Abstract
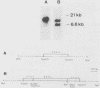
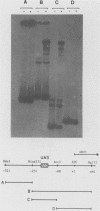
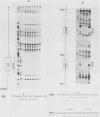
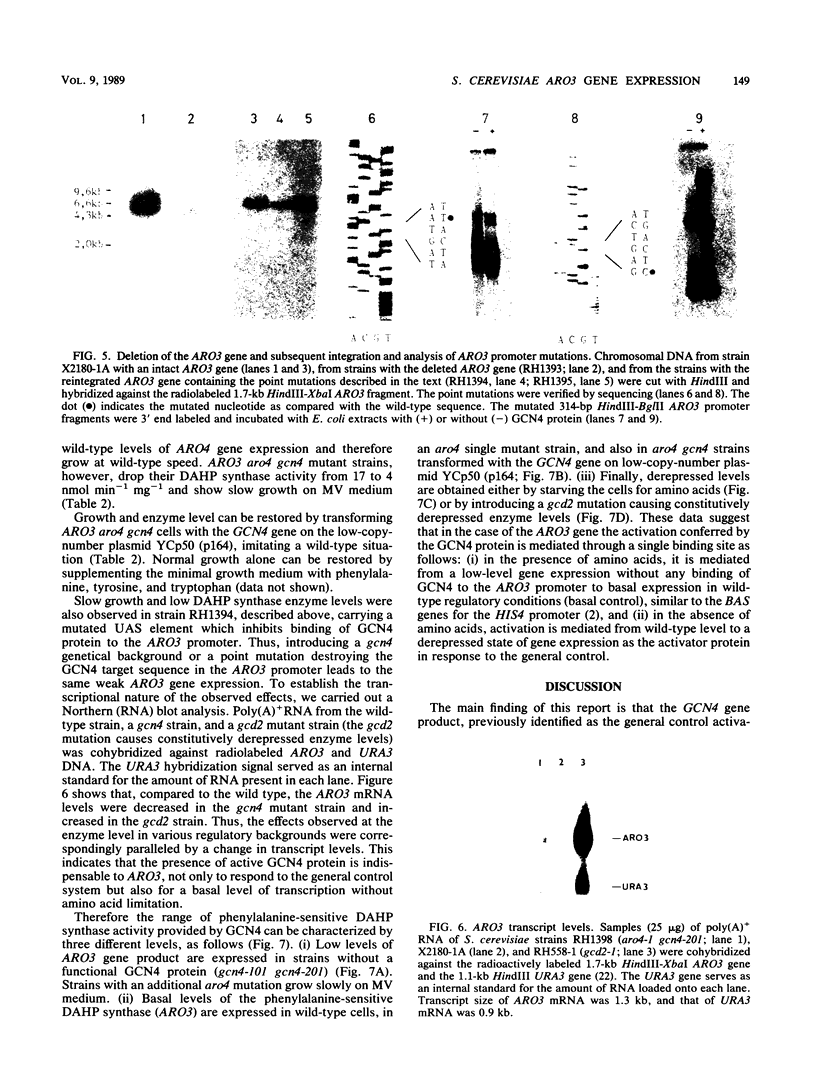
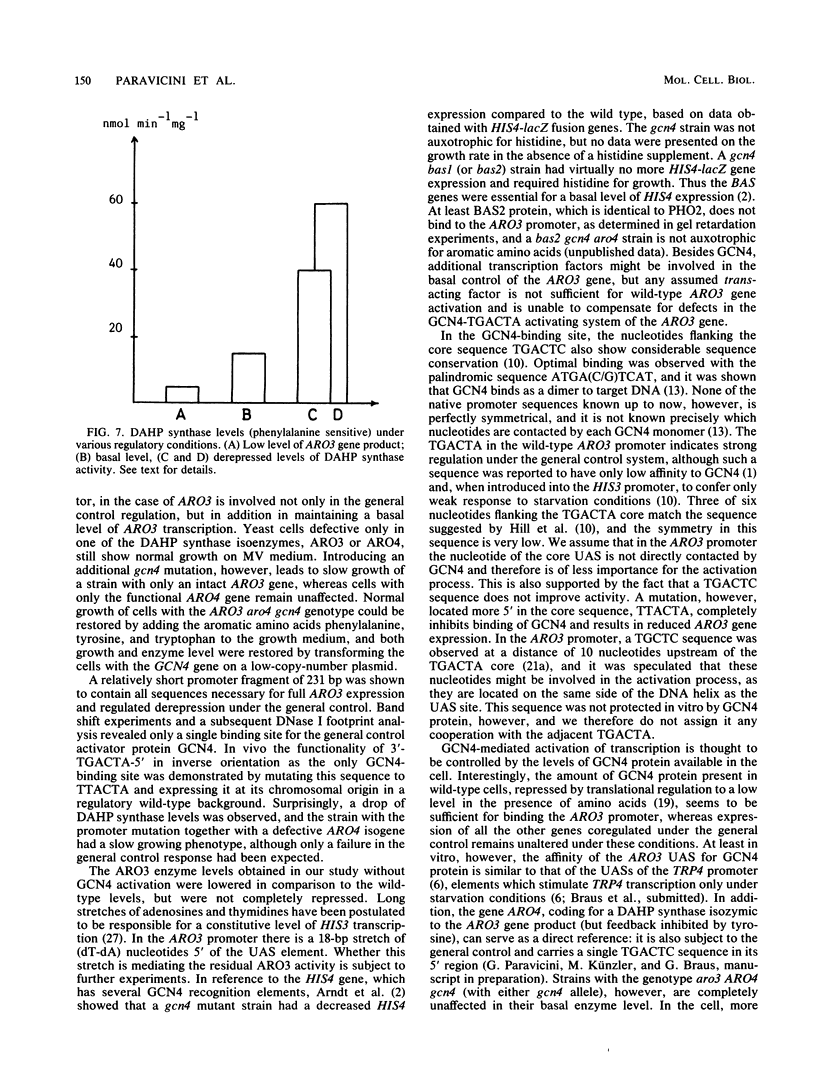
The ARO3 gene encodes one of two 3-deoxy-D-arabino-heptulosonate-7-phosphate isoenzymes in Saccharomyces cerevisiae catalyzing the first step in the biosynthesis of aromatic amino acids. The ARO3-encoded 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (EC 4.1.2.15) is feedback inhibited by phenylalanine; its isoenzyme, the ARO4 gene product, is inhibited by tyrosine. Both genes ARO3 and ARO4 are strongly regulated under the general control regulatory system. Cells carrying only one intact isogene are phenotypically indistinguishable from a wild-type strain when grown on minimal medium. The complete functional ARO3 promoter comprises 231 base pairs and contains only one TGACTA binding site for the general control activator protein GCN4. Mutating this element to TTACTA inhibits binding of GCN4 and results in a decreased basal level of ARO3 gene product and slow growth of a strain defective in its isogene ARO4. In addition, ARO3 gene expression cannot be elevated under amino acid starvation conditions. An ARO3 aro4 strain with gcn4 genetic background has the same phenotype of low ARO3 gene expression and slow growth. The amount of GCN4 protein present in repressed wild-type cells therefore seems to contribute to a basal level of ARO3 gene expression. The general control activator GCN4 has thus two functions: (i) to maintain a basal level of ARO3 transcription (basal control) in the presence of amino acids and (ii) to derepress the ARO3 gene to a higher transcription rate under amino acid starvation (general control).
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt K. T., Styles C., Fink G. R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987 Aug 21;237(4817):874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- Arndt K., Fink G. R. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5' TGACTC 3' sequences. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braus G., Furter R., Prantl F., Niederberger P., Hütter R. Arrangement of genes TRP1 and TRP3 of Saccharomyces cerevisiae strains. Arch Microbiol. 1985 Sep;142(4):383–388. doi: 10.1007/BF00491908. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furter R., Paravicini G., Aebi M., Braus G., Prantl F., Niederberger P., Hütter R. The TRP4 gene of Saccharomyces cerevisiae: isolation and structural analysis. Nucleic Acids Res. 1986 Aug 26;14(16):6357–6373. doi: 10.1093/nar/14.16.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hinnebusch A. G. The general control of amino acid biosynthetic genes in the yeast Saccharomyces cerevisiae. CRC Crit Rev Biochem. 1986;21(3):277–317. doi: 10.3109/10409238609113614. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hütter R., Niederberger P., DeMoss J. A. Tryptophan biosynthetic genes in eukaryotic microorganisms. Annu Rev Microbiol. 1986;40:55–77. doi: 10.1146/annurev.mi.40.100186.000415. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miozzari G., Niederberger P., Hütter R. Tryptophan biosynthesis in Saccharomyces cerevisiae: control of the flux through the pathway. J Bacteriol. 1978 Apr;134(1):48–59. doi: 10.1128/jb.134.1.48-59.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Shore D. Transcriptional regulation in the yeast life cycle. Science. 1987 Sep 4;237(4819):1162–1170. doi: 10.1126/science.3306917. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Paravicini G., Braus G., Hütter R. Structure of the ARO3 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1988 Sep;214(1):165–169. doi: 10.1007/BF00340197. [DOI] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch A., Miozzari J., Hütter R. Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J Bacteriol. 1974 Mar;117(3):1131–1140. doi: 10.1128/jb.117.3.1131-1140.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Chan W. W. Separation and properties of isozymes of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthetase from Saccharomyces cerevisiae. Can J Biochem. 1971 Sep;49(9):1015–1025. doi: 10.1139/o71-149. [DOI] [PubMed] [Google Scholar]
- Teshiba S., Furter R., Niederberger P., Braus G., Paravicini G., Hütter R. Cloning of the ARO3 gene of Saccharomyces cerevisiae and its regulation. Mol Gen Genet. 1986 Nov;205(2):353–357. doi: 10.1007/BF00430450. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Comparison of regulatory and structural regions of genes of tryptophan metabolism. Mol Biol Evol. 1984 Feb;1(2):143–161. doi: 10.1093/oxfordjournals.molbev.a040307. [DOI] [PubMed] [Google Scholar]