering whole or partial segments of alien chromosomes into wheat through chromosome substitution or translocations has been shown to exert significant effects on wheat disease resistance, salt tolerance and other important crop traits (Wang et al. 2010).
The dune grass Leymus mollis (Triticeae; Poaceae, 2n = 28, genome NsNsXmXm) is a wild relative of wheat that grows mainly along sea coasts and in inland dry areas (Fan et al. 2009). It is considered to be a potentially very useful genetic resource for wheat breeding (Kishii et al. 2003), as it is tolerant to salt and drought stress (McGuire and Dvorak 1981), resistant to various diseases, including scab (Mujeeb-Kazi et al. 1983) and powdery mildew (Faith 1983) and highly adaptable to nutrient deprivation and harsh conditions. Recently, to clarify the molecular basis of its high tolerance, we identified AOC among several osmotic stress-responsive genes in L. mollis (Eltayeb Habora et al. 2012). In this paper, we report the molecular cloning of a L. mollis allene oxide cyclase gene (LmAOC), its localization, gene copy number and its expression patterns under various abiotic stresses (salt, drought, cold and heat stresses) and hormone treatments. Moreover, we analyze and discuss LmAOC expression in wheat–Leymus chromosome addition lines in response to salt, drought, heat and cold stresses.
Materials and Methods
Plant materials and growth conditions
Young L. mollis plants were collected from Hamamura beach, Tottori City (35°30′N, 134°14′E), Japan. Plants were washed thoroughly and maintained at 25°C and 60 to 70% relative humidity (RH), with their roots soaked in distilled water for 3 days. They were then transferred into 10-L plastic containers for hydroponic culture in a growth chamber under natural light at 22 to 25°C and 60 to 70% RH. The hydroponic culture consisted of continuously aerated 1/4-strength Hoagland solution (HS), renewed every 2 days and with the pH adjusted to 5.6 using 1 M KOH or NaOH. Wheat–Leymus chromosome addition lines TACBOW0123, 0124 and 0125 harboring chromosome A, G or H, respectively, were provided by the Tottori Alien Chromosome Bank of Wheat, which is supported by the National Bio-Resource Project–Wheat. Seeds of the ‘Chinese Spring’ wheat (CS) and the chromosome addition lines were surface-sterilized in 2% sodium hypochlorite, rinsed thoroughly with sterilized water, and then kept at 4°C for 2 days to break seed dormancy. Seeds were then germinated in petri dishes, transplanted into plastic pots, maintained under controlled conditions (25°C day/20°C night, 50 to 60% RH and a 14-h light cycle) and irrigated with water supplied with 1 mL L−1 nutrient solution (Hyponex 6-10-5, Hyponex, Osaka, Japan). Unless otherwise noted, we used three replicates of eight-week-old plants that were uniform in height and number of leaves in all treatments.
Abiotic stresses and phytohormone treatments
Salt and osmotic stress treatments were carried out using 400 mM NaCl and 15% (w/v) polyethylene glycol (PEG) 6000, respectively, in 1/4-strength HS. Plant hormone treatments were performed using 1/4-strength HS supplied with 10−3 M jasmonic acid or 10−5 M ABA. Control treatments were carried out using only 1/4-strength HS. Drought treatment for CS and the wheat–Leymus chromosome addition lines was carried out by withholding irrigation water from the plants and the salt stress treatment was imposed by irrigating the plants with 250 mM NaCl. Heat stress was imposed by subjecting plants to 37°C day/15°C night temperatures at 80% RH for 4 days. For the cold stress treatment, plants were exposed to 4°C both during the day and at night and at 80% RH. For mechanical wounding treatment, we wounded leaves using a sterile pin to punch small holes into the leaves without damaging the major veins.
Samples were obtained at various intervals after the initiation of treatment (0 to 6 h and 1 d for the wounding, jasmonic acid and ABA treatments and 0, 1, 2, 4, 5 and 6 d for the abiotic stress treatments). The samples were immediately frozen in liquid nitrogen after each treatment and stored at −80°C until they could be analyzed.
Suppressive-subtractive hybridization
Total RNA was isolated from leaf tissues of the control and salt stressed plants using TriPure Isolation Reagent (Roche, Mannheim, Germany) and treated with RNase-free DNase I (Takara, Ohtsu, Japan) to remove any genomic DNA. Poly (A)+ RNA was purified with an Oligotex-dT30 <Super> mRNA purification kit (Takara, Ohtsu, Japan). Suppressive-subtractive hybridization was performed according to the method of Diatchenko et al. (1996) using a PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, CA, USA) according to the manufacturer’s instructions. Forward-subtracted sequences were cloned and verified as described previously (Eltayeb Habora et al. 2012).
Cloning of full-length cDNA and sequence analysis
We used rapid amplification of cDNA ends (RACE) to obtain full-length LmAOC cDNA using the SMART RACE cDNA Amplification Kit (Clontech) following the manufacturer’s instructions. We used the gene-specific primers GSP1 (5′-AGATGCTGTAGATGGCCTCGTAGCGGTC-3′) for the 5′-RACE, GSP2 (5′-CTTTCTACCTCAAGGGCATCCCGGACC-3′) for the 3′-RACE, and the universal primer supplied with the kit. We used the primers GSP3 (5′-ATGGCAGTGCGCCCTTCCTCCGTCTCCGTC-3′) and GSP4 (5′-CTAGTCAGTGAAGTTGTTGAGGCATGCGTG-3′) to obtain the full-length coding region. Sequencing was performed using the BigDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) in an ABI 3130 Genetic Analyzer (Applied Biosystems). Sequences and the phylogenetic analyses were performed using version 9 of the GENETYX software (GENETYX Corporation, Tokyo, Japan). Similarity searches were performed against the NCBI non-redundant database (http://www.ncbi.nlm.nih.gov; National Center for Biotechnology Information, Bethesda, MD, USA) and the TAIR database (http://www.arabidopsis.org) using the default BLAST parameters. Protein alignment was carried using Jalview multiple alignment editor (http://www.jalview.org/). The theoretical isoelectric point (pI) and the molecular weight of the putative protein were calculated using an online version of the ExPASy pI program (http://web.expasy.org/compute_pi). Prediction of subcellular localization was performed using the ChloroP 1.1 (http://www.cbs.dtu.dk/services/ChloroP/) and TargetP (http://www.cbs.dtu.dk/services/TargetP/) prediction servers.
Northern and Southern blot analyses and RT-PCR
Total RNA (20 μg) isolated from control or stress-treated plants was denatured in formaldehyde gel, transferred to Hybond-N+ nylon membranes, and used for northern blot analysis as described previously (Eltayeb Habora et al. 2012). For Southern blot analysis, genomic DNA was isolated using the Isoplant II kit (Nippon Gene, Tokyo, Japan). DNA (10 μg) was digested with restriction enzymes that do not cut within LmAOC coding region (EcoRI, SalI and XhoI), electrophoresed in 1.5% agarose gel and used for Southern blot analysis as described previously (Eltayeb Habora et al. 2012). For RT-PCR, 1 μg of total RNA was used to synthesize first-strand cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche), then 1 μL of the first-strand cDNA was used for PCR using the GSP3 and GSP4 primers, with actin as an internal control, as described previously (Eltayeb et al. 2010).
Construction of LmAOC-GFP and transient gene expression
We used Gateway technology (Invitrogen, Carlsbad CA, USA) to prepare LmAOC-GFP fusion construct. LmAOC cDNA was cloned into pDONR 221 vector and then shuttled via the LR recombination reaction into the destination vector pGWB6 (Nakagawa et al. 2007) using Gateway LR Clonase II enzyme mix (Invitrogen). This construct was introduced into Agrobacterium tumefaciens strain GV310 and used for transient expression by means of agroinfiltration into the lower surface of Nicotiana benthamiana leaves using a 1-mL syringe, as described by Wroblewski et al. (2005). Infiltrated plants were maintained at 25°C for 2 to 4 days, then the infiltrated leaves were examined for GFP fluorescence using a Fluoview FV10i confocal laser-scanning biological microscope (Olympus, Tokyo, Japan).
Results
Isolation of LmAOC and sequence analysis
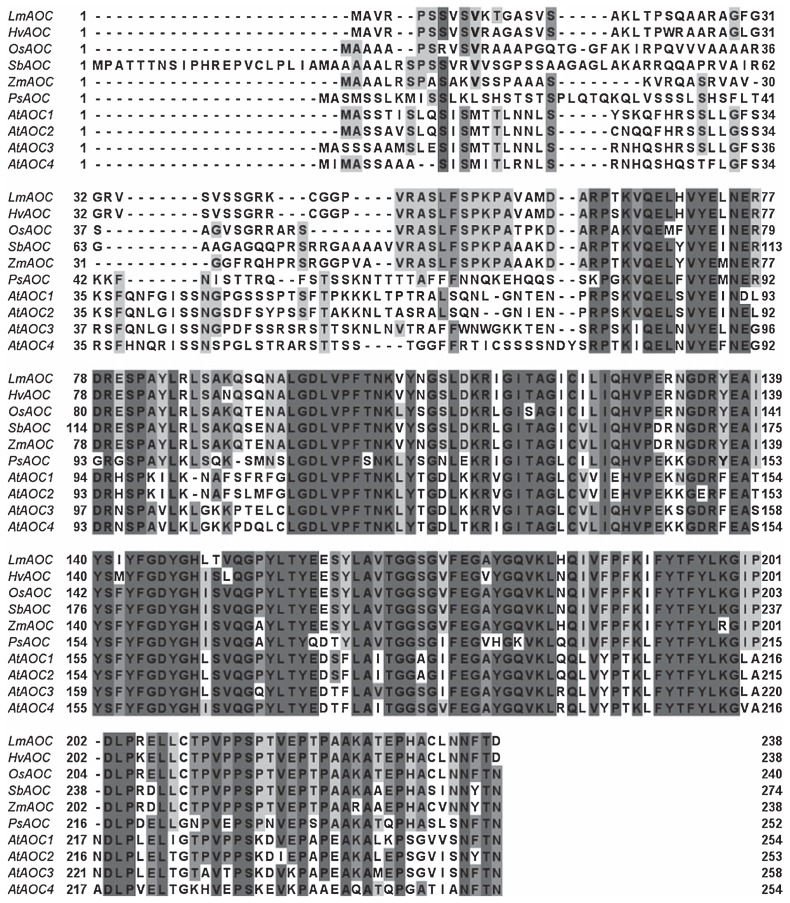
Gene-specific primers were used to clone LmAOC using the RACE procedure. LmAOC consisted of 717 bp, corresponding to a protein containing 238 amino acid residues (accession No. JX218043). Using the pI/Mw software, we found that the putative LmAOC protein has a theoretical isoelectric point (pI) of 8.96 and a molecular weight of 25.8 kDa. Similarity searches performed against the NCBI BLAST database and using Jalview multiple alignment editor showed that LmAOC had high homology with genes from other plant species (Fig. 1). LmAOC showed 94% identity to the sequence in Hordeum vulgare (CAC83766.1), versus 79% in Zea mays (ACG39242.1), 79% in Oryza sativa (NP_001050447.1), 77% in Arabidopsis thaliana (AT3G25760) and 76% in Sorghum bicolor (XP_ 002465087.1).
Fig. 1.
Alignment of the deduced amino acid sequence of LmAOC with other AOCs from different plant species. HvAOC, Hordeum vulgare (CAC83766.1); OsAOC, Oryza sativa (NP_001050447.1); SbAOC, Sorghum bicolor (XP_002465087.1); ZmAOC, Zea mays (ACG39242.1); AtAOC1, Arabidopsis thaliana (AT3G25760); AtAOC2, A. thaliana (AT3G25770); AtAOC3, A. thaliana (AT3G25780) and AtAOC4, A. thaliana (AT1G13280). Identical residues between all species are indicated by black letters on a dark gray background and conserved residues between more than three species are shown by black on light gray background.
LmAOC-GFP fusion protein localized in the chloroplast
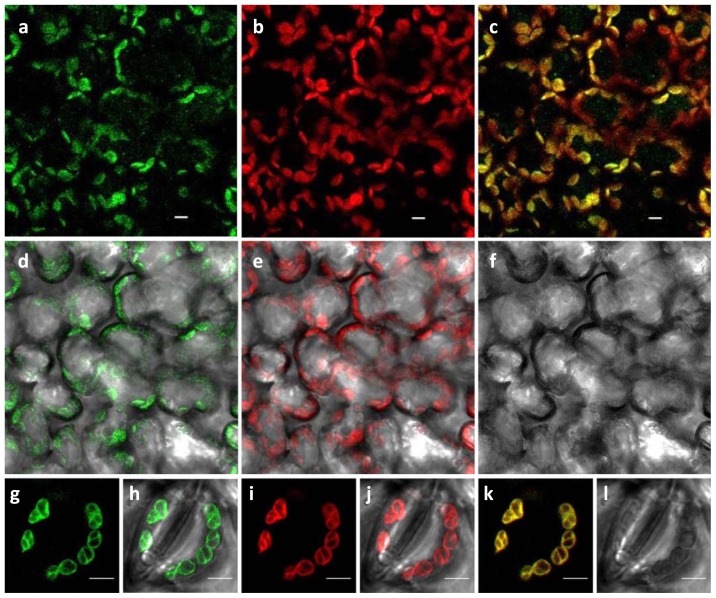
Based on the sequences determined using the ChloroP 1.1 and TargetP prediction servers, both analyses revealed a chloroplastic localization for LmAOC (cTP 0.872), with the putative N-terminal cleavage site at amino acid 47. These results were further confirmed using transient expression of the LmAOC-GFP fusion product in N. benthamiana leaves. Clear GFP fluorescence was observed in the chloroplasts of several cells (Fig. 2a, 2d) and around the stomata (Fig. 2g, 2h), which precisely matched the red autofluorescence patterns of the chloroplasts (Fig. 2b, 2e, 2i, 2j).
Fig. 2.
Subcellular localization of LmAOC in the chloroplast. LmAOC-GFP fusion protein transiently expressed in N. benthamiana leaves. (a–f) shows the localization in leave cells. (g–l) shows close view of the stomatal chloroplast. (a, g) green fluorescence of LmAOC-GFP; (b, i) red chloroplast autofluorescence; (c, k) merge between green fluorescence and the red chloroplast autofluorescence; (d, h) green fluorescence merged with bright field of the cells; (e, j) red chloroplast autofluorescence merged with bright field (f, l) bright field. Scale bars = 5 μm.
Copy number of LmAOC in L. mollis and wheat–Leymus chromosome addition lines
Southern blot analysis was performed to determine the gene copy numbers of LmAOC in the genomes of L. mollis and CS wheat and in the genome of wheat–Leymus chromosome addition lines obtained via wide-hybridization. Ten bands were detected in the genome of L. mollis, and some of these copies were found to be highly conserved in CS wheat and wheat-Leymus additions lines (Fig. 3). CS wheat showed four copies, while wheat-Leymus chromosome addition lines harboring chromosome A, G or H from L. mollis showed 3, 4 and 4 copies, respectively. Particularly, of the four bands detected in the addition lines G and H, one band of LmAOC were found to be derived from L. mollis added chromosome (indicated by arrow, Fig. 3).
Fig. 3.
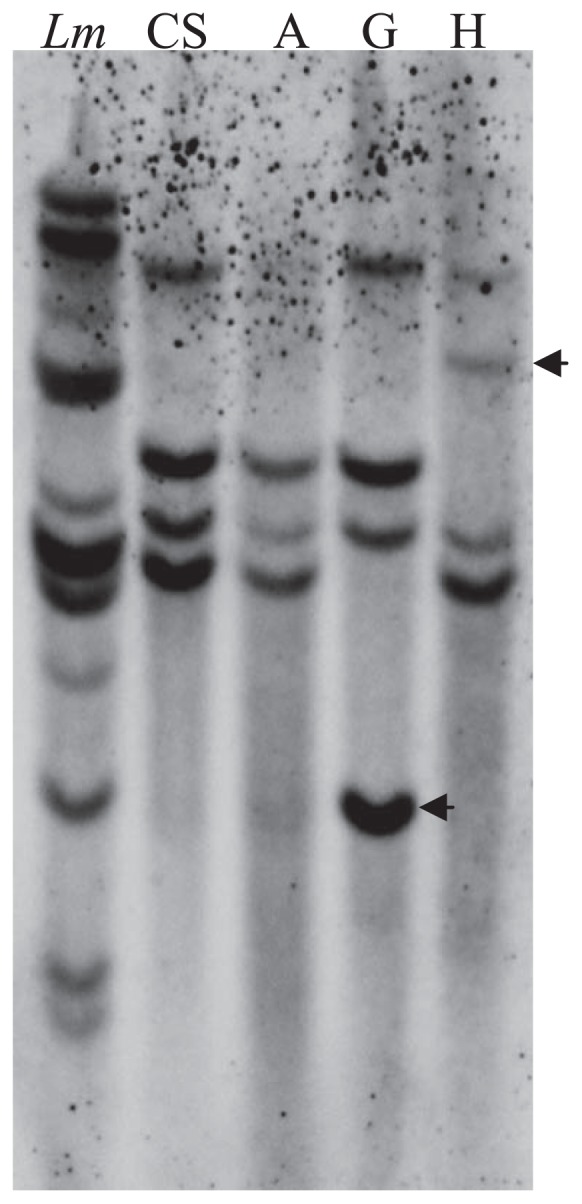
Copy number of LmAOC in L. mollis, CS wheat and wheat-Leymus chromosome addition lines detected by southern blot analysis. DNA (10 μg) was digested with EcoRI, SalI and XhoI restriction enzymes, electrophoresed in 1.5% agarose gel and used for Southern blot. Bands corresponding to LmAOC copies derived from Leymus chromosomes are indicated with black arrow. Lm, L. mollis; CS, Chinese Spring; A, G or H, wheat-Leymus chromosome addition line harboring chromosome A, G or H from L. mollis, respectively.
Expression analysis of LmAOC under abiotic stresses
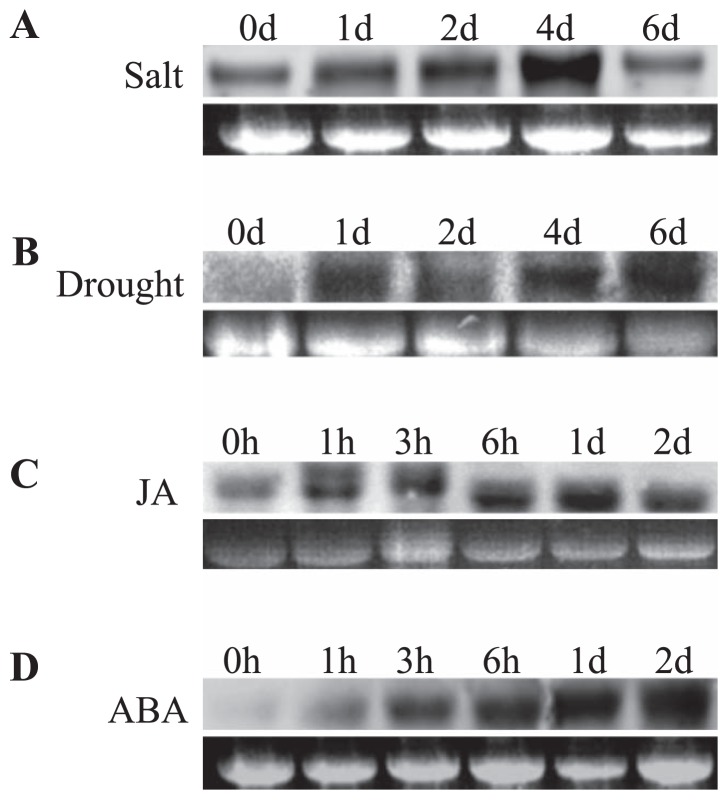
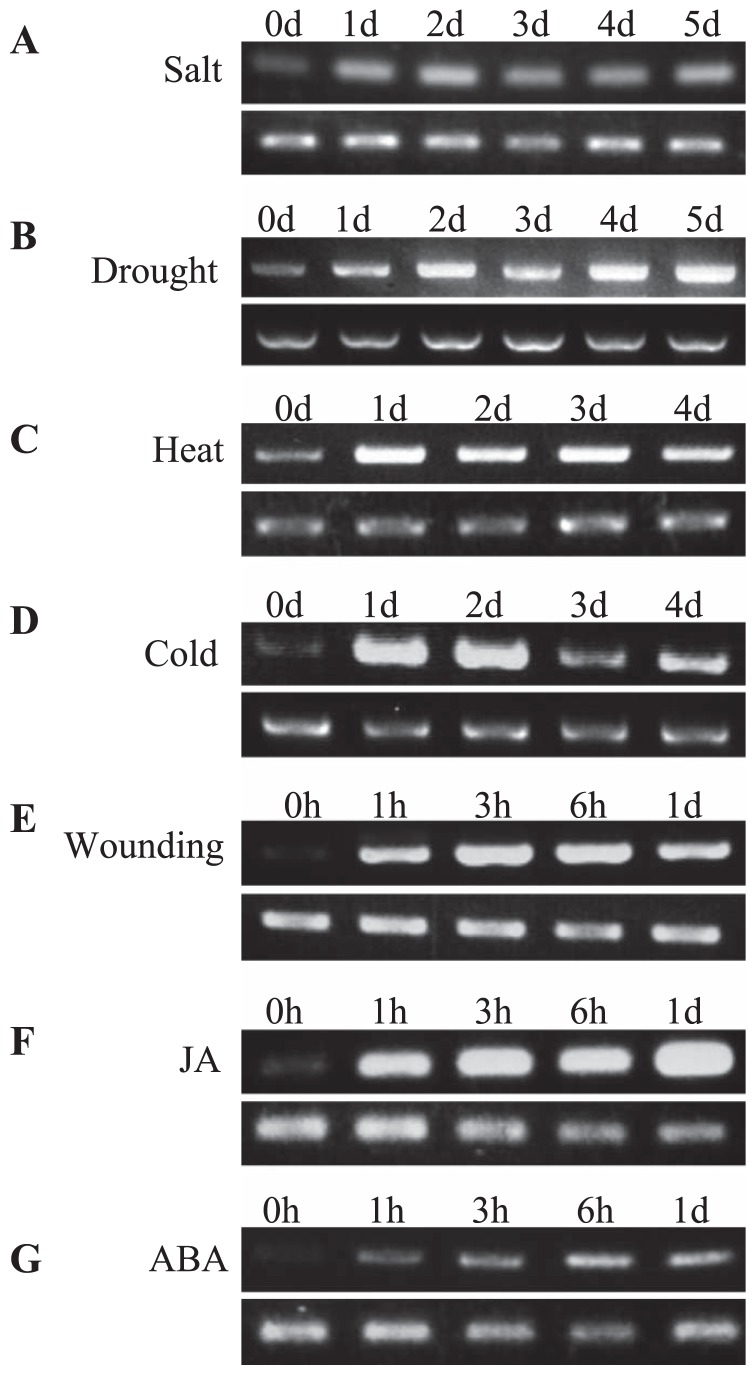
Total RNA isolated from plants subjected to various abiotic stresses (salt, drought, heat, cold and wounding) was used to analyze the expression level of LmAOC. Northern blot and RT-PCR analyses confirmed that LmAOC expression was induced within 1 day of salt stress (Fig. 4A, 5A, respectively), with maximum expression achieved by the fourth day (Fig. 4A) and then decreasing afterwards. Under drought stress, the highest expression was observed on day 6 (Fig. 4B, 5B, respectively). Although strong expression was observed under heat stress (Fig. 5C), LmAOC was strongly induced under cold stress only until day 2 (Fig. 5D), then decreased afterward. LmAOC was rapidly induced within 1 h after the wounding stress, and reached a maximum 6 h after wounding (Fig. 5E).
Fig. 4.
Northern blot analysis for LmAOC expression under some abiotic stresses. (A) salt stress; (B) drought stress; (C) JA, jasmonic acid treatment; (D) ABA, abscisic acid treatments. Total RNA (20 μg) isolated from control or treated plants was denatured in formaldehyde gel, transferred to Hybond-N+ nylon membranes and used for northern blot analysis. Plant subjected to either salt or drought were analyzed following day time course (0, 1 d, 2 d, 4 d and 6 d). Hormone treated plants were analyzed based on hours-day time course (0 h, 1 h, 3 h, 6 h, 1 d and 2 d).
Fig. 5.
RT-PCR analysis for LmAOC expression under various abiotic stresses and hormone treatments. (A) salt stress; (B) drought stress; (C) heat stress; (D) cold stress; (E) wounding stress; (F) JA, jasmonic acid treatment; (G) ABA, abscisic acid treatment. First-strand cDNA was synthesized from one μg total RNA and used to perform RT-PCR using gene specific primers and action as internal control. Plants were analyzed following day time course (0, 1 d, 2 d, 3 d, 4 d, or 5 d) or hours-day time course (0 h, 1 h, 3 h, 6 h, 1 d and 1 d). (Top) Expression of LmAOC. (Bottom) Expression of actin used as internal control.
Expression analysis under phytohormone treatments
Plants were treated with either 10−3 M jasmonic acid or 10−5 M ABA and the time course of LmAOC expression was analyzed during the first 6 hours and after the first day. Northern blot and RT-PCR analyses showed that LmAOC expression was induced within 1 hour after treatment with jasmonic acid (Fig. 4C, 5F, respectively) or ABA (Fig. 4D, 5G, respectively).
Expression of LmAOC in wheat–Leymus chromosome addition lines
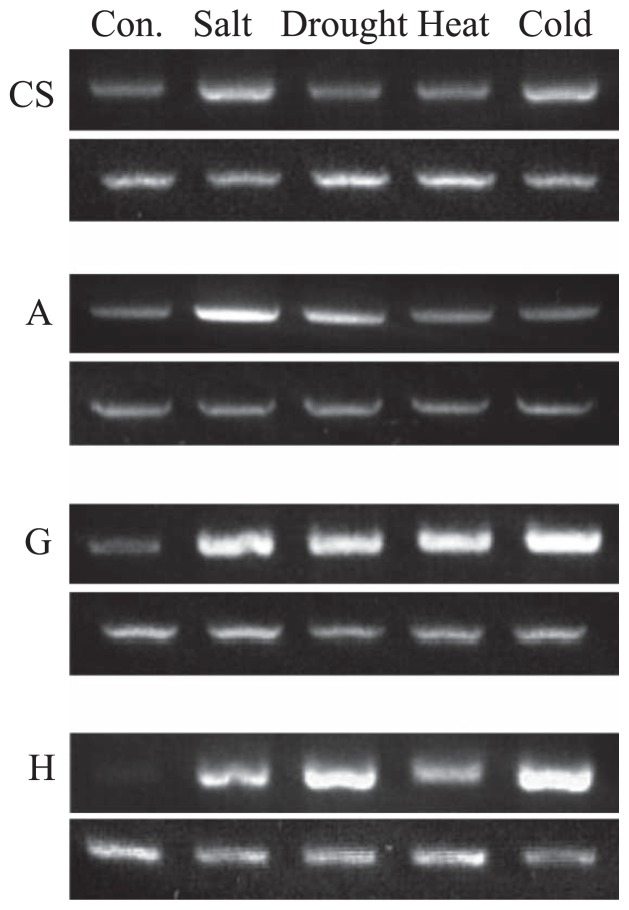
RT-PCR analysis was carried to investigate whether LmAOC is successfully expressed in some wheat–Leymus chromosome addition lines under different abiotic stresses. Chromosome addition lines G and H which integrated one copy of LmAOC showed stronger expression under salt, drought, heat and cold stresses compared CS wheat (Fig. 6).
Fig. 6.
RT-PCR analysis for LmAOC expression in CS wheat and wheat-Leymus chromosome addition lines under various abiotic stresses. One μg total RNA isolated from plants subjected to salt, drought, heat or cold stress was used for first-strand cDNA synthesis and RT-PCR. Cont, control plants, CS, Chinese Spring. A, G and H indicates wheat-Leymus chromosome addition lines harboring chromosome A, G or H from L. mollis, respectively. (Top) Expression of LmAOC. (Bottom) Expression of actin used as internal control.
Discussion
Plants respond to many biotic and abiotic stresses with an endogenous rise of jasmonic acid, an important signaling molecule (Wasternack 2007). Identification and introduction of genes from the jasmonic acid biosynthetic pathway into crop plants by means of wide hybridization or direct gene transfer could represent a potential way to obtain plants with higher tolerance to various stresses. In this study, we cloned the full coding cDNA of the AOC gene LmAOC from L. mollis and characterized its expression in Leymus and wheat–Leymus chromosome addition lines under various abiotic stresses and hormone treatments.
Sequence analysis, subcellular localization and copy number of LmAOC
Analysis of the LmAOC sequence showed high identities to known AOC genes from other plant species, including H. vulgare (Fig. 1). Phylogenetic analysis showed that LmAOC is evolutionarily more closely related to the genes for monocot AOCs, including barley, maize and rice, than to the genes of dicot Arabidopsis (data not shown). LmAOC was predicted to be localized in the chloroplast and these results were confirmed using an LmAOC-GFP fusion product that was localized to the chloroplast of N. benthamiana cells (Fig. 2). Similarly, AOCs carrying chloroplast transit peptides were reported to be localized in the chloroplast of barley (Maucher et al. 2004), rice (Agrawal et al. 2003), potato (Farmaki et al. 2007) and tomato (Ziegler et al. 2000).
Previous studies have revealed the presence of different numbers of copies of the genes encoding AOC in different plant species. A single copy was observed in the genomes of tomato (Ziegler et al. 2000) and barley (Maucher et al. 2004), versus two copies in rice (Agrawal et al. 2003) and up to five copies in Arabidopsis (Stenzel et al. 2003). However, we detected ten strong bands in the genome of L. mollis (Fig. 3), indicating that LmAOC is a multiple-copy gene. Because L. mollis is tetraploid, this species has about five copies in each genome. This is contrasting to the case of diploid barley having one copy and hexaploid wheat having four copies, i. e., 1 or 2 copies in each wheat genome. Banding patterns indicated that one clear band corresponds to LmAOC was detected in wheat–Leymus addition lines G and H, which suggests that LmAOC could be introduced into wheat via these chromosomes. Although these two lines showed four copies similar to that in CS wheat, the fourth band in the line G and the second band in the line H are missing, which indicates high possibility that L. mollis chromosome has substituted one chromosome of CS wheat. Fine mapping of LmAOC in these chromosomes in future studies would be very helpful in generating wheat lines with translocated chromosome segments harboring LmAOC.
Expression patterns of LmAOC under abiotic stresses
Jasmonic acid is a plant growth regulator with signaling properties that is involved in various stress responses and various developmentally regulated processes (Kramell et al. 2000). In the jasmonic acid signaling pathway, perception of the primary stress stimulus leads to the induction of jasmonic acid biosynthesis, which in turn results in induction of defense and stress responses (Turner et al. 2002). Therefore, upregulation of genes involved in jasmonic acid biosynthesis upon plant exposure to abiotic stresses represents a critical intermediate step in jasmonic acid signaling. In our study, analysis of LmAOC expression indicated clear upregulation in response to different abiotic stresses. The upregulation of LmAOC in response to salt stress (Fig. 4A, 5A) is in good agreement with previous reports that emphasized the role of jasmonic acid in adaptation of barley to saline conditions (Maslenkova and Toncheva 1996, Tsonev et al. 1998). Similarly, barley genes corresponding to AOC, lipoxygenase, and allene oxide synthase were upregulated in response to salinity stress (Walia et al. 2006), and expression of an AOC gene was rapidly induced by salinity in Camptotheca acuminata (Pi et al. 2009). Upregulation of LmAOC under drought stress (Fig. 4B, 5B) is consistent with our previous report, which identified AOC and lipoxygenase as differentially upregulated in response to osmotic stresses imposed by polyethylene glycol treatment (Eltayeb Habora et al. 2012).
We observed induced expression of LmAOC in response to heat (Fig. 5C) and cold stress (Fig. 5D), which indicates the importance of AOC in the adaptation of L. mollis to temperature-induced stresses. These results are consistent with the reports of Clarke et al. (2009), who demonstrated the role of jasmonic acid in conferring basal thermotolerance in Arabidopsis and Pi et al. (2009), who reported that AOC improved the tolerance of tobacco and bacteria to low temperature.
Low temperature has been correlated with the jasmonic acid response (Pi et al. 2009, Yoshikawa et al. 2007). Under cold stress, JcAOC from the dicot Jatropha curcas reached its maximum expression at 1 hour (Liu et al. 2010), then expression decreased gradually. In contrast, LmAOC maintained high expression for up to 2 days before expression decreased (Fig. 5D), which suggests that steady expression of LmAOC might be crucial for adaptation to low temperatures in L. mollis.
The role of jasmonic acid in stress-related signaling has been best characterized with respect to the wound-induced expression of defense genes (Sivasankar et al. 2000). Wound stress was found to be effective at inducing the expression of LmAOC (Fig. 5E). Although early induction of OsAOC expression in rice was observed at 3 hours after wounding (Agrawal et al. 2003), LmAOC expression was induced during the first hour after wounding. Rapid induction of AOC production following wounding was also observed in potato plants (Farmaki et al. 2007).
Expression pattern of LmAOC in response to plant phytohormones
Phytohormones such as jasmonic acid and ABA are essential for the ability of plants to adapt to abiotic stresses because these phytohormones mediate a wide range of adaptive responses (Peleg and Blumwald 2011). Jasmonic acid treatment strongly induced LmAOC expression (Fig. 4C, 5F). Similar results were reported for rice (Agrawal et al. 2003) and barley (Maucher et al. 2004), which indicates that transcriptional regulation of LmAOC in response to jasmonic acid treatment is similar to that in other monocots.
The effects of ABA on the expression of AOC genes differ among monocot plant species. ABA treatment strongly but transiently upregulated the expression of rice AOC genes, reaching a maximum at 1 hour (Agrawal et al. 2003), whereas the maximum expression was observed between 12 and 24 hours in barley (Maucher et al. 2004). In our study, induction of LmAOC expression started rapidly, at 1 hour, and the highest expression was observed at 2 days (Fig. 4D). Our results suggest that despite the high conservation of AOC genes in monocots, their transcripts are differently regulated, and that LmAOC regulation appears to be similar to that of barley HvAOC.
Analysis of LmAOC in wheat–Leymus chromosome addition lines
Stronger upregulated expression of LmAOC in response to salt, drought, heat and cold stresses were observed in the wheat–Leymus chromosome addition lines harboring chromosome G and H (Fig. 6), which reflects the successful introduction and expression of LmAOC in the wheat genome. Under cold stress, lower expression was observed in the line harboring chromosome A (Fig. 6) compared to that of CS, which may be due to the first missing band in this line (Fig. 3).
The effects of LmAOC upregulation under abiotic stresses on the phenotypic traits and the physiological gain in abiotic stress tolerance in wheat–Leymus chromosome addition lines remains to be investigated in future studies, which will provide further insights into the role of AOC in abiotic stress tolerance in wheat and its wild relatives.
In conclusion, our results suggest the presence of multiple genes encoding AOCs in L. mollis. The results also suggest that expression of these genes is rapidly induced in response to environmental stimuli and may play a crucial role in the adaptive responses of L. mollis and its tolerance to various abiotic stresses. Furthermore, LmAOC was successfully introduced into and expressed in the Chinese Spring wheat genetic background by means of wide hybridization.
Acknowledgments
This work was supported by a grant from the Global Center of Excellence for Dryland Science of the Arid Land Research Center, Tottori University.
Literature Cited
- Agrawal, G.K., Jwa, N., Agrawal, S.K., Tamogami, S., Iwahashi, H., and Rakwal, R. (2003) Cloning of novel rice allene oxide cyclase (OsAOC): mRNA expression and comparative analysis with allene oxide synthase (OsAOS) gene provides insight into the transcriptional regulation of octadecanoid pathway biosynthetic genes in rice. Plant Sci. 164: 979–992 [Google Scholar]
- Clarke, S.M., Cristescu, S.M., Miersch, O., Harren, F.J.M., Wasternack, C., and Mur, L.A.J. (2009) Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 182: 175–187 [DOI] [PubMed] [Google Scholar]
- Colmer, T.D., Flowers, T.J., and Munns, R. (2006) Use of wild relatives to improve salt tolerance in wheat. J. Exp. Bot. 57: 1059–1078 [DOI] [PubMed] [Google Scholar]
- Diatchenko, L., Lau, Y.F.C., Campbell, A.P., Chenchik, A., Moqadam, F., Huang, B., Lukyanov, K., Gurskaya, N., Sverdlov, E.D., and Siebert, P.D. (1996) Suppression subtractive hybridization: a method of generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93: 6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltayeb Habora, M.E., Eltayeb, A.E., Tsujimoto, H., and Tanaka, K. (2012) Identification of osmotic stress-responsive genes from Leymus mollis, a wild relative of wheat (Triticum aestivum L.). Breed. Sci. 62: 78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltayeb, A.E., Yamamoto, S., Habora, M.E.E., Matsukubo, Y., Aono, M., Tsujimoto, H., and Tanaka, K. (2010) Greater protection against oxidative damages imposed by various environmental stresses in transgenic potato with higher level of reduced glutathione. Breed. Sci. 60: 101–109 [Google Scholar]
- Faith, A.M.B. (1983) Analysis of the breeding potential of wheat-Agropyron and wheat-Elymus derivatives. I. Agronomic and quality characteristics. Hereditas 98: 287–295 [DOI] [PubMed] [Google Scholar]
- Fan, X., Sha, L.N., Yang, R.W., Zhang, H.Q., Kang, H.Y., Ding, C.B., Zhang, L., Zheng, Y.L., and Zhou, Y.H. (2009) Phylogeny and evolutionary history of Leymus (Triticeae; Poaceae) based on a single-copy nuclear gene encoding plastid acetyl-CoA carboxylase. BMC Evol. Biol. 9: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmaki, T., Sanmartín, M., Jiménez, P., Paneque, M., Sanz, C., Vancanney, G., León, J., and Sánchez-Serrano, J.J. (2007) Differential distribution of the lipoxygenase pathway enzymes within potato chloroplasts. Exp. Bot. 58: 555–568 [DOI] [PubMed] [Google Scholar]
- Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Kishii, M., Wang, R.R.C., and Tsujimoto, H. (2003) Characteristics and behaviour of the chromosomes of Leymus mollis and L. racemosus (Triticeae, Poaceae) during mitosis and meiosis. Chromo. Res. 11: 741–748 [DOI] [PubMed] [Google Scholar]
- Kramell, R., Miersch, O., Atzorn, R., Parthier, B., and Wasternack, C. (2000) Octadecanoid-derived alteration of gene expression and the “oxylipin signature” in stressed barley leaves. Implications for different signaling pathways. Plant Physiol. 123: 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León, J., and Sánchez-Serrano, J.J. (1999) Molecular biology of jasmonic acid biosynthesis in plants. Plant Physiol. Biochem. 37: 373–380 [Google Scholar]
- Liu, B., Wang, W., Gao, J., Chen, F., Wang, S., Xu, Y., Tang, L., and Jia, Y. (2010) Molecular cloning and characterization of a jasmonate biosynthetic pathway gene for allene oxide cyclase from Jatropha curcas. Acta Physiol. Plant. 32: 531–539 [Google Scholar]
- Maslenkova, L.T., and Toncheva, S.R. (1996) A methyl-jasmonate induced salinity tolerance in barley seedlings. C. R. Acad. Bulg. Sci. 49: 95–98 [Google Scholar]
- Maucher, H., Stenzel, I., Miersch, O., Stein, N., Prasad, M., Zierold, U., Schweizer, P., Dorer, C., Hause, B., and Wasternack, C. (2004) The allene oxide cyclase of barley (Hordeum vulgare L.)–cloning and organ-specific expression. Phytochemistry 65: 801–811 [DOI] [PubMed] [Google Scholar]
- McGuire, P.E., and Dvorak, J. (1981) High salt-tolerance potential in wheatgrasses. Crop Sci. 21: 702–705 [Google Scholar]
- Mujeeb-Kazi, A., Bernard, M., Bekele, G.T., and Mirand, J.L. (1983) Incorporation of alien genetic information from Elymus giganteus into Triticum aestivum. In: Sakamoto, S. (ed.) Proceedings of the 6th International Wheat Genetic Symposium, Maruzen, Kyoto, pp. 223–231 [Google Scholar]
- Nakagawa, T., Suzuki, T., Murata, S., Nakamura, S., Hino, T., Maeo, K., Tabata, R., Kawai, T., Tanaka, K., and Niwa, Y.et al. (2007) Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotech. Biochem. 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nevo, E., and Chen, G. (2010) Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ. 33: 670–685 [DOI] [PubMed] [Google Scholar]
- Peleg, Z., and Blumwald, E. (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 14: 290–295 [DOI] [PubMed] [Google Scholar]
- Pi, Y., Jiang, K., Cao, Y., Wang, Q., Huang, Z., Li, L., Hu, L., Li, W., and Sun, X. and Tang, K. (2009) Allene oxide cyclase from Camptotheca acuminata improves tolerance against low temperature and salt stress in tobacco and bacteria. Mol. Biotechnol. 41: 115–122 [DOI] [PubMed] [Google Scholar]
- Rajaram, S. (2001) Prospects and promise of wheat breeding in the 21st century. Euphytica 119: 3–15 [Google Scholar]
- Sivasankar, S., Sheldrick, B., and Rothstein, S.J. (2000) Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol. 122: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel, I., Hause, B., Miersch, O., Kurz, T., Maucher, H., Weichert, H., Ziegler, J., Feussner, I., and Wasternack, C. (2003) Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol. Biol. 51: 895–911 [DOI] [PubMed] [Google Scholar]
- Tsonev, T.D., Lazova, G.N., Stoinova, Z.G., and Popova, L.P. (1998) A possible role for jasmonic acid in adaptation of barley seedlings to salinity stress. J. Plant Growth Regul. 17: 153–159 [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002) The jasmonate signal pathway. Plant Cell 14: S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia, H., Wilson, C., Wahid, A., Condamine, P., Cui, X., and Close, T.J. (2006) Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Funct. Integr. Genomics 6: 143–156 [DOI] [PubMed] [Google Scholar]
- Wang, S., Yin, L., Tanaka, H., Tanaka, K., and Tsujimoto, H. (2010) Identification of wheat alien chromosome addition lines for breeding wheat with high phosphorus efficiency. Breed. Sci. 60: 371–379 [Google Scholar]
- Wasternack, C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski, T., Tomczak, A., and Michelmore, R. (2005) Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3: 259–273 [DOI] [PubMed] [Google Scholar]
- Wu, Q., Wu, J., Sun, H., Zhang, D., and Yu, D. (2011) Sequence and expression divergence of the AOC gene family in soybean: insights into functional diversity for stress responses. Biotechnol. Lett. 33: 1351–1359 [DOI] [PubMed] [Google Scholar]
- Yamada, A., Saitoh, T., Mimura, T., and Ozeki, Y. (2002) Expression of mangrove allene oxide cyclase enhances salt tolerance in Escherichia coli, yeast, and tobacco cells. Plant Cell Physiol. 43: 903–910 [DOI] [PubMed] [Google Scholar]
- Yoshikawa, H., Honda, C., and Kondo, S. (2007) Effect of low-temperature stress on abscisic acid, jasmonates, and polyamines in apples. Plant Growth Regul. 52: 199–206 [Google Scholar]
- Ziegler, J., Stenzel, I., Hause, B., Maucher, H., Hamberg, M., Grimm, R., Ganal, M., and Wasternack, C. (2000) Molecular cloning of allene oxide cyclase: The enzyme establishing the stereochemistry of octadecanoids and jasmonates. J. Biol. Chem. 275: 19132–19138 [DOI] [PubMed] [Google Scholar]