Abstract
Purpose.
Myopia, or nearsightedness, is a common ocular genetic disease for which over 20 candidate genomic loci have been identified. The high-grade myopia locus, MYP3, has been reported on chromosome 12q21–23 by four independent linkage studies.
Methods.
We performed a genetic association study of the MYP3 locus in a family-based high-grade myopia cohort (n = 82) by genotyping 768 single-nucleotide polymorphisms (SNPs) within the linkage region. Qualitative testing for high-grade myopia (sphere ≤ −5 D affected, > −0.5 D unaffected) and quantitative testing on the average dioptric sphere were performed.
Results.
Several genetic markers were nominally significantly associated with high-grade myopia in qualitative testing, including rs3803036, a missense mutation in PTPRR (P = 9.1 × 10−4) and rs4764971, an intronic SNP in UHRF1BP1L (P = 6.1 × 10−4). Quantitative testing determined statistically significant SNPs rs4764971, also found by qualitative testing (P = 3.1 × 10−6); rs7134216, in the 3′ untranslated region (UTR) of DEPDC4 (P = 5.4 × 10−7); and rs17306116, an intronic SNP within PPFIA2 (P < 9 × 10−4). Independently conducted whole genome expression array analyses identified protein tyrosine phosphatase genes PTPRR and PPFIA2, which are in the same gene family, as differentially expressed in normal rapidly growing fetal relative to normal adult ocular tissue (confirmed by RT-qPCR).
Conclusions.
In an independent high-grade myopia cohort, an intronic SNP in UHRF1BP1L, rs4764971, was validated for quantitative association, and SNPs within PTPRR (quantitative) and PPFIA2 (qualitative and quantitative) approached significance. Three genes identified by our association study and supported by ocular expression and/or replication, UHRF1BP1L, PTPRR, and PPFIA2, are novel candidates for myopic development within the MYP3 locus that should be further studied.
Association mapping of the MYP3 locus reveals novel candidate genes.
Introduction
Myopia is the most common ocular disorder.1 Recent studies estimate 41.6% of the United States population has some form of myopia.2–4 High-grade myopia, the most extreme manifestation in the refractive error spectrum, has a prevalence of 2% to 3% in the United States, Western Europe, and Australia.2 Although criteria for high-grade myopia vary between studies, a corrective lens of −5.00 diopters (D) or more in the better of the two eyes with onset prior to 12 years of age is one commonly used criterion.5 Sphere (SPH), measured in D, determines the amount of optical correction needed in the axis meridian. Spherical equivalence (SE; sphere + cylinder/2) is also measured in D and is used to determine the optical correction needed to compensate for axial length in addition to curvature of the cornea and/or the lens. The more severe the myopia is, the higher the risks of retinal detachment, glaucoma, cataract, myopic degeneration, and other ocular morbidities.2,6 As the prevalence of high-grade continues to increase, so does its overall contribution to vision loss.2
Emmetropization, an active childhood regulatory process of ocular growth, aims to match the optical power of the cornea and lens to the axial length of the eye.7 Animal models have demonstrated that visual cues interpreted by the retina and signaled through the choroid and sclera locally control the shape and size of the eye.8–11 Failure of emmetropization in postnatal ocular development results in refractive error.7 Myopic refractive error is characterized by a focal point of parallel light rays converging in front of the retinal fovea plane, resulting in blurred vision, frequently caused by increased axial length.2 In high-grade myopia the axial elongation is localized primarily to the posterior or central pole of the globe, suggesting that changes in those tissues (central retinal, retinal pigment epithelium [RPE], and scleral) are responsible for myopic development.12
Linkage studies have identified over 20 loci for myopia and high-grade myopia, most with candidate gene screenings of limited success.13,14 Relatively few association studies have been performed for these linkage peaks, especially for the larger intervals, such as the high-grade myopia locus on chromosome 12, MYP315 (MIM 603221). Young et al.15 first established the MYP3 locus for familial high-grade nonsyndromic myopia. Using microsatellite markers with a maximum logarithim of odds (LOD) score of 3.8, a linkage study involving a single, large, multigenerational family of German/Italian descent mapped the MYP3 locus to chromosome 12q21–23.15 The MYP3 locus was replicated by Farbrother et al.16 using 51 British myopia families with a maximum LOD score of 2.54. Nürnberg et al.17 replicated the MYP3 locus using a single high-grade myopia family of German descent with a maximum LOD score of 3.9. Most recently, Li et al.18 performed a whole genome SNP linkage scan using 254 high-grade myopia families in an international cohort with a peak LOD score of 3.48 for MYP3. The four independent linkage peaks in the 12q region from these studies are shown in Figure 1. Although the linkage peaks do not exactly overlap with one another, linkage is often an imprecise tool and the origin of the signal may be found anywhere within megabases of the most linked markers.
Figure 1.
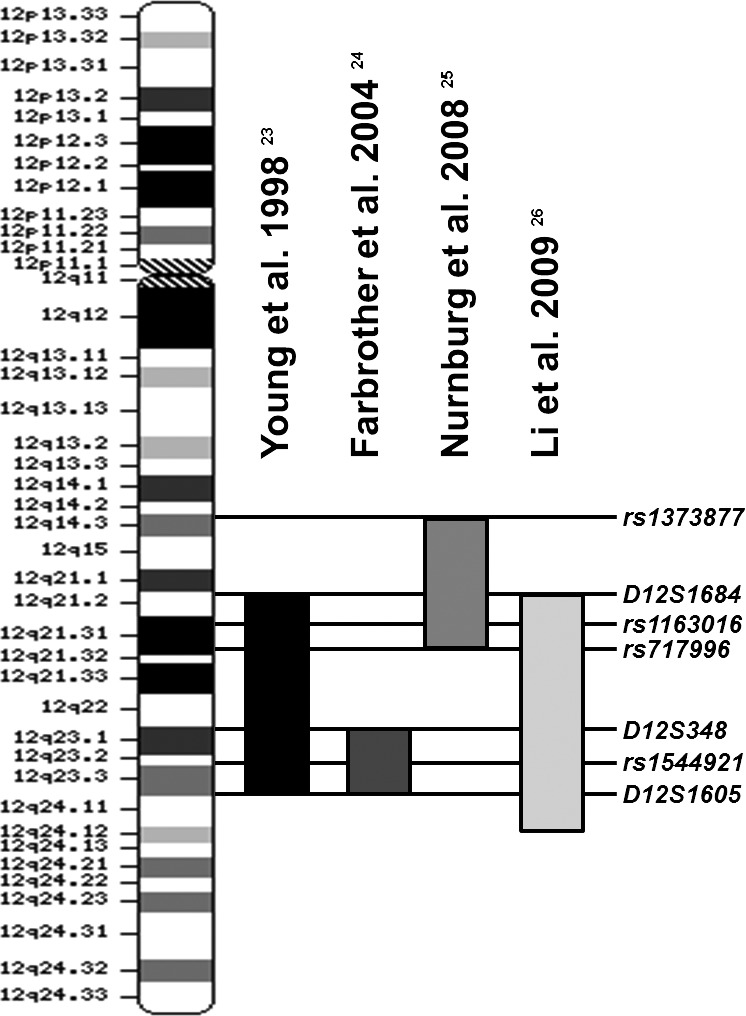
Overlap of linkage studies implicating MYP3. Four linkage studies, indicated by horizontal bars, have found linkage of the MYP3 locus on chromosome 12.15–18 (adapted from Nürnberg et al. 200817).
In addition to evidence for linkage with the qualitative trait of high-grade myopia, quantitative trait linkage analyses also revealed nominally significant LOD scores in the MYP3 locus for SPH.15–19 The region implicated by the independent linkage studies, MYP3, spans nearly 44 Mb (Fig. 1), making candidate sequencing studies impractical in narrowing the region. Finer mapping, such as SNP association, may help to prioritize candidates within the region. MYP3 may contain variants that associate with high-grade myopia as well as those with quantitative traits used to measure the degree of refractive error. However, the size of this linkage peak limits the number of SNPs that can be cost- and resource-effective. Thus, further prioritization of candidate genes after association is necessary.
To fine-map the chromosome 12 linkage peak, MYP3, we conducted a peakwide SNP qualitative and quantitative association mapping study in our high-grade myopia family cohort. Additionally, independently collected expression data were used as supporting evidence of association (F. Hawthorne, S. Feng, E. St. Germain, M. Wang, T. Young, and R. Metlapally, unpublished observations, 2012). Genes that were within 100 kb of the most significantly associated SNPs were assessed for differential expression during and after growth in the central retina, RPE, choroid, and sclera of normal individuals.
Materials and Methods
Family-Based Genetic Association Analyses
Patient Cohort.
The cohort consisted of Caucasian families of three or more individuals previously collected by T. L. Young and used in whole genome linkage analyses replicating the MYP3 locus.18 Phenotypic information was collected for each family member included in the study by ocular examination. Affected individuals were characterized with high-grade nonsyndromic myopia using the same criteria as in linkage analyses (dioptric SPH measurement of less than or equal to −5.00 in the better of two eyes and an onset by 12 years of age).18 Families with highly myopic phenotypes were preferentially selected and they were included in the analyses on the exclusive basis of the presence of at least one high-grade myopia family member. The average SPH for the individuals in the cohort was −4.78 D (affected: −10.44 D; others: −1.36 D). Patients with ophthalmologic conditions that might predispose them to high-grade myopia or syndromic disorders, such as Stickler, Ehler-Danlos, or Marfan syndromes, were excluded from this study. Previous evidence of positive linkage to MYP3 was not considered when choosing families. The cohort consisted of 530 (affected = 235) individuals from 67 multigeneration families of various sizes with at least two affected individuals per family. The average family size was 7.91 individuals, with a range of 3 to 35 individuals per family. The cohort was composed of an equal number of males and females (n = 265 each). The cohort was composed entirely of self-reported Caucasians to reduce ethnic stratification issues. All participants provided written informed consent that was approved by Duke University's Institutional Review Board and adhered to the tenets of the Declaration of Helsinki guidelines. The patient cohort description is summarized in Table 1.
Table 1. .
Patient Cohort Description
| Number of patients | 530 | Affected | 235 |
| Unaffected or unknown | 295 | ||
| Males | 265 | ||
| Females | 265 | ||
| Number of families | 67 | Average size | 7.91 |
| Size range | 3 to 35 | ||
| Family type | Multigenerational | ||
| Self-reported ethnicity | Caucasian | ||
| Affection criteria | Spherical diopters (D) | Affected | ≤−5 D (both eyes) |
| Unaffected | ≥−0.50 D (both eyes) | ||
| Unknown | All others | ||
| Average SPH | −4.78 D | Affected | −10.44 D |
| Unaffected | −1.36 D | ||
| Range | −32.00 D to +9.25 D | ||
| Median | −3.25 D |
SNP Selection.
A list of all potential SNPs was generated by genetic analysis research (Illumina; San Diego, CA) upon submission of the requested 44 Mb region. The region was covered using nonuniform SNP density. In all, 768 SNPs were distributed across the region at a base density of one SNP every 60 kb with regions of a higher density SNP placement of one every 40 kb. The regions of higher density SNP placement were: (1) the region of overlap between Young et al.15 and Nürnberg et al.17; (2) the region of overlap between Young et al.15 and Farbrother et al.16; and (3) the highest peak region of Li et al.18 (Fig. 1). SNPs were selected using the following criteria: (1) location (as described above); (2) minor allele frequency (MAF) > 0.05; (3) Illumina designability score > 0.7 (with preference to higher scores); (4) location relative to genes (coding nonsynonymous [n = 36] > coding synonymous [n = 38] > UTR [n = 48] > intronic [n = 207] > intergenic [n = 439]).
Genotyping and Quality Control.
Genomic DNA was extracted from each individual from either blood or saliva as previously described.20 Family members were genotyped using custom-designed 768-plex Oligo Pool Assay (OPA; GoldenGate Universal 32 Beadchip Kits; Illumina). Samples were directly hybridized to the chips and detected by array-based genetic analysis (Illumina iScan system). The genotyping assays (GoldenGate; Illumina) were run with 672 samples on seven 96-well plates that included 28 quality controls. These samples included an additional 114 individuals from 15 families of Hispanic, Latino, Asian, or African American descent that were excluded from these analyses but are included in the supplementary materials (see Supplementary Material, http://www.iovs.org/content/54/3/2076/suppl/DC1). Each genotyping plate included two Centre d'Etude du Polymorphisme Humain (CEPH) DNA samples as well as two internal replicates with masked identity during analysis. The internal replicates were selected from samples in the cohort, which had sufficient quantities of DNA for duplicate genotyping. The CEPHs and internal replicate genotypes were checked for concordance. The data were cleaned using a commercial system (GenomeStudio; Illumina). Mendelian inconsistencies and relationships within families were screened using a program for identification of genotype incompatibilities in linkage analysis (PedCheck; Division of Statistical Genetics, University of Pittsburgh, PA).21 Samples with a call rate below 95% and parent-to-child Mendelian errors above 1% were removed (n = 59 and 12, respectively) from further analyses. SNPs with a call rate below 95% (n = 15) and Mendelian errors over 95% (n = 3) were also removed from further analyses. Deviations from Hardy-Weinberg equilibrium (HWE) were checked using genetic data analysis.22 Markers were removed from analyses if the control population deviated from HWE (P < 0.05) (n = 32). Pairwise linkage disequilibrium (LD) within the population was checked using graphical overview of linkage disequilibrium (GOLD) and LD Select.23,24 The final number of SNPs (n = 702) was used to calculate the significance threshold (for all association analyses) after correction for multiple testing of 7.12 × 10−5.
Qualitative Association Testing.
The association in the presence of linkage test (APL)25 was used in qualitative association tests because it infers absent parental genotypes using estimates of identity by descent, allowing for single and multilocus haplotype analysis. Individuals were classified as affected for high-grade myopia if both of their eyes were ≤ −5 spherical D; individuals with ≥ −0.50 spherical D in both eyes were classified as unaffected; all other individuals including those with missing phenotype data were classified as unknown affection status. Consistency of the data was checked with the same affection status classifications using the additional non-Caucasian individuals (used in previous linkage analyses18 but excluded from these association tests) as well as with another common biometric parameter used in determining myopic refraction, SE (see Supplementary Material and Supplementary Fig. S1, http://www.iovs.org/content/54/3/2076/suppl/DC1). Additionally, consistency of the data was checked using −5 D and −6 D affection status cutoffs (see Supplementary Material and Supplementary Fig. S1, http://www.iovs.org/content/54/3/2076/suppl/DC1).
Quantitative Association Testing.
The quantitative trait linkage disequilibrium test (QTLD)26 in sequential oligogenic linkage analysis routines (SOLARs)27 was conducted by treating refractive errors as quantitative traits. Quantitative tests were run using the SPH phenotype by taking the average (avg) of the two eyes. The same affection status classifications using the additional non-Caucasian individuals as well as SE were again tested for consistency (see Supplementary Material and Supplementary Fig. S1, http://www.iovs.org/content/54/3/2076/suppl/DC1). The QTLD tests were run using the default model, including identity by descent calculations of our study population. The QTLD test calculations of identity by descent were obtained using SOLAR.27
Independent Validation Cohorts.
Five (DNA quantities prohibited additional candidate screening; refer to Results section for selection criteria) of the most significantly associated SNPs from qualitative and quantitative testing were genotyped in an independent collection of Caucasian high-grade myopia pedigrees recruited from four international collection sites. These pedigrees have been described previously.18 Genotyping was performed using allelic discrimination assays (Taqman; Foster City, CA), with quality control measures as previously described (see Supplementary Material and Supplementary Table S2, http://www.iovs.org/content/54/3/2076/suppl/DC1).28 Qualitative and quantitative tests, described above, for SPH were used for each SNP.
Qualitative and quantitative tests for SPH were previously carried out in a cohort comprised of a sample of twins (UK Twin Study) that were not recruited on the basis of their refractive error.29 These test results were used as a second independent validation cohort. As described,29 this cohort of twins had been SNP genotyped at high density across the entire genome. All of the SNPs selected for the discovery association tests, including some of our most significantly associated candidates, were not genotyped in this cohort. All of the most significantly associated SNPs from both the qualitative and quantitative discovery analyses were screened in the twin test results. SNPs in high linkage disequilibrium with the most significantly associated SNPs not genotyped in the cohort were also screened, but none was genotyped in the cohort.
Ocular Tissue Expression
Whole Genome Expression Analysis.
Independently designed, collected and reported ocular expression data (F. Hawthorne et al., unpublished observations, 2012) were used to prioritize candidate genes from association testing. Genes that were within 100 kb of the most significantly associated SNPs in MYP3 were screened and prioritized by differential ocular expression. Given that myopia can be considered a failure of normal growth, genes involved in normal growth were considered likely candidates, and those with differential expression in central retina, RPE, choroid, and/or sclera between fetal and adult eyes were prioritized.
To compare gene expression between rapidly growing and fully grown ocular tissue types, normal samples from two age groups were used: fetal eyes and adult eyes. The group of fetal eyes consisted of late prenatal fetal eyes of approximately 24-weeks gestational age from elective abortions with no known defects or abnormalities. Nine fetal donor eyes (four male and five female samples), and six fully grown adult donor eyes (three of each sex) were used for microarray analyses. Whole globes were preserved (RNAlater; Qiagen, Hilden, Germany) within 6.5 hours of collection and shipped overnight on ice for immediate tissue isolation (Table 2). The retina, RPE, and choroid and scleral tissues were isolated at the posterior pole using a circular, double-embedded technique using round 7 and 5 mm biopsy punches to reduce contamination of retina to the other tissue samples. RNA was extracted from each tissue sample using a total RNA extraction kit (mirVana; Ambion, Austin, TX) following the manufacturer's protocol. RNA samples were hybridized (HumanHT-12 v4 Expression BeadChips; Illumina), following the manufacturer's protocol recommendations.
Table 2. .
Donor Information for Adult Ocular Whole Globes Used in Tissue Expression Analyses
|
Individual |
Race |
Sex |
Age, y |
Preservation Time Interval, h |
Cause of Death |
| 1658* | Caucasian | Male | 71 | 5:05 | Respiratory failure |
| 1812* | Caucasian | Female | 70 | 4:28 | Liver failure |
| 2809 | Caucasian | Female | 76 | 4:00 | Leukemia/acute myeloid leukemia |
| 2835 | Caucasian | Female | 55 | 4:55 | Dementia |
| 2828 | Caucasian | Male | 80 | 5:20 | Pneumonia |
| 2834 | Caucasian | Male | 56 | 5:22 | Abdominal aortic aneurysm rupture/heart disease |
| 2217 | Caucasian | Female | 77 | 6:24 | Chronic obstructive pulmonary disease/emphysema |
| 2836 | Caucasian | Male | 67 | 5:10 | Heart disease |
Indicates samples that were used for RT-qPCR but not microarray analysis.
Background noise was subtracted from the intensity values prior to exportation on log2 transformation (GenomeStudio program; Illumina). Sample outliers were determined by principal component analyses using the Hotelling T2 test30 (at 95% confidence interval) and removed from further analyses. The data intensity was normalized by quantile normalization followed by multichip averaging31 to reduce chip effects. The exact Wilcoxon rank-sum test32 was used to identify differentially expressed genes. Fetal ocular tissues were compared with their adult counterparts, and assessed for relative fold changes per probe. Adult retina and RPE samples were averaged for comparison to fetal retina/RPE. The Benjamin and Hochberg False Discovery Rate33 (FDR) procedure was applied, and FDR was controlled at 0.05 to determine statistical significance for all comparisons.
Quantitative Real-Time PCR Validation.
Candidate genes within 100 kb of the most significantly associated SNPs in the MYP3 association results were compared with genes identified by microarray analysis as significantly differentially expressed. Genes of interest in the expression analysis were determined by their relation to candidate genes from the association analyses, their degree of differential expression, concordance of probes within the same genes, and their level of expression (see Supplementary Material and Supplementary Table S1, http://www.iovs.org/content/54/3/2076/suppl/DC1). Genes that were the same or directly, functionally related to candidate genes in at least one tissue type were validated using RT-qPCR (Taqman) gene expression assays (see Supplementary Material and Supplementary Table S2, http://www.iovs.org/content/54/3/2076/suppl/DC1). RT-qPCR for each gene was performed for all tissue types, rather than just the tissue(s) of known overlap. Each gene was validated with four biological replicates of each adult central and 24-week central tissue, in addition to one nontemplate control. Each biological replicate contained equal amounts of total RNA pooled from the tissue of two individuals. In total, RNA from the tissue of eight individuals was used for each gene per tissue type. Each biological replicate was carried out in four technical replicates of RT-qPCR. Expressions of adult retina and RPE were averaged and compared with 24-week retina/RPE since they could not be separated. Each plate containing quantitative real-time assays contained the two control genes GAPDH (MIM 138400) and 18S for each biological replicate of each tissue type. Reverse-transcription reactions were performed with a commercial reverse transcription kit (Applied Biosystems [ABI] High Capacity cDNA RT kit (Carlsbad, CA) per manufacturer's protocol (Carlsbad, CA). RT-qPCR was carried out using a commercial fast real-time PCR system (ABI 7900), per gene expression assays protocol (Taqman). Fold changes and SD errors in each tissue type were calculated by the 2−ΔΔCT method with normalization by the two housekeeping genes.34 Technical replicates were removed if the SD was >0.5 and no biological replicates were removed.
Results
Qualitative Association
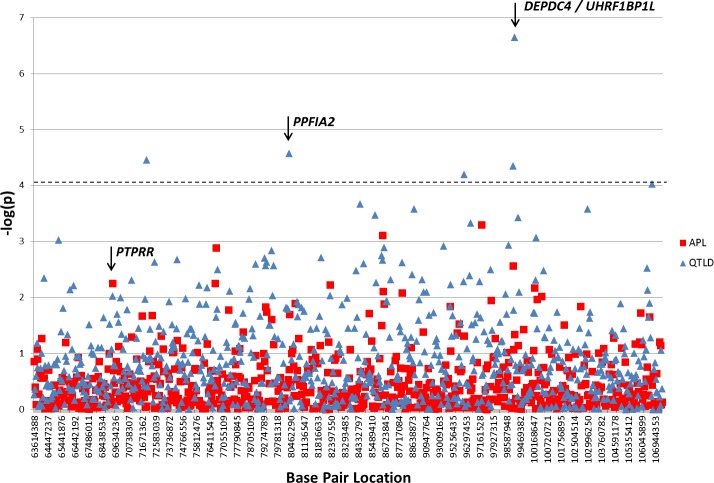
Testing for association with high-grade myopia status was carried out classifying individuals with a dioptric SPH ≤−5 D as affected and those with ≥−0.50 D as unaffected. After stringent quality (refer to the Materials and Methods section) control, 702 SNPs remained. Although no SNPs passed Bonferroni correction significance levels, the most associated SNPs were evaluated as possible candidates (Fig. 2). Table 3 contains the five SNPs with the lowest qualitative test P values (P < 6 × 10−3). The three SNPs with the highest probability of association are located over 50 kb from the nearest genes: rs741525 (P = 7.80 × 10−4), rs790436 (P = 5.09 × 10−4), and rs1358228 (P = 1.29 × 10−3). The two SNPs with the next highest association probabilities are located within coding genes: rs7134216 (P = 2.74 × 10−3) is intronic to the UHRF1BP1L gene (UHRF1 [MIM 607990] binding protein 1-like isoform b), and rs3803036 (P = 5.65 × 10−3), is a nonsynonymous coding variant in the PTPRR gene (protein tyrosine phosphatase receptor type R; MIM 602853).
Figure 2.
APL and QTLD association results by −log(p). P values for each SNP are stacked in the same vertical position and indicated by separate symbols. The blue triangles indicate average of two eyes (avg) SPH QTLD results. The red squares and square indicate the high myopia by SPH APL results. The dashed line indicates the Bonferroni significance threshold by number of SNPs. Base-pair locations are given in human genome build 36.2, chromosome 12.
Table 3. .
The Most Significantly Associated SNPs from Qualitative and Quantitative Association Testing
|
Base Pair Location |
SNP |
Gene Location |
MAF |
Qualitative SPH
P
Value |
Quantitative SPH
P
Value |
| 69425930 | rs3803036 | Coding nonsyn. in PTPRR | 0.258 | 5.65 × 10−3 | 9.39 × 10−3 |
| 71970854 | rs1520562 | >600 kb from nearest gene | 0.271 | 0.875 | 3.51 × 10−5 |
| 76672201 | rs1358228 | 76 kb upstream of NAV3 (MIM 611629) | 0.415 | 1.29 × 10−3 | 2.22 × 10−2 |
| 80351222 | rs17306116 | Intronic PPFIA2 | 0.119 | 0.195 | 2.65 × 10−5 |
| 86528808 | rs790436 | >150 kb from nearest gene | 0.425 | 7.80 × 10−4 | 2.12 × 10−3 |
| 96200365 | rs1558726 | >150 kb from nearest gene | 0.108 | 4.86 × 10−2 | 6.36 × 10−5 |
| 97316149 | rs741525 | 55 kb from SLC9A7P1 | 0.325 | 5.09 × 10−4 | 0.698 |
| 99043879 | rs7134216 | Intronic UHRF1BP1L | 0.233 | 2.74 × 10−3 | 4.49 × 10−5 |
| 99155927 | rs4764971 | 3′ UTR of DEPDC4, intronic ACTR6 | 0.133 | 4.62 × 10−2 | 2.25 × 10−7 |
Values of P that withstood Bonferroni correction are in bold. Base pairs are based on human genome build 36.2, chromosome 12. MAF is for Caucasians, the ethnicity used in association analyses.
Quantitative Association
Quantitative tests were performed to test each SNP for association with the average SPH endophenotype. Quantitative traits were normally distributed and removal of phenotypic extremes did not affect the results (data not shown). The quantitative tests showed markers with overlapping significance with the qualitative results. Figure 2 shows both the qualitative and quantitative association results for the 702 SNPs that passed quality control measures. The five most significantly associated SNPs that met the Bonferroni threshold with 702 SNPs of P < 7.12 × 10−5 (α = 0.05). The first and fourth most significant SNPs, rs4764971 (P = 2.25 × 10−7) and rs7134216 (P = 4.49 × 10−5), are over 100 kb apart, although not in LD (Table 3). The first SNP, rs4764971, is located in the 3′ untranslated region (UTR) of the longest isoform of the gene DEPDC4 (DEP domain containing 4) and is also intronic in ACTR6 (actin-related protein 6), whereas rs7134216 is the same intronic SNP located in the gene UHRF1BP1L as in APL results. The second-best SNP intronic in PPFIA2 (PTPRF interacting; MIM 603143), rs17306116 (P = 2.65 × 10−5), was also statistically significant. Although rs1520562 and rs1558726 also withstood Bonferroni correction, they are in regions that are at least 600 and 150 kb, respectively, from the nearest genes. The region containing rs1520562 is conserved in higher primates, rhesus, marmoset, and guinea pig, but not in rats, mice, other mammals, or lower vertebrates (see Supplementary Material and Supplementary Table S3, http://www.iovs.org/content/54/3/2076/suppl/DC1).
Validation of Variants in Two Independent Cohorts
Three candidate SNPs were selected from both the qualitative and from the quantitative test results, one of which overlapped, giving five SNPs in total (Table 4). SNPs were selected based on their association probabilities and overlap between qualitative and quantitative test results. The SNPs selected from qualitative test results were located in or near PTPRR, UHRF1BP1L, and UTP20. The SNPs selected from quantitative test results were located in or near DEPDC4, UHRF1BP1L (same SNP from qualitative test results), and PPFIA2.
Table 4. .
Validation of Candidate SNPs in an Independent Cohort
|
Base Pair Location |
SNP |
Gene Location |
Qualitative SPH
P
Value |
Quantitative SPH
P
Value |
| 69425930 | rs3803036 | Coding nonsyn. in PTPRR | 0.881 | 1.50 × 10−2 |
| 80351222 | rs17306116 | Intronic PPFIA2 | 7.37 × 10−2 | 6.59 × 10−2 |
| 99043879 | rs7134216 | Intronic UHRF1BP1L | 0.114 | 2.63 × 10−3 |
| 99155927 | rs4764971 | Exonic DEPDC4 | 0.555 | 0.172 |
| 100195248 | rs824311 | 3 kb upstream UTP20 | 0.509 | 0.806 |
Base pairs are based on human genome build 36.2, chromosome 12.
As in the discovery cohort, none of the SNPs tested in the independent Caucasian, family-based, high-grade myopia cohort was significant in tests using the qualitative SPH trait definition. One of the two SNPs in PTP genes approached significance: rs17306116 intronic in PPFIA2 (P = 7.37 × 10−2). In tests using the quantitative SPH trait definition, two SNPs had values of P < 0.05, but only one survived multiple testing correction criteria: rs7134216 in UHRF1BP1L (P = 2.63 × 10−3). SNPs in/near the two PTP genes approached significance: rs3803036 (PTPRR; 1.50 × 10−2) and rs17306116 (PPFIA2; 6.59 × 10−2).
We also sought evidence of replication of our most strongly associated SNPs in an independent cohort of twins not ascertained on the basis of refractive error, for whom genomewide genotyping and imputation had previously been carried out.29 As shown in Table 5, none of our SNPs demonstrated evidence of replication in this cohort; however, one of our most significantly associated quantitative SNPs that was not selected for validation in the high-grade myopia validation cohort, rs1520562, had SPH values of P = 2.4 × 10−2 for quantitative, 7.5 × 10−2 for qualitative testing, and was directionally consistent with our discovery data.
Table 5. .
Validation of the Most Significantly Associated SNPs in a Previously Genotyped Independent Cohort
|
Base Pair Location |
SNP |
Gene Location |
Qualitative SPH
P
Value |
Quantitative SPH
P
Value |
| 69425930 | rs3803036 | Coding nonsyn. in PTPRR | 0.73 | 0.15 |
| 71970854 | rs1520562 | >600 kb from nearest gene | 0.075 | 0.024 |
| 76672201 | rs1358228 | 76 kb upstream of NAV3 | 0.17 | 0.32 |
| 78976016 | rs7315130 | >100 kb from nearest gene | 0.86 | 0.97 |
| 80351222 | rs17306116 | Intronic PPFIA2 | 0.82 | 0.99 |
The most significantly associated SNPs from our data not included in this table were not present in the independent data set. Base pairs are based on human genome build 36.2, chromosome 12. Methods of this testing and cohort information have been previously published.29
Differential Expression in Fetal versus Adult Ocular Tissues
Independently designed and performed whole genome expression analysis comparing normal rapidly growing fetal ocular samples to normal fully grown adult samples revealed a number of differentially expressed genes (Hawthorne F et al., unpublished observations, 2012). All candidate genes from Table 3 were expressed in all tissues and age groups tested; as such, none could be eliminated for lack of expression in the relevant tissues. All fold changes are presented below as fetal relative to adult expression. When filtered by candidate genes from the qualitative and/or quantitative association results we found only two genes (PPFIA2 and PTPRR) to select from for validation by real-time quantitative PCR (RT-qPCR). Additionally, the related gene, PTPRF (MIM 179590), which interacts with PPFIA2, was also selected for validation. Fold changes for all genes selected for validation are displayed in Figure 3 with biological variation as error bars (SD of technical variation < 0.5; data not shown). All significant or nominally significant SNPs were considered when creating lists of potential candidate genes (genes within 100 kb of SNP). All genes from the association under consideration were included on the microarray but were not statistically significant for differential expression.
Figure 3.
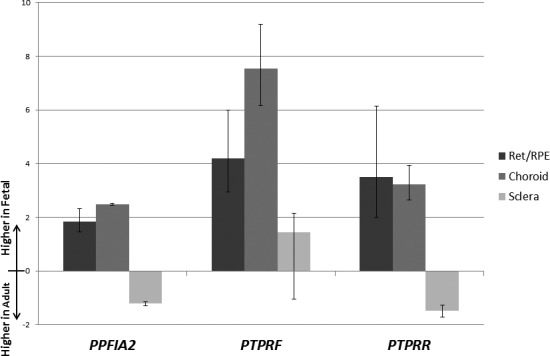
Fetal versus adult fold changes of real-time quantitative PCR validated genes. The columns from left to right (and darkest to lightest) for each gene represent the retina/RPE, choroid, and sclera. Fold changes were calculated by the 2−ΔΔCT method. Biological variation is shown by error bars for each tissue/gene.
The three genes selected for RT-qPCR validation (PTPRR, PPFIA2, and PTPRF) were all PTP-related genes. Upon validation using RT-qPCR, fold changes were generally consistent with the microarray data, except in cases where the expression was very low and/or fold changes were small (Fig. 3; also see Supplementary Material and Supplementary Table S1, http://www.iovs.org/content/54/3/2076/suppl/DC1). In the retina/RPE and choroid all three genes were found to be upregulated in fetal relative to adult tissue-specific expression. In the sclera only PTPRF was upregulated, whereas PTPRR and PPFIA2 were slightly downregulated in fetal relative to adult expression. PTPRR, a candidate from the qualitative high-grade myopia association data, did not show expression in the adult RPE. As a result the retina/RPE fold change could not be calculated. Instead a fold change comparing only the adult retina to the fetal retina/RPE is presented. The change for retina/RPE (without adult RPE) was a 3.49-fold higher expression in fetal tissue; however, the absence in adult RPE limits conclusions based on these data. The fold change for the choroid was 3.22 higher expression in the fetal tissue and the sclera showed a fold change in the opposite direction of −1.49, where the adult was more highly expressed. PPFIA2, a candidate gene from the quantitative SPH association testing, showed a 2.48 higher expression fold change in the fetal choroid relative to the adult, whereas the −1.22 lower expression fold change in the fetal sclera was opposite the direction of the array data (possibly explained by the low overall level of expression and biological variation between samples). The retina/RPE was not significant in array data, but showed a 1.83 higher expression fold change in fetal tissue in RT-qPCR. Interestingly, PTPRF, which interacts with PPFIA2, showed larger fold changes: 4.20 higher expression in fetal retina/RPE, 7.53 higher expression in fetal choroid, and 1.43 higher expression in fetal sclera. The retina/RPE fold changes for PTPRF were much larger; however, the choroid and sclera were consistent with the microarray data.
Discussion
The discovery of qualitative high-grade myopia and quantitative SPH analyses within the MYP3 locus both yielded potentially interesting candidate SNPs. Although none of the SNPs in the qualitative analyses was statistically significant after correction for multiple testing, SNPs approaching significance were further evaluated as candidates. The high number of significant SNPs in quantitative analyses can likely be attributed to their location in a well-established linkage region, as well as the increased power of using quantitative data rather than qualitative classifications. Strict affection status cutoffs and consequently low numbers of affected individuals in the qualitative data may have contributed to reduced power, resulting in higher P values.
Validations
Independent validation, using a Caucasian high-grade myopia cohort, of our most significant association SNPs for both qualitative and quantitative testing, validated one quantitative SNP, rs7134217 within UHRF1BP1L. The significance of this SNP overlapped between both qualitative and quantitative discovery cohort results, but was replicated for quantitative association only in this second Caucasian high-grade myopia validation cohort. One of the two PTP genes genotyped approached significance in qualitative testing: rs71306116 in PPFIA2. However, this SNP was not one of the most significantly associated candidates in our qualitative association and none of our qualitative results in the discovery cohort reached statistical significance criteria. Both of the two SNPs in PTP candidate genes had suggestive association in quantitative association assessment in this high-grade myopia validation cohort: rs3803036 (PTPRR) and rs1730611 (PPFIA2). Only the SNP within PPFIA2 was statistically significant in the discovery quantitative testing, although PTPRR did approach statistical significance in our data as well. In total, three of the five SNPs chosen for follow-up genotyping were nominally significant or statistically significant in a second high-grade myopia cohort, supporting their association with myopia.
In a second independent twin cohort that did not contain data for all of our most significantly associated SNPs, one of the most significant SNPs from the discovery cohort showed potentially interesting results. The SNP rs1520562 is not near any known genes; however, it is in a region that is conserved largely by higher mammals including primates. The conservation pattern suggests that the area should be thoroughly screened for regulatory elements that may affect vision in this subset of mammals. Since this second replicate cohort was not collected with preferential selection for highly myopic individuals (twin cohort average of −0.40 D compared with discovery cohort average of −4.78 D) and used a higher cutoff in qualitative testing (−6 D compared with our −5 D cutoff), it is possible that this twin cohort lacked the power to identify some of the variants from our study. Additionally, since these data were previously collected, we were not able to screen all of our candidate SNPs. Considering these limitations, it is highly encouraging that this SNP nominally replicated in the independent twin cohort. The overlap suggests that results obtained in the discovery cohort may be representative of others not tested.
PTPRR and PPFIA2 as Candidate Genes by Genomic Convergence
Qualitative and quantitative analyses had some overlap in their most associated SNPs (Table 3) and biological connections between some candidate genes. The same intronic SNP located in UHRF1BP1L was one of the most associated SNPs in both tests. Four of the five most associated SNPs in the qualitative analyses had values of P < 0.05 for the same SNPs in quantitative analyses, including the SNP located in the PTP gene PTPRR, which is functionally related to another most associated SNP from the quantitative data, PPFIA2.
One of our candidate genes from the qualitative association data, PTPRR, was found to be more highly expressed in the rapidly growing fetal retina/RPE (not detected in adult RPE) and choroid, although slightly more highly expressed in the grown adult sclera. From our quantitative association data, PPFIA2 and its namesake interacting gene, PTPRF, both were found to be differentially expressed in fetal relative to adult ocular tissues. Like PTPRR, PPFIA2 showed higher expression in the rapidly growing fetal retina/RPE and choroid, and higher expression in the adult sclera. PTPRF showed similar, although more pronounced, patterns with retina/RPE and choroid expression but showed higher expression in the rapidly growing fetal sclera (Fig. 3). Overall, in the microarray data and in all three PTP genes evaluated, the tissue with the highest fold changes was the choroid. The lack of high fold changes in the retina/RPE may have been caused by the averaging methods used since the fetal tissues could not be separated.
Insights into Possible Biological Mechanisms of Refractive Error
Interestingly, the two MYP3 candidate genes from the association data (one from the qualitative and one from the quantitative) that were found to be differentially expressed in ocular tissues, were both PTP-related genes. PTPs are enzymes that remove a phosphate group from the amino acid tyrosine. These proteins play a role in the regulation of signal transduction by relaying signals from outside the cell that regulate cell growth, division, maturation, and function.35,36 Although PTP genes seem ubiquitous, several studies have implicated them in tissue-specific diseases and functions as well as various cancers.35–39 There has also been one PTP-related gene previously associated with rapid ocular growth that lies within the MYP3 locus; microarray work validated by RT-qPCR of choroid/RPE expression in marmoset eyes undergoing lens treatment to induce rapid ocular growth showed differential expression of PTP receptor type B (PTPRB; MIM 176882).40
PTPRR shares a gene symbol (considered synonymous) with PTPRQ (MIM 603317) that, when overexpressed, inhibits cell proliferation and induces apoptosis.41 PTPRQ has been identified as belonging to a subgroup of PTP receptors, which have different primary biological activities.41 This subgroup acts as active phosphoinositide phosphatases that regulate cell proliferation,41 making genes, such as PTPRR, an early and frequent target of silencing in some forms of cancer.42 PTPRR has also been shown to be a key negative regulator of MAPK signaling in androgen-regulated expression.43 Ptprr in mice has distinct localization of each of the isoforms, all of which contain the kinase interacting motif necessary to bind and inactivate MAPK by dephosphorylation.44 In mice, two of these isoforms of Ptprr localize on the membranes of endocytotic vesicles like those used for glutamate or other cell signaling molecules.45,46 Mutations in PTPRR could be involved in the development of high-grade myopia by a change in its ability to bind to or dephosphorylate MAPK, leading to increased MAPK growth signals. Increased expression of PTPRR in rapidly growing fetal retina/RPE (not detected in adult RPE) and choroid may be the result of normal developmental or growth regulators, suggesting it plays a role in reigning in ocular growth. Disruption of this protein function may account for the rapid growth seen in highly myopic individuals during early childhood that slows into adulthood. As normal growth regulators slow, the need for this protein would also decrease.
PPFIA2 is a member of the leukocyte common antigen-related (LAR) PTP interacting protein family liprin. Liprins are proposed to act as scaffolding proteins for recruitment and anchoring of LAR PTPs.47 Vertebrates have four homologs of liprin-α (1–4), each enriched in different synaptic and nonsynaptic cell populations.48 Liprin-αs are thought to be essential elements of the active zone at the presynaptic plasma membrane by guiding synaptic vessels to their fusion sites and regulating neurotransmitter releases.48 Liprin-α2 in the brain is preferentially localized in excitatory synapses, like the hippocampal mossy fiber nerve endings that release glutamate via synaptic vesicles.48 PPFIA2 is a liprin-α2 known to be downregulated by androgens in prostate cancer cell lines.49 Mutations in PPFIA2 could affect its ability to guide glutamate-filled synaptic vesicles to their fusion sites, thereby disrupting the signaling cascade throughout the retina and possibly onto other ocular tissues. Alterations in the normal signaling cascade may disrupt the normal process of emmetropization. Converse to PTPRR, the function of PPFIA2 seems more likely to be a positive regulator of cell growth that fits with its moderate increase in expression in rapidly growing retina/RPE and choroid.
Although this study focuses on qualitative high-grade myopia determined by SPH measurements and the quantitative trait SPH due to their relation to the ocular tissues discussed, it should be noted that other tests were performed in the discovery cohort, but not reported. Concordance was found in our candidates selected for follow-up in qualitative association using a higher affection criteria cutoff of −6 D (see Supplementary Material and Supplementary Fig. S1, http://www.iovs.org/content/54/3/2076/suppl/DC1). In addition, SE qualitative high-grade myopia as well as quantitative testing were performed. Qualitative tests for SE revealed the same candidate SNPs and genes as those in SPH tests (see Supplementary Material and Supplementary Fig. S1, http://www.iovs.org/content/54/3/2076/suppl/DC1). Quantitative tests for SPH and SE were also concordant for both the average of both eyes as well as the better of the two eyes, although there was some shuffling of the order of most significantly associated SNPs (data not shown). Because no SNPs in the qualitative data were statistically significant, the additional tests do not drop any SNPs from statistical significance. In the quantitative testing, the SNPs in the candidate genes UHRF1BP1L and PPFIA2 were both statistically significant after correction for multiple testing including those not reported (4212 tests, Table 3). Interestingly, in the complete quantitative association results, a pattern was detected in SNPs within functionally related PTP genes (PPFIA2, PPP1R12A [MIM 602021], PTPRB, PTPRR, PTPRQ, DUSP6 [MIM 602748], and DYRK2 [MIM 603496]), two of which are our candidate genes. Whereas most of these SNPs were not individually statistically significant, in the quantitative association testing this subset of SNPs was significantly more likely to have values of P < 0.5 than the rest (P = 1.2 × 10−5) (see Supplementary Material and Supplementary Fig. S2, http://www.iovs.org/content/54/3/2076/suppl/DC1). In addition to PTPRR and PPFIA2, some or all of these genes may warrant further investigation into their possible roles in the variation of traits associated with myopia. It is also potentially interesting that many other known myopia loci contain PTP-related genes (see Supplementary Material and Supplementary Table S4, http://www.iovs.org/content/54/3/2076/suppl/DC1), which may be investigated further for insights into a possible pathway.
Although UHRF1BP1L was not directly supported by our ocular expression data, it still may be an interesting candidate gene. UHRF1BP1L binds to UHRF1, which regulates topoisomerase II alpha (TOP2A; MIM 126430) and retinoblastoma (RB1; MIM 180200) gene expression,50 suggesting a possible link to regulation of cell growth and proliferation within the retina. In the independent expression array data, TOP2A had significantly lower expression in the adult retina/RPE, choroid, and sclera (−1.88-, −56.64-, and −5.43-fold changes, respectively). UHRF1BP1L could be involved in myopic development by regulating the expression of other genes that contribute to axial growth. Pathway analysis of differentially expressed genes may help uncover a more precise possible mechanism for this gene's role in myopic progression.
Conclusions
One of the largest limitations in our discovery cohort was the SNP coverage, given the large size of the MYP3 locus. It is possible that variants within the spacing of our coverage have been missed; however, cost limitations prohibited higher density coverage of the region. Our association data were also limited by the number of samples in the discovery genotyping cohort. Despite these limitations, quantitative association testing yielded several strong candidates withstanding conservative significance cutoffs. The expression data were limited by the sample number and the tissues, which could not be separated in fetal samples. Our estimation of tissues in a complex compared with separate tissues is likely imprecise. The low fold change differences we found in the retina/RPE may be an artifact of our testing. Better tissue isolation techniques will need to be developed for future expression analyses. However, despite these limitations, two of our candidate genes from the association data were supported by the expression data. PTPRR, PPFIA2, and its namesake interacting gene, PTPRF, were found to be differentially expressed in central ocular tissues. This overlap supports our model of eye growth for myopic development and use of these expression data both in other known myopia loci and also in detailed pathway analyses to look for targets within the known loci. However, our high-grade myopia validation cohort most strongly supported UHRF1BP1L, which was not differentially expressed in our data. Differential expression may have been missed by the microarray data, or it may not be differentially expressed. Although the expression data have been shown to be useful in prioritizing candidates, we have shown that it cannot be used to exclude candidates. Further validation and refinement of these three novel MYP3 candidate genes is necessary to make claims as to their involvement in myopic progression.
Supplementary Material
Acknowledgments
The authors thank the families/twins who participated in this study; Catherine Bowes-Rickman for technical advice; Janeen Morgan for assistance in data acquisition; Diana Abbott for help cleaning and formatting the data for statistical analyses; Christina Markunas for statistical assistance in the association study.
Online Web Resources
Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/omim/ National Institutes of Health.
Footnotes
Supported by the National Eye Institute/National Institutes of Health (NIH) Grants R01-EY014685 (TLY) and R01-EY018246 (TLY); Research to Prevent Blindness Inc. (TLY); the Wellcome Trust; the NIH Center for Inherited Disease Research; and the National Institute for Health Research Comprehensive Biomedical Research Centre award (to Guy's and St. Thomas' National Health Service Foundation Trust partnering with King's College London).
Disclosure: F. Hawthorne, None; S. Feng, None; R. Metlapally, None; Y.-J. Li, None; K.-N. Tran-Viet, None; J.A. Guggenheim, None; F. Malecaze, None; P. Calvas, None; T. Rosenberg, None; D.A. Mackey, None; C. Venturini, None; P.G. Hysi, None; C.J. Hammond, None; T.L. Young, None
References
- 1. Pararajasegaram R. VISION 2020—the right to sight: from strategies to action. Am J Ophthalmol. 1999; 128: 359–360 [DOI] [PubMed] [Google Scholar]
- 2. Kempen JH, Mitchell P, Lee KE, et al. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004; 122: 495–505 [DOI] [PubMed] [Google Scholar]
- 3. Javitt JC, Chiang YP. The socioeconomic aspects of laser refractive surgery. Arch Ophthalmol. 1994; 112: 1526–1530 [DOI] [PubMed] [Google Scholar]
- 4. Vitale S, Sperduto RD, Ferris FL III. Increased prevalence of myopia in the United States between 1971–1972 and 1999–2004. Arch Ophthalmol. 2009; 127: 1632–1639 [DOI] [PubMed] [Google Scholar]
- 5. Shih YF, Ho TC, Hsiao CK, Lin LL. Long-term visual prognosis of infantile-onset high myopia. Eye. 2006; 20: 888–892 [DOI] [PubMed] [Google Scholar]
- 6. Burton TC. The influence of refractive error and lattice degeneration on the incidence of retinal detachment. Trans Am Ophthalmol Soc. 1989; 87: 143–155 [PMC free article] [PubMed] [Google Scholar]
- 7. Gordon RA, Donzis PB. Refractive development of the human eye. Arch Ophthalmol. 1985; 103: 785–789 [DOI] [PubMed] [Google Scholar]
- 8. Wallman J, Winawer J. Homeostasis of eye growth and the question of myopia. Neuron. 2004; 43: 447–468 [DOI] [PubMed] [Google Scholar]
- 9. Wildsoet C, Wallman J. Choroidal and scleral mechanisms of compensation for spectacle lenses in chicks. Vision Res. 1995; 35: 1175–1194 [DOI] [PubMed] [Google Scholar]
- 10. Faulkner AE, Kim MK, Iuvone PM, Pardue MT. Head-mounted goggles for murine form deprivation myopia. J Neurosci Methods. 2007; 161: 96–100 [DOI] [PubMed] [Google Scholar]
- 11. Tkatchenko TV, Shen Y, Tkatchenko AV. Mouse experimental myopia has features of primate myopia. Invest Ophthalmol Vis Sci. 2010; 51: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Atchison DA, Jones CE, Schmid KL, et al. Eye shape in emmetropia and myopia. Invest Ophthalmol Vis Sci. 2004; 45: 3380–3386 [DOI] [PubMed] [Google Scholar]
- 13. Young TL. Molecular genetics of human myopia: an update. Optom Vis Sci. 2009; 86: E8–E22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hornbeak DM, Young TL. Myopia genetics: a review of current research and emerging trends. Curr Opin Ophthalmol. 2009; 20: 356–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Young TL, Ronan SM, Alvear AB, et al. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998; 63: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farbrother JE, Kirov G, Owen MJ, Pong-Wong R, Haley CS, Guggenheim JA. Linkage analysis of the genetic loci for high myopia on 18p, 12q, and 17q in 51 U.K. families. Invest Ophthalmol Vis Sci. 2004; 45: 2879–2885 [DOI] [PubMed] [Google Scholar]
- 17. Nürnberg G, Jacobi FK, Broghammer M, et al. Refinement of the MYP3 locus on human chromosome 12 in a German family with Mendelian autosomal dominant high-grade myopia by SNP array mapping. Int J Mol Med. 2008; 21: 429–438 [PubMed] [Google Scholar]
- 18. Li YJ, Guggenheim JA, Bulusu A, et al. An international collaborative family-based whole-genome linkage scan for high-grade myopia. Invest Ophthalmol Vis Sci. 2009; 50: 3116–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abbott D, Li YJ, Guggenheim JA, et al. An international collaborative family-based whole genome quantitative trait linkage scan for myopic refractive error. Mol Vis. 2012; 18: 720–729 [PMC free article] [PubMed] [Google Scholar]
- 20. Oliveira SA, Li YJ, Noureddine MA, et al. Identification of risk and age-at-onset genes on chromosome 1p in Parkinson disease. Am J Hum Genet. 2005; 77: 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998; 63: 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lewis PO, Zaykin D. Genetic Data Analysis. V1.1 2001. May be accessed at http://www.eeb.uconn.edu/people/plewis/software.php [Google Scholar]
- 23. Abecasis GR, Cookson WO. GOLD—graphical overview of linkage disequilibrium. Bioinformatics. 2000; 16: 182–183 [DOI] [PubMed] [Google Scholar]
- 24. Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004; 74: 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin ER, Bass MP, Hauser ER, Kaplan NL. Accounting for linkage in family-based tests of association with missing parental genotypes. Am J Hum Genet. 2003; 73: 1016–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Havill LM, Dyer TD, Richardson DK, Mahaney MC, Blangero J. The quantitative trait linkage disequilibrium test: a more powerful alternative to the quantitative transmission disequilibrium test for use in the absence of population stratification. BMC Genet. 2005; 6 (suppl 1): S91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998; 62: 1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yanovitch T, Li YJ, Metlapally R, Abbott D, Viet KN, Young TL. Hepatocyte growth factor and myopia: genetic association analyses in a Caucasian population. Mol Vis. 2009; 15: 1028–1035 [PMC free article] [PubMed] [Google Scholar]
- 29. Hysi PG, Young TL, Mackey DA, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010; 42: 902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hotelling H. The generalization of Student's ratio. Ann Math Statist. 1931; 2: 360–378 [Google Scholar]
- 31. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003; 4: 249–264 [DOI] [PubMed] [Google Scholar]
- 32. Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bull. 1945; 1: 80–83 [Google Scholar]
- 33. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995; 57: 289–300 [Google Scholar]
- 34. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001; 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 35. Andersen JN, Jansen PG, Echwald SM, et al. A genomic perspective on protein tyrosine phosphatases: gene structure, pseudogenes, and genetic disease linkage. FASEB J. 2004; 18: 8–30 [DOI] [PubMed] [Google Scholar]
- 36. Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006; 7: 833–846 [DOI] [PubMed] [Google Scholar]
- 37. Ozaltin F, Ibsirlioglu T, Taskiran EZ, et al. Disruption of PTPRO causes childhood-onset nephrotic syndrome. Am J Hum Genet. 2011; 89: 139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen X, Yoshida T, Sagara H, Mikami Y, Mishina M. Protein tyrosine phosphatase σ regulates the synapse number of zebrafish olfactory sensory neurons. J Neurochem. 2011; 119: 532–543 [DOI] [PubMed] [Google Scholar]
- 39. Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004; 304: 1164–1166 [DOI] [PubMed] [Google Scholar]
- 40. Shelton L, Troilo D, Lerner MR, Gusev Y, Brackett DJ, Rada JS. Microarray analysis of choroid/RPE gene expression in marmoset eyes undergoing changes in ocular growth and refraction. Mol Vis. 2008; 14: 1465–1479 [PMC free article] [PubMed] [Google Scholar]
- 41. Oganesian A, Poot M, Daum G, et al. Protein tyrosine phosphatase RQ is a phosphatidylinositol phosphatase that can regulate cell survival and proliferation. Proc Natl Acad Sci U S A. 2003; 100: 7563–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Menigatti M, Cattaneo E, Sabates-Bellver J, et al. The protein tyrosine phosphatase receptor type R gene is an early and frequent target of silencing in human colorectal tumorigenesis. Mol Cancer. 2009; 8: 124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shi C, Zhang K, Xu Q. Gender-specific role of the protein tyrosine phosphatase receptor type R gene in major depressive disorder. J Affect Disord. 2012; 136: 591–598 [DOI] [PubMed] [Google Scholar]
- 44. Hendriks WJ, Dilaver G, Noordman YE, Kremer B, Fransen JA. PTPRR protein tyrosine phosphatase isoforms and locomotion of vesicles and mice. Cerebellum. 2009; 8: 80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dilaver G, Schepens J, van den Maagdenberg A, et al. Colocalisation of the protein tyrosine phosphatases PTP-SL and PTPBR7 with beta4-adaptin in neuronal cells. Histochem Cell Biol. 2003; 119: 1–13 [DOI] [PubMed] [Google Scholar]
- 46. Van Den Maagdenberg AM, Bachner D, Schepens JT, et al. The mouse Ptprr gene encodes two protein tyrosine phosphatases, PTP-SL and PTPBR7, that display distinct patterns of expression during neural development. Eur J Neurosci. 1999; 11: 3832–3844 [DOI] [PubMed] [Google Scholar]
- 47. Wei Z, Zheng S, Spangler SA, Yu C, Hoogenraad CC, Zhang M. Liprin-mediated large signaling complex organization revealed by the liprin-alpha/CASK and liprin-alpha/liprin-beta complex structures. Mol Cell. 2011; 43: 586–598 [DOI] [PubMed] [Google Scholar]
- 48. Spangler SA, Jaarsma D, De Graaff E, Wulf PS, Akhmanova A, Hoogenraad CC. Differential expression of liprin-alpha family proteins in the brain suggests functional diversification. J Comp Neurol. 2011; 519: 3040–3060 [DOI] [PubMed] [Google Scholar]
- 49. Fujinami K, Uemura H, Ishiguro H, Kubota Y. Liprin-alpha2 gene, protein tyrosine phosphatase LAR interacting protein related gene, is downregulated by androgens in the human prostate cancer cell line LNCaP. Int J Mol Med. 2002; 10: 173–176 [PubMed] [Google Scholar]
- 50. Bonapace IM, Latella L, Papait R, et al. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J Cell Biol. 2002; 157: 909–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.