Abstract
Until recently, actin was thought to act merely as a passive track for its motility partner, myosin, during actomyosin interactions. Yet a recent report having observed dynamical conformational changes in labeled skeletal muscle α-actin suggests that actin has a more active role. Because the labeling technique was still immature, however, conclusions regarding the significance of the different conformations are difficult to make. Here, we describe the preparation of fully active α-actin obtained from a baculovirus expression system. We developed α-actin recombinants, of which subdomains 1 and 2 have specific sites for fluorescent probes. This specific labeling technique offers to significantly expand the information acquired from actin studies.
Keywords: α-actin, recombinant protein, fluorescent imaging, actomyosin, polymerization, in vitro motility assay
Introduction
Actins are highly conserved cytoplasmic proteins found in all eukaryotes and are involved in many critical physiological functions including muscle contraction. There exist many different actin isoforms, several of which have been purified from various animal, plant, and fungal species.1) Overall, three main actin isoforms have been identified: α, β and γ. All three have high amino acid conservation (>90 percent) and take similar structures. In cells, actin normally takes one of two filament types: microfilaments, which are one of the three major components in the cytoskeleton, are made up of β- and γ-actin; and thin filaments, which are an essential component of the contractile apparatus in muscle, are made up of α-actin.
Striated muscle myofibrils (actomyosin filament bundles) represent the most extensively studied contractile network in eukaryotic cells. In vitro studies of myofibrils at low ionic strength found actin takes its monomeric form, while at high ionic strength actin is filamentous. The change in actin state is important as various physiological processes like chemotaxis and cytokinesis require that actin be kept in dynamical equilibrium, constantly polymerizing and depolymerizing.
Among actin, the most well studied is the vertebrate α-skeletal muscle isoform, which makes up twenty percent or more of the total protein in muscle. Because techniques for highly purified actin and myosin from muscle are readily available, many physiological and biochemical studies investigating the actomyosin interaction mechanism have been reported,2) although most of these reports took a greater interest in myosin. This is because actin has often been thought to have a passive role in myosin motility, acting merely as a track for the myosin to transverse.
However, by using different techniques, Oda et al. and we separately reported that actin is not passive. Noting that the structure of an actin monomer unit is different between G-actin (monomeric form) and F-actin (filamentous form), Oda et al. showed in a model that there was a conformational transition state from G-actin to F-actin.3) We showed actin dynamic states conform in the span of seconds.4) These results suggest that actin may play a more active role in the actomyosin complex. Yet the necessary experiments needed to confirm this theory have to date proven impractical. One reason is the difficulty in binding a fluorescent label to specific actin positions in order to detect intra-molecule fluctuations without a loss in actin activity. Recombinant actin has the potential to overcome this problem. However, despite multiple reports describing the expression of recombinant β-actins in Dictyostelium discoideum, Drosophila, Arabidopsis thaliana and Yeast,5)–7) very few techniques have proven useful for the expression and purification of mammalian α-actin recombinants and their mutants, and even then with limited success.8)–10) For example, techniques that used myoblast cells and an in vitro expression system had recombinants that had low purification yields, mostly because high ion concentrations had to be used resulting in recombinant and endogenous actin mixing to form heterogeneous filaments.9) Furthermore, techniques that expressed recombinants from the E. coli system often suffer from inclusion bodies.11)
Here we describe a baculovirus expression system that overcomes these problems by constructing two types of α-actin recombinants connected to a Flag-tag for purification at low ionic strength. Thymosin β4, an actin monomer binding protein, was fused to α-actin via a flexible peptide in order to obstruct actin filament formation. We also made cysteine substitution α-actin mutants for fluorescent labeling at specific actin sites. We are currently testing the viability of these recombinant α-actins in single molecule fluorescence resonance energy transfer (smFRET) studies regarding the actomyosin complex with the expectation that these new recombinant mammalian actins will be applicable to an array of new actin studies.
Materials and methods
Rat α-actin cDNA
α-actin cDNA was obtained from a rat skeletal muscle full-length cDNA library (Seegene Inc., Korea) using PCR amplification. Primers for the PCR amplification were as follows.
Rat α-actin Forward Nco I (5’ GACACCATGGGTGACGAGGACGAGAC 3’) and rat α-actin Reverse (5’ GTGCGCCTAGAAGCATTTGCGGTGC 3’).
Construction of baculovirus expression plasmids
3*Flag-α-actin: the obtained actin coding sequence (see Rat α-actin cDNA, Materials and methods) was inserted into pFastBac1 (Invitrogen, USA) between the Bam HI and Eco RI sites with a 3*Flag-tag sequence at the 5’ terminal of the rat αactin gene. The 3*Flag-tag sequence was created by annealing the two oligonucleotides, 3*Flag-tag Forward (5’ GATCGATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGCGATCCATG 3’) and 3*Flag-tag Reverse (5’ AATTCATGGATCGCTTGTCATCGTCATCCTTGTAATCGATG TCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCCATC 3’) together (Fig. 1A-1).
Fig. 1.
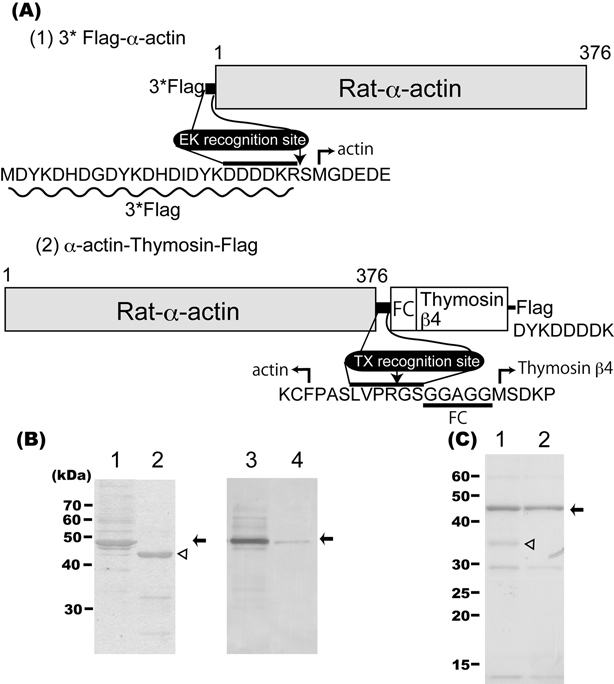
(A) Schematic representation of the two rat α-actin recombinant proteins. FC: glycine rich flexible chain. EK: Enterokinase. TX: Thrombin. Flag: FLAG epitope.31) Details in text. (B) Purified α-actins with CBB staining before and after enterokinase treatment (lanes 1, 2) and immunoblots using a Flag-tag antibody of the same samples (lanes 3, 4) were loaded on SDS-PAGE (10 percent concentration). Arrows show the position of the 3*Flag-α-actin; arrowhead shows the position of the α-actin after the tag was removed. (C) Purification of α-actin before and after treatment with thrombin protease was checked using SDS-PAGE (12.5 percent concentration) and CBB staining (lanes 1, 2). Arrow and arrowhead point to α-actin and biotinylated thrombin, respectively. For both B and C, approximately 3 μg were loaded onto each lane.
α-actin-thymosin-Flag: the obtained α-actin coding sequence and Flag-tag sequence (the gene corresponding to the D-Y-K-D-D-D-D-K peptide) were inserted into pFastBac1 in series. The thrombin recognition-thymosin-β4 fragment, which is a G-actin binding peptide, was inserted between actin and the Flag-tag. The thrombin recognition-thymosin-β4 fragment was created by first annealing separately thrombin-FC Forward (5’ CTAGCCTGGTTCCGCG TGGATCCGGAGGAGCAGGAGGAATGTCTGACAAACCCGATATGGCTGAGATCGAGAAATTCGATAAGT 3’) with thrombin-FC Reverse (5’ GTTTCGACTTATCGAATTTCTCGATCTCAGCCATATCGGGTTTGTCAGACATTCCTCCTGCTCCTCCGGATCCACGCGGAACCAGG 3’) and FC-thymosin β4 Forward (5’ CGAAACTGAAGAAGACAGAGACGCAAGA GAAAAATCCACTGCCTTCCAAAGAAACGATTGAACAGGAGAAGCAAGCAGGCGAATCGTAACTGCA 3’) with FC-thymosin β4 Reverse (5’ GTTACGATTCGCCTGCTTGCTTCTCCTGTTCAATCGTTTCTTTGGAAGGCAGTGGATTTTTCTCTTGCGTCTCTGTCTTCTTCA 3’), and then ligating the two annealed sets together. The thrombin recognition-thymosin-β4 fragment contained a glycine-rich flexible chain (FC: 5 mer, G-G-A-G-G) peptide between the thrombin recognition site and thymosin (Fig. 1A–2).
Mutant constructs
Mutant rat α-actin constructs (Q41C, C374A, Q41CC374A) were created using site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit, Strategene, USA) using appropriately designed oligonucleotides and 3*Flagα- actin parental constructs.
Cell culture
Sf9 insect cells were maintained in a monolayer culture in ventilated 175 cm2 flasks at 28 °C with 10 % FBS (Invitrogen)-GIBCO™ Antibiotic- Antimycotic liquid (Invitrogen)/EX-Cell 420 (SAFC Biosciences, USA). The cells were routinely sub-cultured every 2–3 days to maintain log growth phase. Cells were replaced with frozen stocks approximately every 2 months.
Protein purification
All steps were performed at less than 4 °C. Cells were collected for 60 hrs post-infection by centrifugation at 6,000*g for 5 min and stored at −80 °C. Frozen cells were suspended in 10 ml/g cells of Lysis solution (10 mM HEPES-NaOH pH 7.5, 10 mM NaCl, 1 mM EGTA, 0.1 mM MgCl2, 1 mM ATP, 0.05 mM DTT containing the protease inhibitor mix, Complete, EDTA-free (Roche Diagnostics, Switzerland)) and sonicated immediately afterward. After ultra centrifugation at 10,000*g for 20 min, soluble fractions were run through 200 μl/g cell in an anti-Flag M2 Affinity Gel (Sigma Aldrich, USA), and then washed thoroughly with Wash solution (10 mM HEPES-NaOH pH 7.5, 10 mM NaCl, 0.1 mM EGTA, 0.1 mM MgCl2, 1 mM ATP). We applied two different methods to purify rat actin. In the first method, the 3*Flag-αactin was eluted from the resin in competition with the 3*Flag peptide (Sigma Aldrich). Rat α-actin recombinant and its mutants were separated from the 3*Flag-tag sequence by treatment with Enterokinase (Novagen, Germany) treatment for 30 hrs at 20 °C. In the second method, rat α-actin recombinant was also obtained from α-actin-thymosin-Flag, which was bound to an anti-Flag M2 affinity Gel via biotinylated human thrombin (Novagen, Germany) treatment for 2 hrs at room temperature without Flag peptide competition. The biotinylated thrombin was then removed from actin by using Streptavidin Agarose (Novagen).
Recombinant protein analysis
SDS-PAGE: Purified α-actin recombinant and its mutants were analyzed by SDS-polyacrylamide gel electorophoresis. The gels were stained by the CBB staining solution Quick-CBB (Wako, Japan). Immunoblot analysis was done by using an anti-Flag polyclonal antibody (Sigma Aldrich), VECTASTEIN Elite ABC Kit and DAB Substrate Kit (Vector lab. Inc., USA). In addition, purified α-actin recombinant and its mutants were labeled by Alexa Fluor 555 C2 maleimide (Invitrogen). The relative band intensities of CBB and Alexa Fluor 555 were determined with Molecular Imager FX (Bio-Rad, USA).
In vitro motility assay: Purified α-actin recombinant, which was separated from α-actin-Thymosin-Flag using thrombin protease, polymerized at high potassium concentrations (>100 mM). Actin filaments were labeled with a phalloidin-tetramethylrhodamine B isothiocyanate conjugate (Fluka, USA). Rabbit skeletal muscle actin was used as a control. In addition, actin filaments were made by copolymerizing Alexa Fluor 555-labeled α-actin recombinant with unlabeled rabbit skeletal muscle actin at a 1 to 50 ratio at high ionic strength. Motility assays were performed in solution containing 20 mM HEPES-KOH (pH 7.8), 25 mM KCl, 5 mM MgCl2, 1 mM ATP and an oxygen scavenging system to reduce photo-bleaching at 25 °C using rabbit skeletal muscle Heavy Mero-Myosin (HMM) with some modifications.12) Skeletal muscle HMM and actin were obtained from rabbit skeletal muscle and purified according to Margossian et al.13) and Spudich & Watt,14) respectively.
Results and discussion
Production of α-actin recombinants
Conventional actins play an important role in cell function in all eukaryotes, often through polymerization-depolymerization dynamics. In in vitro experiments, polymerization-depolymerization equilibrium can be controlled by manipulating the concentration of cations like Ca2+ and Mg2+.
In general, recombinants are a popular tool for the study of proteins, including actin. All actin have a highly conserved amino acid sequence and form filaments (filamentation) at high ionic concentrations. Filamentation complicates actin studies since this property makes the purification of tag-fused actin recombinant from a host cell using specific affinity chromatography difficult. Here we discuss the construction of two types of α-actin recombinants. For both our recombinants, the rat α-actin gene was obtained from a rat skeletal muscle cDNA library using PCR amplification methods. The sequence of the obtained α-actin gene was in accordance with information from Gene Bank (accession no. BC061974.1). Initially, we tried to express His-tag fused α-actin recombinant using the method from Anthony et al.,9) but could not elute it from Ni2+-affinity columns at high imidazole ion levels. To resolve this, we constructed Flag-tag fused α-actin recombinant which could be purified at low ionic strengths. Fig. 1A-1 and 2 show two α-actin recombinant constructs. The Flag-tag is an artificial and sensitive epitope that can be used for affinity chromatography. 3*Flag-tag is a more sensitive version and contains an enterokinase cleavage site between the 3*Flag and α-actin (Fig. 1A). After enterokinase treatment, an Arg-Ser peptide was fused to α-actin recombinant at the N-terminus. We also included methionine and glycine residues, both of which exist at the N-terminus in skeletal muscle actin but are removed during translation to form the mature terminus.15) Consequently, our obtained α-actin recombinant has 4 additional amino acid residues (Arg, Ser, Met, Gly) at the N-terminus relative to mature skeletal muscle actin. In the other α-actin construct, thymosin β4, which is a monomeric actin specific binding protein,16) was fused to α-actin at the C terminus in order to disturb polymerization in SF9 cells. We also inserted the specific protease thrombin recognition site and a flexible Gly-rich peptide, which supports the binding of α-actin to thymosin, between α-actin and thymosin β4 (Fig. 1A–2). This α-actin construct is monomeric until the cell extract is attached to the anti-Flag M2 Affinity Gel.
From both purifications, we obtained about 0.3 mg of soluble peptide α-actin recombinant from 1 g of wet SF9 cells using a flask culture. However, the suspension culture accelerated the formation of insoluble inclusion bodies. Therefore, we substituted the Cys from the additional 4 residues at the N-terminus with Gly. However, the actin N-terminus acidic cluster and especially the exchanged Cys residue are thought to be important for actin myosin interactions. 17)–19) In fact, the Cys residue is well conserved in most α-actin types. There are exceptions, though. It is absent in some β-actin and replaced with Gly in non-muscle and amoeba.20),21) Furthermore, it is deleted post modification in mature muscle actin. Figure 1B shows SDS-PAGE and immunoblot results of purified recombinants. Using CBB staining, 3*Flagα-actin was detected as the main band around 45 kDa (Fig. 1B, lane1 arrow). Following enterokinase treatment, the main band migrated to 43 kDa, which is equivalent to the molecular weight of monomeric α-actin recombinant (Fig. 1B, lane2 arrowhead). The 45 kDa band was detected by immunoblot analysis using specific anti-Flag polyclonal antibody (Fig. 1B, lane3) but became much weaker after enterokinase treatment, suggesting the Flag-tag was removed (Fig. 1B, lane4). Skeletal muscle α-actin exists as a polymer at physiological conditions in cells and forms filaments at high ionic strength in vitro. The polymerization of 3*Flag-α-actin and α-actin, respectively, was examined at 0.1 M KCl. Actin polymers were easily seen using fluorescence microscopy following labeling with a phalloidin-tetramethyl rhodamine B isothiocyanate conjugate. The α-actin recombinant then formed filaments of similar length to rabbit skeletal muscle actin (data not shown). On the other hand, obvious filaments were not detected from enterokinase untreated 3*Flag-α-actin (data not shown). The actin N terminus, where the 3*Flag-tag is placed, is important for both the formation of the actomyosin complex19) and polymerization.22) Enterokinase treatment separates the 3*Flag-tag from the α-actin recombinant. Despite having two additional amino acids, Arg and Ser, α-actin recombinant polymerization was unperturbed.
The second α-actin recombinant construct we used was an α-actin-thymosin fused protein. This recombinant is easy to separate from endogenous actins and can be purified in a short time. Thymosin fused-α-actin cannot form filaments. Figure 1C shows an SDS-PAGE following α-actin purification from the thymosin fused recombinant. After biotinylated thrombin digestion (Fig. 1C, lane1) and Streptavidin Agarose treatment, α-actin appeared as the main band (Fig. 1C, lane2). The purified α-actin recombinant, which had no additional amino acid residues at the N-terminus but 7 additional amino acid residues (P-A-S-L-V-P-R) at the C-terminus, had sufficient polymerization activity (data not shown). If the digestive treatment is applied for only a short period, then thrombin can be used whereas enterokinase cannot. Furthermore, unlike enterokinase, biotinylated thrombin treatment can be applied in the presence of ATP for a short time without compromising polymerization.23) Biotinylated thrombin can be separated from α-actin recombinant easily using Streptavidin Agarose.
In vitro motility assay
The in vitro motility assay24) is a common assay applied to examine the interaction between purified myosin and recombinant actin. In this assay, myosin molecules bound to the coverslip bind tightly to fluorescently labeledactin filaments in the absence of ATP. When ATP is added to the system, myosin begins to move along the actin filaments. Figure 2A shows the sliding velocities of α-actin filaments from α-actin-Thymosin-Flag propelled by rabbit skeletal muscle myosin HMM bound to the surface of a nitrocellulose-coated coverslip. The velocity was 6.25±1.82 μm/s (arithmetic mean±SD, n = 684) at 25 °C. This result is consistent with the velocity of wild type skeletal muscle actin filament (Fig. 2B, 6.31±1.86, μm/s (arithmetic mean±SD, n = 667)) indicating that our recombinant α-actin interacts similarly to wild type with rabbit skeletal muscle myosin. Similar results were found using α-actin recombinant treated with enterokinase (data not shown). These data show that substituting Cys for Gly does not affect velocity.
Fig. 2.
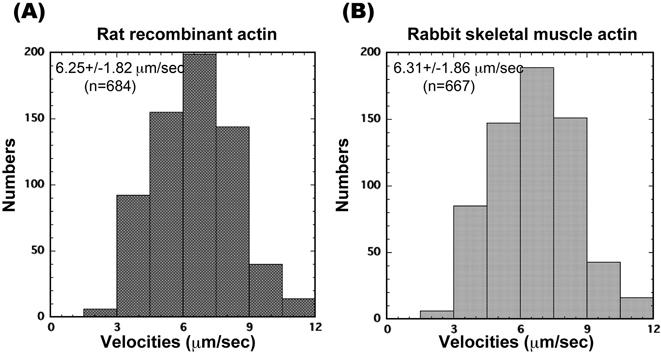
(A, B) Histograms of actin filament sliding velocities ((A) rat recombinant α-actin; (B) rabbit skeletal muscle actin) propelled by rabbit HMM directly bound to a nitrocellulose-coated surface. Each histogram shows sliding velocities for >100 individual actin filaments. Assay medium: 20 mM HEPES-KOH (pH 7.8), 25 mM KCl, 5 mM MgCl2, 1 mM ATP and an oxygen scavenger system. Temperature 25 °C.
α-actin Cys mutants and specific labeling
In general, actin filaments have often been thought to be nothing more than passive rails for myosin to move and generate force in many necessary cell activities like muscle contraction. However, a recent report found that actin filaments make dynamical changes between conformational states.4) To pursue this matter further, we did smFRET experiments using actin mutants with fluorescent labeling at specific positions (Morimatsu M. et al. unpublished data). Figure 3A shows the crystal structure of rabbit αactin, which has four subdomains. The red and green indicate subdomains 1 and 2, respectively. We constructed actin Cys-mutants using 3*Flag-α-actin recombinant as a parental construct for fluorescent labeling. Gln41 (yellow ball) and Cys374 (orange ball where the thiol group of Cys374 is shown) were chosen as the fluorescent labeling positions since the region around Gln41 is considered to be important for actomyosin recognition25) and because of Cys 374’s known reactivity.26),27)Figure 3B shows the main band (approximately 43 kDa) of the SDS-PAGE fluorescent image using Alexa Fluor 555 (upper) and CBB staining (lower). WT, Q41C and Q41CC374A were detected by Alexa Fluor 555 fluorescence, but C374A was not. There are five cysteine residues in skeletal muscle actin. According to G-actin crystal structure analysis, the thiol groups from Cys10, Cys217 and Cys257 are always directed into the actin molecule whereas that from Cys 374 points outward and that from Cys285 is unknown. Because Q41C but not C374A was successfully labeled by Alexa Fluor 555 C2 maleimide, we concluded that Q41C was labeled with fluorescent dyes at positions 41 and 374, while WT was labeled at position 374. It should be noted, the bands of all actins except C374A mutant showed a mobility shift by Alexa Fluor 555 labeling on SDS-PAGE (data not shown).
Fig. 3.
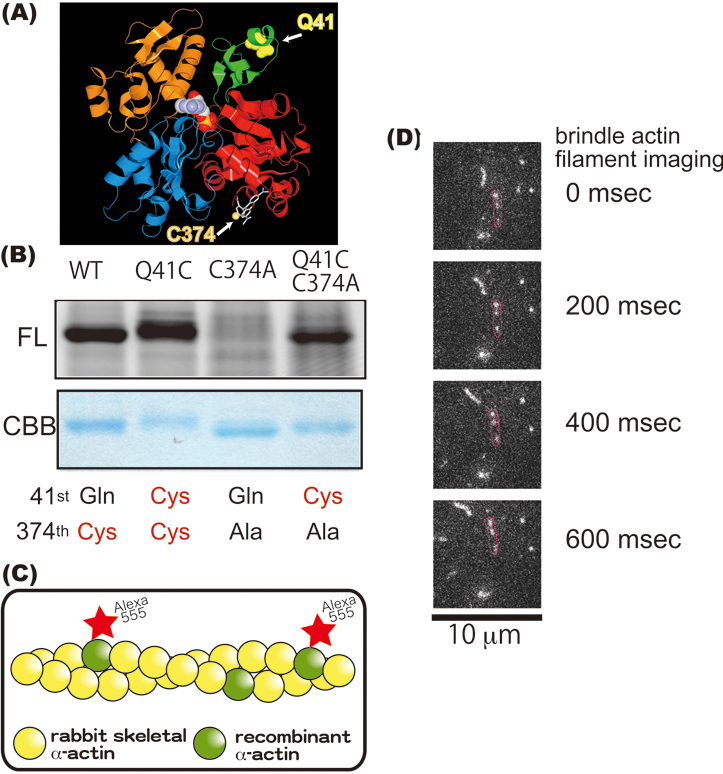
(A) Ribbon representation of the crystal structure of monomeric rabbit α-actin in the ADP state (PDB ID: 1J6z). Four α-actin subdomains are shown (red, subdomain1; green, subdomain2; blue, subdomain3; orange, subdomain4). Residue 41 is seen as a yellow ball while residue 374 is seen as an orange ball via rhodamine. In F-actin, subdomains 1 and 3 point toward the barbed end of the actin filament while subdomains 2 and 4 point toward the pointed end. (B) Analysis of the main band from the SDS-PAGE in Fig. 1. Approximately 2 μg of protein were loaded onto each lane. Upper half gels are fluorescent images of Alexa Fluor 555 C2 maleimide α-actin recombinants; lower half are images stained with CBB. WT is actually our α-actin recombinant. Q41C, C374A and Q41CC374A are α-actin mutants. (C) Schematic drawing of a brindle actin filament. Alexa Fluor 555-labeled recombinant α-actin and rabbit skeletal muscle actin formed a co-filament. (D) Sliding movement of actin co-filaments every 200 miliseconds. The red circles enclose a brindle actin filament.
Finally, we tried to co-polymerize and observe Alexa Flour 555 labeled Q41C mutants with unlabeled rabbit skeletal muscle actin at a ratio of 1 to 50 using fluorescence microscopy (Fig. 3D). Brindle actin filaments (Fig. 3C) were seen to successfully interact with rabbit myosin HMM in in vitro motility assays (Fig. 3D). We confirmed that the obtained αactin recombinant could form not only filaments by itself but also co-filaments with rabbit skeletal muscle actin.
During filamentation, the transition from ATP to ADP bound actin is accompanied by a conformational change in actin subdomain 2, or more specifically in the D loop (DNase I binding loop) located at the top of subdomain 2.25) Within the D loop exist several structures including an α-helical structure. Moreover, large conformational changes around Q41 have also been observed in molecular dynamics simulations. 28) Using the specific actin mutants described here and smFRET techniques, the dynamic conformational fluctuations of actin filaments can potentially be analyzed (detailed analysis to be published) and thus elucidate more information regarding the active role of actin filaments in muscle contraction.29),30)
Acknowledgments
We thank T. Komori, S. Nishikawa and Y. Togashi for valuable discussion; P. Karagiannis for reading the manuscript; T. M-Ichinose and R. Ogawa for generating the actin constructs. This work was supported by Grant-in-Aid for Scientific Research(C) and Technology program of the Japan Science and Technology Agency and the Yuragi Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan. MM is supported by a young research fellowship of JSPS.
References
- 1).Mounier N. and Sparrow J.C. (1997) Structural comparisons of muscle and nonmuscle actins give insights into the evolution of their functional difference. J. Mol. Evo. 44, 89–97 [DOI] [PubMed] [Google Scholar]
- 2).Sellers J.R. (1999) Myosins Second Edition. Oxford University Press, Oxford [Google Scholar]
- 3).Oda T., Iwasa M., Aihara T., Maeda Y. and Narita A. (2009) The nature of the globular- to fibrous-actin transition. Nature 457, 441–445 [DOI] [PubMed] [Google Scholar]
- 4).Kozuka J., Yokota H., Arai Y., Ishii Y. and Yanagida T. (2006) Dynamic polymorphism of single actin molecules in the actin filament. Nat. Chem. Biol. 2, 83–86 [DOI] [PubMed] [Google Scholar]
- 5).Sutoh K., Ando M. and Toyoshima Y.Y. (1991) Site-directed mutations of Dictyostelium actin: disruption of a negative charge cluster at the N terminus. Proc. Natl. Acad. Sci. USA 88, 7711–7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Noguchi T.Q., Kanzaki N., Ueno H. and Hirose K., Uyeda T.Q. (2007) A novel system for expressing toxic actin mutants in Dictyostelium and purification and characterization of a dominant lethal yeast actin mutant. J. Biol. Chem. 282, 27721–27727 [DOI] [PubMed] [Google Scholar]
- 7).Aspenström P. and Karlsson R. (1991) Interference with myosin subfragment-1 binding by site-directed mutagenesis of actin. Eur. J. Biochem. 200, 35–41 [DOI] [PubMed] [Google Scholar]
- 8).Joel P.B., Fagnant P.M. and Trybus K.M. (2004) Expression of a nonpolymerizable actin mutant in Sf9 cells. Biochemistry 43, 11554–11559 [DOI] [PubMed] [Google Scholar]
- 9).Anthony Akkari P., Nowak K.J., Beckman K., Walker K.R., Schachat F. and Laing N.G. (2003) Production of human skeletal α-actin proteins by the baculovirus expression system. Biochem. Biophys. Res. Commun. 307, 74–79 [DOI] [PubMed] [Google Scholar]
- 10).Miller B.M. and Trybus K.M. (2008) Functional effects of nemaline myopathy mutations on human skeletal alpha-actin. J. Biol. Chem. 283, 19379–19388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Hitchcock-DeGregori S.E. (1989) Structure-function analysis of thin filament proteins expressed in Escherichia coli. Cell Motil. Cytoskeleton 14, 12–20 [DOI] [PubMed] [Google Scholar]
- 12).Harada Y., Noguchi A., Kishino A. and Yanagida T. (1987) Sliding movement of single actin filaments on one-headed myosin filaments. Nature 326, 805–808 [DOI] [PubMed] [Google Scholar]
- 13).Margossian S.S. and Lowey S. (1982) Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 85Pt B, 55–71 [DOI] [PubMed] [Google Scholar]
- 14).Spudich J.A. and Watt S. (1971) The regulation of rabbit skeletal muscle concentration. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 15).Vandekerckhove J. and Weber K. (1978) At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J. Mol. Biol. 126, 783–802 [DOI] [PubMed] [Google Scholar]
- 16).Huff T., Zerzawy D. and Hannappel E. (1995) Interactions of β-thymosins, thymosin β4-sulfoxide, and N-terminally truncated thymosin β4 with actin studied by equilibrium centrifugation, chemical cross-linking and viscometry. Eur. J. Biochem. 230, 650–657 [DOI] [PubMed] [Google Scholar]
- 17).Aspenstrom P., Lindberg U. and Karlsson R. (1992) Site-specific amino-terminal mutants of yeast-expressed beta-actin. Characterization of the interaction with myosin and tropomyosin. FEBS Lett. 303, 59–63 [DOI] [PubMed] [Google Scholar]
- 18).Cook R.K., Blake W.T. and Rubenstein P.A. (1992) Removal of the amino-terminal acidic residues of yeast actin. Studies in vitro and in vivo. J. Biol. Chem. 267, 9430–9436 [PubMed] [Google Scholar]
- 19).Cook R.K., Root D., Miller C., Reisler E. and Rubenstein P.A. (1993) Enhanced stimulation of myosin subfragment 1 ATPase activity by addition of negatively charged residues to the yeast actin NH2 terminus. J. Biol. Chem. 268, 2410–2415 [PubMed] [Google Scholar]
- 20).Bergsma D.J., Chang K.S. and Schwartz R.J. (1985) Novel chicken actin gene: third cytoplasmic isoform. Mol. Cell. Biol. 5, 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Redman K.L., Martin D.J., Korn E.D. and Rubenstein P.A. (1985) Lack of NH2-terminal processing of actin from Acanthamoeba castellanii. J. Biol. Chem. 260, 14857–14861 [PubMed] [Google Scholar]
- 22).Bertrand R., Chaussepied P. and Audemard E., Kassab R. (1989) Functional characterization of skeletal F-actin labeled on the NH2-terminal segment of residues 1–28. Eur. J. Biochem. 181, 747–754 [DOI] [PubMed] [Google Scholar]
- 23).Pardee J.D. and Spudich J.A. (1982) Purification of muscle actin. Methods Enzymol. 85Pt B, 164–181 [DOI] [PubMed] [Google Scholar]
- 24).Kron S. J. and Spudich J. A. (1986) Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. USA 83, 6272–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Graceffa P. and Dominguez R. (2003) Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J. Biol. Chem. 278, 34172–34180 [DOI] [PubMed] [Google Scholar]
- 26).Takashi R. (1979) Fluorescence energy transfer between subfragment-1 and actin points in the rigor complex of actosubfragment-1. Biochemistry 18, 5164–5169 [DOI] [PubMed] [Google Scholar]
- 27).Otterbein L.R., Graceffa P. and Dominguez R. (2001) The crystal structure of uncomplexed actin in the ADP state. Science 293, 708–711 [DOI] [PubMed] [Google Scholar]
- 28).Pfaendtner J., Branduardi D., Parrinello M., Pollard T.D. and Voth G.A. (2009) Nucleotide-dependent conformational states of actin. Proc. Natl. Acad. Sci. USA 106, 12723–12728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Nishikawa M., Takagi H., Iwane A.H. and Yanagida T. (2008) Fluctuation analysis of mechanochemical coupling depending on the type of biomolecular motors. Phy. Rev. Lett. 101, 128103. [DOI] [PubMed] [Google Scholar]
- 30).Kitamura K., Tokunaga M., Esaki S., Iwane A. H. and Yanagida T. (2005) Mechanism of muscle contraction based on stochastic properties of single actoymosin motors obserbed in vitro. Biophysics 1, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Brizzard B.L., Chubet R.G. and Vizard D.L. (1994) Immunoaffinity purification of FLAG epitopetagged bacterial alkaline phosphatase using a novel monoclonal antibody and peptide elution. Biotechniques 16, 730–735 [PubMed] [Google Scholar]


