Abstract
Plants and animals carry specific receptors that recognize invading pathogens and respond by activating an immune response. The rice XA21 receptor confers broad-spectrum immunity to the Gram-negative bacterial pathogen, Xanthomonas oryzae pv. oryzae upon recognition of a small protein, Ax21, that is conserved in all Xanthomonas species and related genera. Here we demonstrate that XA21 is cleaved to release the intracellular kinase domain and that this intracellular domain carries a functional nuclear localization sequence. Bimolecular fluorescence complementation assays indicate that the XA21 intracellular domain interacts with the OsWRKY62 transcriptional regulator exclusively in the nucleus of rice protoplasts. In vivo cleavage of XA21 and translocalization of the intracellular kinase domain to the nucleus is required for the XA21-mediated immune response. These results suggest a new model for immune receptor function: on receptor recognition of conserved microbial signatures, the associated kinase translocates to the nucleus where it directly interacts with transcriptional regulators.
 The rice pattern recognition receptor—XA21—confers immunity against the Gram-negative bacterial pathogen, Xanthomonas oryzae pv. oryzae. This study shows that the intracellular kinase domain of XA21 translocates to the nucleus and that this translocation is essential for the XA21-mediated immune response.
The rice pattern recognition receptor—XA21—confers immunity against the Gram-negative bacterial pathogen, Xanthomonas oryzae pv. oryzae. This study shows that the intracellular kinase domain of XA21 translocates to the nucleus and that this translocation is essential for the XA21-mediated immune response.
Plants and animals perceive conserved microbial signatures via plasma membrane and cytoplasmically localized receptors1. Such immune receptors, also called pattern recognition receptors, include animal TLRs (Toll-like receptors) and plant receptor kinases (RKs). These receptors often carry serine-threonine kinases of the non-arginine aspartate (non-RD) class that are either integral to the receptor (plants) or associated with it (animals)2,3. Whereas RD kinases are regulated by autophosphorylation of the activation segment, a centrally located loop that sits close to the catalytic centre, very little is known about non-RD kinase activation.
In plants, well-studied immune receptors that carry the non-RD kinase motif include rice XA21 (Xanthomonas resistance 21), Arabidopsis FLS2 (flagellin sensitive 2) and the Arabidopsis elongation factor Tu receptor (EFR). All plant RKs characterized to date that carry the non-RD kinase motif are involved in recognition of conserved microbial signatures2. Animal immune receptors include TLR1, 3, 5, 6, 7, 8 and 9, which signal via non-RD interleukin-1 receptor-associated kinases 1 (IRAK1), and TLR3 and TLR4, which signal through non-RD receptor interacting protein 1 kinases1,3,4. A general theme that has emerged from these studies is that non-RD kinase activity is at least partially dispensable for the innate immune response in both plants and animals1 and that the kinases function partly as phosphorylation-mediated scaffold proteins that recruit different signaling components4. In rice, the XB24 ATPase physically associates with the XA21 juxtamembrane domain and uses ATP to promote phosphorylation of certain Ser/Thr sites on XA21, keeping the XA21 protein in an inactive state5. Together these results suggest that non-RD kinases are activated in a manner distinctly different from the well-characterized RD kinases.
Similar to the plant immune receptors, all members of the epidermal growth factor receptor (EGFR) family have an extracellular ligand-binding domain, a transmembrane domain, and a cytoplasmic kinase domain. Many of these receptors require a nuclear translocation step for their signal transductions. For example, in response to binding their corresponding ligands, the intact protein or the intracellular domain of the EGFR family members, ErbB-1 (v-erb-a erythroblastic leukemia viral oncogene homologue 1), ErbB-2, ErbB-3 and ErbB-4 are translocated to the nucleus6,7. ErbB-2 and ErbB-4 carry proline-rich carboxyl termini that contain intrinsic transcription activity and function as transcriptional regulators in the nucleus8,9,10. ErbB-1 interacts with the transcription factors STAT3 (signal transducer and activator of transcription 3), STAT5 and E2F transcription factor-1. Each of these transcription factors then regulates expression of target genes11,12,13. Such nuclear translocation events have not been reported for receptor kinases governing the innate immune response.
Here we show that XA21 is cleaved to release the intracellular kinase domain and that this intracellular domain carries a functional nuclear localization sequence. Bimolecular fluorescence complementation (BiFC) assays indicate that the XA21 intracellular domain interacts with the OsWRKY62 transcriptional regulator exclusively in the nucleus of rice protoplasts. In vivo cleavage of XA21 and translocalization of the intracellular kinase domain to the nucleus is required for the XA21-mediated immune response. These results suggest a new model for immune receptor function where, upon receptor recognition of conserved microbial signatures, the associated kinase translocates to the nucleus and directly interacts with transcriptional regulators.
Results
XA21 is cleaved to release the intracellular kinase domain
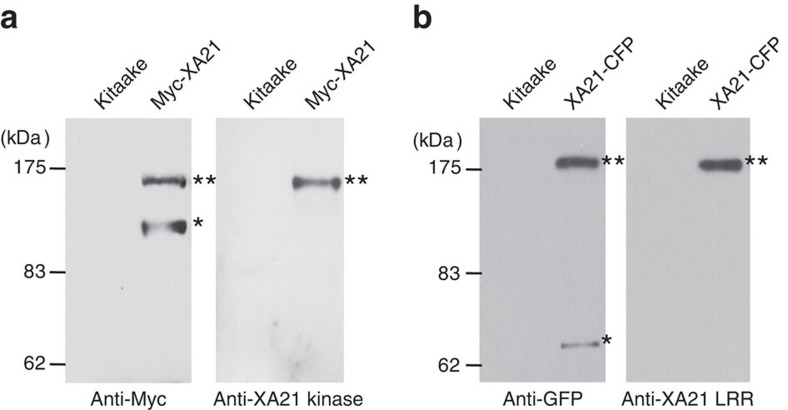
Rice XA21 confers immunity to the Gram-negative bacterium Xanthomonas oryzae pv. oryzae (Xoo)14, XA21 binds a conserved sulphated peptide, called AxYS22 derived from the Xoo Ax21 (activator of XA21-mediated immunity) protein15. We previously reported that on binding to AxYS22, XA21 accumulates, releasing a 110-kDa amino-terminal cleavage product in transgenic rice plants expressing a N-terminal Myc-tagged XA21 (Myc-XA21)15,16,17. Here we show that a 70-kDa carboxy-terminal cleavage product, corresponding to the kinase domain fused to cyan fluorescent protein (CFP), is also detected after infection of transgenic rice plants carrying a C-terminal CFP-tagged XA21 (XA21-CFP)18 (Supplementary Fig. S1 and Fig. 1). To further characterize the XA21 cleavage product, we used rice lines expressing Myc-XA2118 and XA21-CFP18. A 110-kDa N-terminal cleavage product was immunoprecipitated using an agarose-conjugated anti-Myc antibody. This 110 kDa product did not cross-react with the anti-XA21 kinase antibody (Fig. 1a). The 70-kDa C-terminal cleavage product immunoprecipitated using the anti-green fluorescent protein (GFP) antibody did not react with the anti-XA21 LRR antibody (Fig. 1b). These results indicate that 110 kDa C-terminal cleavage product consists of the XA21 intracellular domain.
Figure 1. XA21 cleavage releases the intracellular kinase domain.
(a) Protein extracts from Kitaake and Myc-XA21 (T4, 20-4-2) plants were immunoprecipitated with agarose-conjugated anti-Myc antibody. The immunoprecipitates were separated on duplicate gels and transferred to membranes for western blot analysis. Two bands were detected using anti-Myc antibody (left) and a single band was detected using anti-XA21 kinase antibody (right). The upper 140 kDa band (marked with '**') corresponds to the full-length Myc-XA21 fusion protein and the lower 110 kDa band (marked with '*') corresponds to the N-terminal cleavage product15,16,17,42. (b) Protein extracts from Kitaake and XA21-CFP (T3, 15A-1-1-4) plants were immunoprecipitated with agarose-conjugated anti-GFP antibody. The immunoprecipitates were separated on duplicate gels and transferred to membranes for western blot analysis. Two bands were detected using anti-GFP antibody, (left) and one band was detected using anti-XA21 LRR antibody (right). The upper 180 kDa band (marked with '**') corresponds to the full-length XA21-CFP fusion protein. The lower 70 kDa band (marked with '*') corresponds to the C-terminal cleavage product.
The XA21 intracellular kinase translocates to the nucleus
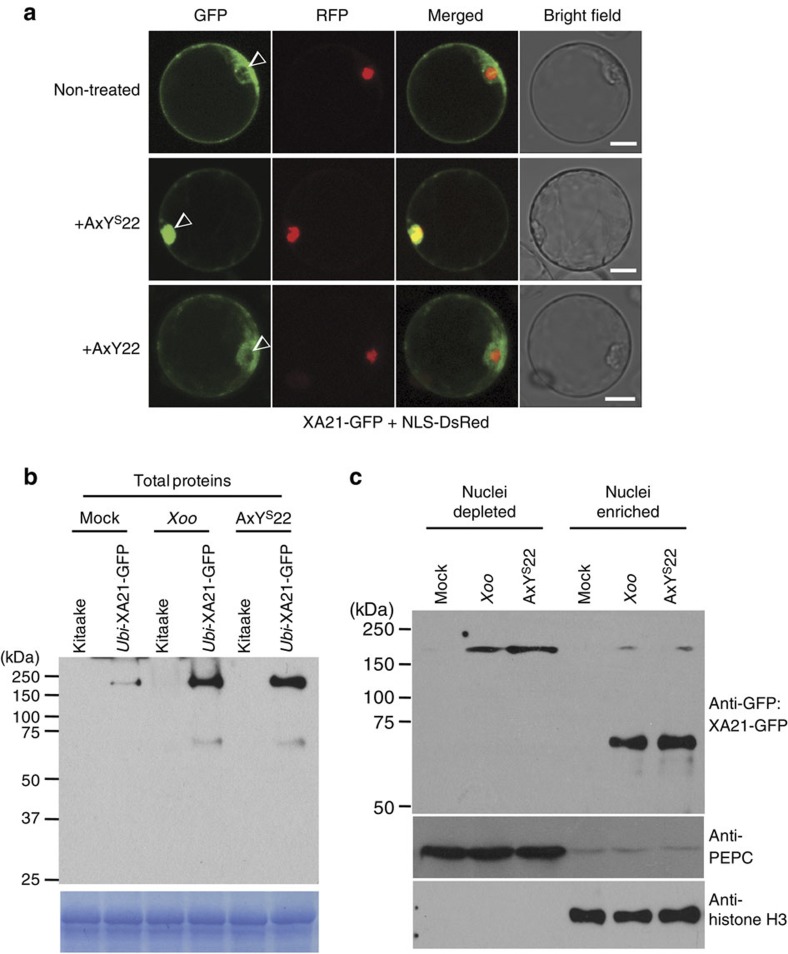
To localize the XA21 intracellular domain in vivo, we investigated the cellular distribution of C-terminal GFP-tagged XA21 (XA21-GFP) in rice protoplasts. Consistent with our previous results16, XA21-GFP is localized both to the plasma membrane and the cortical and perinuclear endoplasmic reticulum in the absence of AxYS22 (Fig. 2a and Supplementary Fig. S2). GFP alone localizes to both the nucleus and cytoplasm (Supplementary Fig. S3a). In contrast, treatment with AxYS22 (Fig. 2a) or with supernatants prepared from Xoo15 (Supplementary Figs S2 and S4) triggers significant accumulation of the XA21-GFP inside the nucleus (24.1±3.8% of the protoplasts accumulated XA21-GFP protein in the nucleus, Supplementary Table S1). Nuclear localization was not observed in protoplasts treated with water or the biologically inactive peptide, AxY2215. (1.2±0.1% and 1.8±0.7%, respectively; Fig. 2a and Supplementary Table S1). These results indicate that the XA21 intracellular domain translocates to the nucleus in protoplasts after treatment with biologically active AxYS22.
Figure 2. Nuclear translocation of XA21-GFP after treatment with AxYS22 peptide or Xoo.
(a) XA21-GFP was expressed in rice protoplast cells using polyethylene glycol-mediated transformation. AxYS22 or AxY22 peptides were applied 16 h after transformation. Expression of the introduced genes was observed 1 h after peptide treatment. NLS-DsRed (nuclear localization signal-Discosoma sp. red fluorescent protein) was co-transformed as a nucleus marker. Images were collected using a Leica True Confocal Scanner SPE confocal microscope and coded in green and red for GFP and DsRed, respectively. Two confocal z-section images were merged to have a 2D image. Arrowheads mark the nuclei. Scale bar, 10 μm. (b) Western blot analysis of XA21-GFP proteins extracted from transgenic rice plants expressing XA21-GFP under the control of the Ubi promoter (Ubi-XA21-GFP, homozygous 5-5-4). Equal amount of total proteins (75 μg) were extracted from leaf discs of Kitaake and Ubi-XA21-GFP plants 3 h after Mock, Xoo or AxYS22 treatment. XA21-GFP and C-terminal cleavage product, detected with an anti-GFP antibody, displayed bands of approximately 170 and 70 kDa, respectively. Equal loading of total proteins was confirmed by Coomassie blue staining of proteins (lower panel). (c) Western blot analysis of XA21-GFP extracted from nuclei-depleted and nuclei-enriched fractions from Ubi-XA21-GFP line (homozygous 5-5-4) detected by the anti-GFP antibody. Nuclei-enriched (100 μg) and nuclei-depleted (100 μg) fractions were prepared from total proteins in b. XA21-GFP and C-terminal cleavage product-GFP displayed bands at approximately 170 and 70 kDa, respectively. Cytolsolic phosphoenolpyruvate carboxylase (PEPC) and nuclear histone H3 protein were used as cytosolic and nuclear markers, respectively.
To investigate whether the XA21 intracellular domain translocates to the nucleus in mature plants, we generated stable transgenic rice plants expressing biologically active XA21-GFP (Ubi-XA21-GFP, Supplementary Fig. S5; Native-XA21-GFP, Supplementary Fig. S6). After Xoo infection or AxYS22 treatment of Ubi-XA21-GFP plants, the XA21-GFP protein accumulates (Fig. 2b). This result is consistent with our published studies showing that Xoo treatment triggers enhanced XA21 protein accumulation without a corresponding increase in transcription16. We also observed a significant increase of the XA21 intracellular domain in the nuclei-enriched fraction (Fig. 2b,c, and Supplementary Fig. S7). In contrast, the intracellular domain was not detected in nuclei-enriched fractions after mock infection. These results indicate that AxYS22 recognition triggers accumulation of XA21 and that the XA21 intracellular domain is translocated to the nucleus in planta.
Because XA21 does not carry a proline-rich domain indicative of intrinsic transcription activity, we hypothesized that the nuclear-localized XA21 interacts with transcription factors to control transcriptional reprogramming. To test this hypothesis, we assessed whether the rice transcription factor OsWRKY62, which negatively regulates XA21-mediated immunity19, interacts with the XA21 intracellular domain in plant cells. Indeed, using BiFC assays, we showed that the XA21 intracellular domain and OsWRKY62 interact exclusively in the nucleus of rice protoplasts (Supplementary Fig. S8).
XA21 kinase nuclear translocation is required for immune function
We next assessed whether the nuclear localization of the XA21 intracellular domain is critical for XA21-mediated immunity. For this purpose, we generated transgenic rice plants expressing XA21 fused to a C-terminal nuclear export signal (NES)20 (Ubi-XA21-GFP-NES) and inoculated with Xoo (Supplementary Fig. S9). Out of 12 independently transformed rice plants expressing Ubi-XA21-GFP-NES, all displayed significantly enhanced susceptibility to Xoo compared with the control transgenic rice plants expressing Ubi-XA21-GFP, lacking the NES. The NES did not affect the expression or stability of XA21-GFP (Supplementary Figs S3b, S9a and S10).
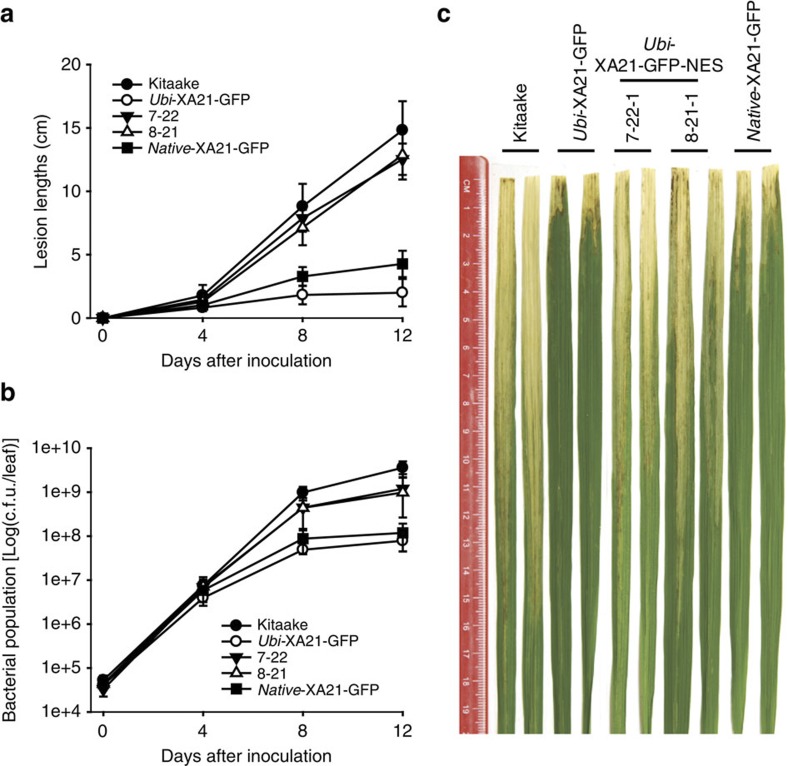
To quantify bacterial growth in the Ubi-XA21-GFP-NES lines, we measured lesion lengths and bacterial growth in the T2 progeny using well-established techniques14 (Fig. 3 and Supplementary Fig. S11). At 4 days after inoculation (DAI), slight increases in the lesion lengths and bacterial populations were detected in the Ubi-XA21-GFP-NES lines compared with the Ubi-XA21-GFP control. At 12 DAI, the Ubi-XA21-GFL-NES displayed significantly enhanced susceptibility to Xoo, with lesions of approximately 10–13 cm. In contrast, the Ubi-XA21-GFP control displayed lesion lengths of 2–3 cm (Fig. 3a). The bacterial growth curve correlated well with lesion lengths (Fig. 3b). At 12 DAI, Xoo populations in Ubi-XA21-GFP transgenic rice reached approximately 7.8×107 colony-forming units (c.f.u.) per leaf (cfu/leaf), whereas populations in Kitaake plants reached to more than 3.6×109 c.f.u./leaf. In Ubi-XA21-GFP-NES line 7–22, Xoo populations grew to 1.2×109 c.f.u./leaf, a greater than 15-fold increase compared with the Ubi-XA21-GFP control. In Fig. 3c, we show two typical leaves from each of the inoculated rice plants at 12 days after Xoo inoculation. These results demonstrate that the nuclear translocation of the XA21 intracellular domain is required for its immune function. Rice plants expressing XA21-GFP-NES under the control of its native promoter (Native-XA21-GFP-NES) were also susceptible compared with resistant Native-XA21-GFP, lacking NES (Supplementary Fig. S12). These results demonstrate that nuclear translocation of the XA21 intracellular domain is required for XA21-mediated immunity.
Figure 3. Nuclear export of XA21 disrupts XA21-mediated immunity.
(a) Lesion length measurements of Xoo-inoculated Kitaake, Ubi-XA21-GFP (homozygous 5-5-4), Ubi-XA21-GFP-NES (progeny from 7-22 and 8-21) or Native-XA21-GFP (progeny from 9 to 7) rice plants. Each data point represents the average and standard deviation of at least four samples. (b) Xoo populations were monitored in Kitaake, Ubi-XA21-GFP (homozygous 5-5-4), Ubi-XA21-GFP-NES (progeny from 7 to 22 and 8-21) or Native-XA21-GFP (progeny from 9 to 7) rice plants. For each time point, bacterial populations were determined in three separate leaves for each genotype. Capped vertical bars represent standard deviation values (c.f.u./leaf) obtained from the three samples. (c) Transgenic plants expressing XA21 carrying a nuclear export signal (Ubi-XA21-GFP-NES) developed long lesion lengths after Xoo inoculation. Rice leaves were photographed 12 days after inoculation (DAI) with Xoo. From left to right: Kitaake, Ubi-XA21-GFP (homozygous 5-5-4), Ubi-XA21-GFP-NES (7-22-1 and 8-21-1) and Native-XA21-GFP (9-7-2) rice plants.
The XA21 intracellular domain carries a functional nuclear localization sequence
Many animal receptor kinases, including ErbB-4, Notch1 and β-amyloid precursor protein, undergo proteolytic cleavage by membrane-anchored proteases21,22,23. The released intracellular domains often carry a basic amino acid-rich nuclear localization sequence (NLS)21,24,25, XA21 also harbours a putative NLS in the intracellular domain (Supplementary Fig. S13a). To assess whether the putative XA21 NLS is functional, we investigated the cellular distribution of the XA21 intracellular domain. For this purpose, we generated a C-terminal GFP-tagged XA21 intracellular domain constructs containing a putative NLS (XA21NLS-GFP), a nonfunctional NLS with alanine substitutions (XA21nls-GFP), or a deletion of the putative NLS (XA21NLS-GFP) (Supplementary Fig. S13a). Whereas wild-type XA21NLS primarily localizes to the nucleus, alanine substitutions in the predicted NLS (XA21nls) or deletion of the NLS (XA21NLS) prevent XA21 nuclear localization (Supplementary Fig. S13b). These results suggest that the basic amino acid-rich region of the XA21 intracellular domain functions as an NLS. Whole plant studies with a full-length XA21 carrying a mutation in the NLS are needed to confirm this result.
Discussion
This study demonstrates that nuclear translocation of the XA21 intracellular domain is required for XA21-mediated immunity. Whereas C-terminal-tagged XA21 is primarily localized to the plasma membrane in the absence of AxYS22 treatment with biologically active AxYS22 or with supernatants prepared from Xoo triggers significant accumulation of XA21 inside the nucleus. We hypothesize that chaperone proteins may also be involved in the nuclear translocation of XA21. In support of this hypothesis, we have previously shown that the co-chaperones, RAR1 and SGT1, interact with the XA21 kinase domain in yeast and plant cells26. The HSP90/RAR1/SGT1 chaperone complex has previously been demonstrated to regulate the plant immune response27,28. These proteins are localized in both the cytoplasm and the nucleus, shuttling between these two compartments28,29,30,31.
The observation that the non-RD kinase domain of a plant immune receptor is cleaved and localized to the nucleus where it interacts with a transcriptional regulator has not previously been reported. However, studies of animal TLRs and their associated non-RD kinases have demonstrated a similar nuclear localization requirement for activation of TLR-mediated immune responses. For example, on challenge with lipopolysaccharide or interleukin-1, IRAK1 localizes to the nucleus and binds to nuclear factor-κB-responsive elements located within the inhibitor of κB (IκB)-α promoter to regulate IκB-α transcription32,33,34,35. Nuclear-localized IRAK1 also binds to the promoters of STAT3 and interleukin-10 upon lipopolysaccharide challenge and subsequently regulates interleukin-10 gene expression34,36.
Together these results suggest a model for function of non-RD kinases associated with immune receptors. On receptor recognition of conserved microbial signatures, the associated non-RD kinase translocates to the nucleus where it directly interacts with transcription factors to regulate the immune response.
Methods
Plant material and growth conditions
Rice (Oryza sativa L) plants (cultivar Kitaake) were maintained in the green house. Growth chambers were set on 14 h light/10 h dark photoperiod, 28/26 °C temperature cycle, and 85/90% humidity. Healthy and well-expanded leaves from 6-week-old rice plants were used for Xoo strain PXO99 inoculation (designated Xoo in text).
Plasmid constructs
Construction of Ubi-pCAMBIA-1300 has been described before37. For construction of XA21 driven by the XA21 native (Native) promoter, a 2,204-nt fragment from the XA21 promoter was amplified from rice genomic DNA extracted from Native-XA2118 transgenic plants using the following primers: 5′-AAGCTTGGTACCCAACTTTTTGTGCTCCTA-3′ and 5′-GGATCCCGTGCAGAAGTGCAAGGCAAGACA-3′. This construct was named XA21-pCAMBIA-1300. The XA21 promoter, was replaced with the maize ubiquitin (Ubi) constitutive promoter38 using HindIII and BamHI enzymes to generate the Ubi-pCAMBIA-1300 construct.
The NES was generated by annealing the following oligonucleotides: 5′-GAACGAGCTTGCTCTTAAGTTGGCTGGACTTGATATTAACAAGACTGGAGGATAGCCCGGGCTGCA-3′ and 5′-GCCCGGGCTATCCTCCAGTCTTGTTAATATCAAGTCCAGCCAACTTAAGAGCAAGCTCGTTCTGCA-3′, encoding the amino-acid residues LQNELALKLAGLDINKTGG (stop)20. To amplify the XA21-GFP-PstI site, PCR was performed using an XA21-specific oligonucleotide primer and a GFP-specific oligonucleotide primer, 5′-CACCATGATATCACTCCCATTATTGCTC-3′/5′-CTGCAGTTTGTATAGTTCATCCATGCC-3′. Using PCR, the termination codon of XA21-GFP was replaced with PstI site. The PCR-amplified products were cloned into pENTR/D-TOPO/D vector (Invitrogen). To generated XA21-GFP-NES, NES fragment was ligated into PstI-digested XA21-GFP in pENTR/D-TOPO vector. Using Gateway LR Clonase (Invitrogen), XA21-GFP-NES and XA21-GFP moved into the Ubi-pCAMBIA-1300 (ref. 37) and Native-CAMBIA-1300.
The XA21NLS-GFP was PCR amplified using oligonucleotides, 5′-CACCATGCACAAGAGAACTAAAAAGGGA-3′/5′-TTATTTGTATAGTTCATCCATGCCATG-3′ and template Ubi-XA21-GFP. The XA21nls-GFP and XA21ΔNLS variants were amplified using oligonucleotides, 5′-CACCATGACCTGGGCAGCCGCAACTGCGGCAGGAGCCCCTTCAAGAACTTCCATGAAA-3′ and 5′-CACCATGGGAGCCCCTTCAAGAACTTCCATGAAA-3′, respectively. The PCR-amplified products were cloned into pENTR/D-TOPO vector. Using Gateway LR Clonase XA21NLS-GFP, XA21nls-GFP and XA21ΔNLS moved into the Ubi-pCAMBIA-1300 (ref. 37).
Rice transformation
Rice transformation was constructed as described previously37. Agrobacterium strain EHA105 was used to infect rice callus from cultivar Kitaake for transformation. Transgenic lines were selected using hygromycin.
Expression analysis. For reverse transcriptase-PCR (RT–PCR) analysis, total RNAs were extracted from leaves using TRIzol reagent (Invitrogen) after each treatment. The RT–PCR reaction was performed following the manual for QuantumRNA 18S Internal Standards (Ambion). PCR analyses were performed with primers pairs, 5′-TCATCACTCTACTTGCTTATA-3′/5′-TCAGAATTCAAGGCTCCCACCTTC-3′ (for XA21-GFP). The amplified products were resolved by gel electrophoresis after 28 cycles.
In vivo immunoprecipitation of XA21
Immunoprecipitation was conducted as described previously16. Total proteins were extracted from 5 g of leaf tissue in 25 ml of ice-cold extraction buffer II (0.15 M NaCl, 0.01 M Na-phosphate pH 7.2, 2 mM EDTA, 1% Triton X-100, 10 mM β-mercaptoethanol, 20 mM NaF, 1 mM phenylmethylsulphonyl fluoride, 1% protease cocktail (Sigma), 2 mg ml−1 leupeptin, 2 mg ml−1 antipain and 2 mg ml−1 aprotinin). After filtering through Miracloth (Calbiochem) followed by centrifugation twice at 13,000 g for 20 min at 4 °C, the supernatant was mixed with 50 ml of agarose-conjugated anti-Myc antibody (Santa Cruz) for Myc-XA21 and anti-GFP antibody (Santa Cruz) for XA21-GFP and incubated at 4 °C for 2 h. The beads were then washed four times in 1 ml of extraction buffer II without proteinase inhibitors. The proteins were eluted with 4× Laemmli-loading buffer. Protein blot analyses were performed.
Immunodetection
For immunoblot analysis, proteins were separated by an 8% SDS–polyacrylamide gel. The proteins were then blotted onto a Hybond-P membrane (Amersham Pharmacia Biotech) using a SemiPhor Semi-Dry Transfer Unit (Amersham Pharmacia Biotech). For XA21-GFP detection, anti-GFP mouse monoclonal IgG (Santa Cruz) and anti-mouse IgG, horseradish peroxidase-linked whole antibody were used as primary and secondary antibodies at a final dilution of 1:1,000 and 1:5,000 for 2 h, respectively. Bands were visualized using the SuperSignal West Pico Chemiluminescent Substrate (Pierce) according to standard protocol.
For the anti-XA21 kinase and XA21 LRR antibodies, synthetic peptides and monospecific antibodies were generated. Epitope selection was directed to hydrophobic, flexible regions of the proteins. The epitopes, RNLFQTREQDDWGFI in the LRR domain and KALKSFTAECEALRN in the kinase domain were used for anti-XA21 LRR and XA21 kinase antibodies, respectively. Peptides were synthesized and conjugated to keyhole limpet haemocyanin. Rabbits were immunized with peptide in complete Freund's adjuvant, followed by three boosts in incomplete Freund's adjuvant. Monospecific antibodies were affinity-purified to the synthetic peptides bound to C3-SEP-PAK cartridges. Antibodies were conjugated to horseradish peroxidase for use in western blots.
Xoo inoculation and determination of bacterial populations
For Xoo inoculation, rice plants were grown in the greenhouse until they were 6-week-old and then transferred to the growth chamber. Xoo was used to inoculate rice by the scissors-dip method14,37. Xoo was grown on a PSA plate (peptone sucrose agar, 10 g l−1 peptone, 10 g l−1 sucrose, 1 g l−1 glutamic acid, 16 g l−1 agar and pH 7.0) containing cephalexin (100 μg l−1) for 3 days and suspended with water at OD=0.5 (600 nm) for inoculation. Only the top two to three expanded leaves of each tiller were inoculated. Xoo colony count was conducted as described previously14,16,37. Twenty centimeter of leaf tissue from the top, including lesions and tissue showing no lesions, was ground up and resuspended in 10 ml water to collect bacteria. The extract was diluted accordingly and plated out on PSA plates.
Xoo and AxYS22 treatments of rice leaf discs
Leaf disc assays39 were modified for Xoo and AxYS22 treatments of rice. Briefly, fully expanded rice leaves were shredded into 2–3 mm with scissors. Because previous studies have demonstrated that leaf discs floated overnight do not show symptoms of wounding39,40,41, the shredded leaves were floated and maintained in a growth chamber under 14 h light/10 h dark conditions with 28 °C for 1 day to recover from wounding. After 3 h incubation with 1 μM AxYS22, 1 μM AxY22 or Xoo (OD=0.5 at 600 nm), the shredded leaves were immediately frozen in liquid nitrogen for protein extractions.
Nuclei isolation
Nuclear fractionation was performed using CellLytic PN Plant Nuclei Isolation/Extraction Kit (Sigma) with minor modifications. Briefly, tissue was homogenized in 1× nuclear isolation buffer (NIB) containing 1 mM DTT and protease inhibitor cocktail for plant cell (Sigma) using a mortar and pestle and then filtered through a 100-μm (pore-size) nylon mesh. Triton X-100 was added to a final concentration of 1.0%, and the mixture was incubated on ice for 15 min. The solution was centrifuged at 12,000 g for 10 min, and the pellet was washed with 1× NIB containing 1 mM DTT and protease inhibitor cocktail. Nuclei-containing lysate was applied to 1.7 M sucrose cushion and centrifuged at 12,000 g for 10 min. Nuclei pellet was washed twice with 1× NIB containing 1 mM DTT and protease inhibitor cocktail and stored in nuclei storage buffer (Sigma).
XA21-GFP and XA21-GFP-NES transient expression analyses
Plasmids, Ubi-XA21-GFP, Ubi-XA21-GFP-NES, XA21NLS-GFP, XA21nls-GFP and XA21NLS-GFP driven by Ubi promoter38, were used for transient expression analyses. Images were collected with a Leica True Confocal Scanner SPE confocal microscope. GFP was imaged under the following conditions: excitation, 488 nm and emission, 500–530 nm. DsRed was imaged under the following conditions: excitation, 532 nm and emission, 560–620 nm. Images were analysed using Leica LAS AF software (Ver 2.3.5) and coded green (for GFP) or red (for DsRed).
BiFC assays
Plasmids, 735-YFPC−term (YC)-XA21K668, 736-YFPN−term (YN)-OsWRKY62 and Ubi-YFP, were described previously26. As negative controls, we included the both empty vectors (735-YC-empty vector and 736-YN-empty vector) for each pairwise test. All images were taken 1 day after transformation. Yellow fluorescent protein (YFP) was imaged under the following conditions: excitation, 488 nm and emission, 515–565 nm. Images were analysed using Leica LAS AF software (Ver 2.3.5) and coded yellow for YFP.
Author contributions
C.-J.P. and P.C.R. designed the project and wrote the paper. C.-J.P. performed experiments.
Additional information
How to cite this article: Park, C.-J. & Ronald P.C. Cleavage and nuclear localization of the rice XA21 immune receptor. Nat. Commun. 3:920 doi: 10.1038/ncomms1932 (2012).
Supplementary Material
Supplementary Figures S1-S13, Supplementary Table S1 and Supplementary Reference.
Acknowledgments
We thank Dr. Benjamin Schwessinger, Dr. Ofir Bahar, Dr. Rita Sharma, Dr. David de Vleesschauwer and Dr. Benoit Lefebvre for critical reading of the manuscript, Dr. Taeyun Oh for statistical analysis, Patrick E. Canlas for generation of transgenic rice plants and Randy Ruan for lab and green house management. This work was supported by NIH GM59962.
References
- Ronald P. C. & Beutler B. Plant and animal sensors of conserved microbial signatures. Science 330, 1061–1064 (2010). [DOI] [PubMed] [Google Scholar]
- Schwessinger B. & Ronald P. C. Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482 (2012). [DOI] [PubMed] [Google Scholar]
- Dardick C. & Ronald P. Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog. 2, e2 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton G. M. & Medzhitov R. Toll signaling: RIPping off the TNF pathway. Nat. Immunol. 5, 472–474 (2004). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. An ATPase promotes autophosphorylation of the pattern recognition receptor XA21 and inhibits XA21-mediated immunity. Proc. Natl. Acad. Sci. USA 107, 8029–8034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. N., Yamaguchi H., Hsu J. M. & Hung M. C. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene 29, 3997–4006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter G. & Liao H. J. Trafficking of receptor tyrosine kinases to the nucleus. Exp. Cell Res. 315, 1556–1566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. Y. et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3, 802–808 (2001). [DOI] [PubMed] [Google Scholar]
- Ni C. Y., Murphy M. P., Golde T. E. & Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294, 2179–2181 (2001). [DOI] [PubMed] [Google Scholar]
- Wang S. C. et al. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell 6, 251–261 (2004). [DOI] [PubMed] [Google Scholar]
- Hanada N. et al. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol. Carcinog. 45, 10–17 (2006). [DOI] [PubMed] [Google Scholar]
- Hung L. Y. et al. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 36, 4337–4351 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H. W. et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell 7, 575–589 (2005). [DOI] [PubMed] [Google Scholar]
- Song W. Y. et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806 (1995). [DOI] [PubMed] [Google Scholar]
- Lee S. W. et al. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326, 850–853 (2009). [DOI] [PubMed] [Google Scholar]
- Park C. J. et al. Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS One 5, e9262 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. H. et al. The autophosphorylated Ser686, Thr688, and Ser689 residues in the intracellular juxtamembrane domain of XA21 are implicated in stability control of rice receptor-like kinase. Plant J. 45, 740–751 (2006). [DOI] [PubMed] [Google Scholar]
- Park C. J. et al. Ectopic expression of rice Xa21 overcomes developmentally controlled resistance to Xanthomonas oryzae pv. oryzae. Plant Sci. 179, 466–471 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y. et al. OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 1, 446–458 (2008). [DOI] [PubMed] [Google Scholar]
- Matsushita T., Mochizuki N. & Nagatani A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424, 571–574 (2003). [DOI] [PubMed] [Google Scholar]
- Vidal G. A., Naresh A., Marrero L. & Jones F. E. Presenilin-dependent gamma-secretase processing regulates multiple ERBB4/HER4 activities. J. Biol. Chem. 280, 19777–19783 (2005). [DOI] [PubMed] [Google Scholar]
- Sastre M. et al. Presenilin-dependent gamma-secretase processing of beta-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2, 835–841 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodia S. S. & St George-Hyslop P. H. Gamma-Secretase, Notch, Abeta and Alzheimer?s disease: where do the presenilins fit in? Nature reviews. Neuroscience 3, 281–290 (2002). [DOI] [PubMed] [Google Scholar]
- Fortini M. E. Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat. Rev. Mol. Cell Biol. 3, 673–684 (2002). [DOI] [PubMed] [Google Scholar]
- Cai J. et al. Gamma-Secretase and presenilin mediate cleavage and phosphorylation of vascular endothelial growth factor receptor-1. J. Biol. Chem. 286, 42514–42523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y. S. et al. Towards establishment of a rice stress response interactome. PLoS Genet. 7, e1002020 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K. & Guerois R. NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem. Sci. 35, 199–207 (2010). [DOI] [PubMed] [Google Scholar]
- Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60, 139–164 (2009). [DOI] [PubMed] [Google Scholar]
- Noel L. D. et al. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19, 4061–4076 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q. H. & Schulze-Lefert P. Rumble in the nuclear jungle: compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J. 26, 4293–4301 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. OsRAR1 and OsSGT1 physically interact and function in rice basal disease resistance. Mol. Plant Microbe. Interact. 21, 294–303 (2008). [DOI] [PubMed] [Google Scholar]
- Bol G., Kreuzer O. J. & Brigelius-Flohe R. Translocation of the interleukin-1 receptor-associated kinase-1 (IRAK-1) into the nucleus. FEBS Lett. 477, 73–78 (2000). [DOI] [PubMed] [Google Scholar]
- Liu G., Park Y. J. & Abraham E. Interleukin-1 receptor-associated kinase (IRAK) -1-mediated NF-kappaB activation requires cytosolic and nuclear activity. FASEB J. 22, 2285–2296 (2008). [DOI] [PubMed] [Google Scholar]
- Huang Y., Li T., Sane D. C. & Li L. IRAK1 serves as a novel regulator essential for lipopolysaccharide-induced interleukin-10 gene expression. J. Biol. Chem. 279, 51697–51703 (2004). [DOI] [PubMed] [Google Scholar]
- Su J., Richter K., Zhang C., Gu Q. & Li L. Differential regulation of interleukin-1 receptor associated kinase 1 (IRAK1) splice variants. Mol. Immunol. 44, 900–905 (2007). [DOI] [PubMed] [Google Scholar]
- Huang Y. S., Misior A. & Li L. W. Novel role and regulation of the interleukin-1 receptor associated kinase (IRAK) family proteins. Cell. Mol. Immunol. 2, 36–39 (2005). [PubMed] [Google Scholar]
- Chern M., Canlas P. E., Fitzgerald H. A. & Ronald P. C. Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J. 43, 623–635 (2005). [DOI] [PubMed] [Google Scholar]
- Christensen A. H., Sharrock R. A. & Quail P. H. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18, 675–689 (1992). [DOI] [PubMed] [Google Scholar]
- Heese A. et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl Acad. Sci. USA 104, 12217–12222 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A., Ludwig A. A. & Jones J. D. Rapid phosphorylation of a syntaxin during the Avr9/Cf-9-race-specific signaling pathway. Plant Physiol. 138, 2406–2416 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Park C. J. & Ronald P. C. Quantitative measurements of Xanthomonas Oryzae pv. Oryzae distribution in rice using fluorescent-labeling. J. Plant Biol. 54, 269–274 (2011). [Google Scholar]
- Wang Y. S. et al. Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18, 3635–3646 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S13, Supplementary Table S1 and Supplementary Reference.