Abstract
In this study, we investigate a new approach for examining the separation of the brain into resting-state networks (RSNs) on a group level using resting-state parameters (amplitude of low-frequency fluctuation [ALFF], fractional ALFF [fALFF], the Hurst exponent, and signal standard deviation). Spatial independent component analysis is used to reveal covariance patterns of the relevant resting-state parameters (not the time series) across subjects that are shown to be related to known, standard RSNs. As part of the analysis, nonresting state parameters are also investigated, such as mean of the blood oxygen level-dependent time series and gray matter volume from anatomical scans. We hypothesize that meaningful RSNs will primarily be elucidated by analysis of the resting-state functional connectivity (RSFC) parameters and not by non-RSFC parameters. First, this shows the presence of a common influence underlying individual RSFC networks revealed through low-frequency fluctation (LFF) parameter properties. Second, this suggests that the LFFs and RSFC networks have neurophysiological origins. Several of the components determined from resting-state parameters in this manner correlate strongly with known resting-state functional maps, and we term these “functional covariance networks”.
Key words: amplitude of low-frequency fluctuation (ALFF), fractional ALFF (fALFF), functional MRI (fMRI), Hurst exponent, resting state, spontaneous neuronal activity
Introduction
Resting-state functional connectivity (RSFC) has become an increasingly popular technique of functional magnetic resonance imaging (fMRI). As first proposed by Biswal et al. (1995), in the absence of external stimulus, low-frequency fluctuations (LFFs) in the blood oxygenation level-dependent (BOLD) MR signal can be used to reveal brain networks (Greicius et al., 2003; Lowe et al., 1998; Xiong et al., 1999). Several resting-state networks (RSNs) have since been identified, such as the sensorimotor, executive control, visual, and default mode networks (DMN) (Beckmann et al., 2005; Damoiseaux et al., 2006; Raichle et al., 2001; Seeley et al., 2007). Methods for determining these networks include seed-based correlations and independent component analysis (ICA) of voxel time series (Calhoun et al., 2002; Kiviniemi et al., 2003; McKeown et al., 1998).
Though RSFC has been widely used, to date no complete theoretical or physiological explanations for the importance of these specific low frequencies (which correspond to lengthy timescales of 10–100 sec) have been given. Moreover, in some cases, it has been suggested that signal sources are significantly cardiological, respiratory, or vascular in nature (Birn et al., 2006; Chang et al., 2009; Shmueli et al., 2007; van Buuren et al., 2009; Wise et al., 2004). However, important studies have shown that RSFC cannot be explained solely by these sources: (1) filtering for cardiac and respiratory components leaves a strong LFF signal, from which RSNs are detected (Biswal et al., 1995; De Luca et al., 2006); (2) in disease studies, clinical subjects and healthy controls with similar cardiac and respiratory patterns have shown significant RSFC differences in specific ROIs (Jones et al., 2010).
Several RSFC parameters have been developed, such as the amplitude of LFFs (ALFF) (Zang et al., 2007), fractional ALFF (fALFF) (Zou et al., 2008), and the Hurst exponent (Bullmore et al., 2004; Maxim et al., 2005). These have been shown to be useful in representing resting-state MRI properties on a voxel- and region-wise basis in several studies of healthy and diseased populations. The test–retest reliabilities of ALFF and fALFF have also been shown to be moderate to high, particularly in gray matter (GM) (Zuo et al., 2010). While the significantly higher performance of each parameter in gray matter compared to white matter (WM) is suggestive of their important neuronal content, however, the exact information of these parameters (such as in regards to neuronal firing) remains undetermined. Consequently, the relative efficacy of each parameter in elucidating RSFC information is also uncertain, and, for example, Zuo et al. (2010) found that the test–retest reliability of ALFF was slightly higher than for fALFF, whereas the latter was found to have higher specificity and determined to be more effective at minimizing cardiac and respiratory contributions. Additionally, the BOLD signal standard deviation, σ, has also been used in studying the resting state, along with the closely related standard deviation of BOLD signal filtered to LFFs only, σLFF (see, e.g., Kannurpatti and Biswal, 2008). The relations of these parameters are given, for reference, in the Appendix.
Another means of utilizing these RSFC parameters would be on a global basis to investigate the presence of underlying, extended spatial structures across populations (as opposed to their local properties). Using the intersubject variation of properties, for example, Mechelli et al. (2005) found network patterns in how regional gray matter volume (GMV) covaried across subjects and suggested that structural variations were linked to underlying differences in physiological factors and neural circuitry. Such structural connectivity networks have been found to be useful in investigating development (Zielinski et al., 2010) and aging (Bergfield et al., 2010). The appearance of networks due to intersubject covariance was attributed to the presence of some common, underlying influence within the brain of each subject, rather than to a null case of the given structural property occurring independently per region within the brain.
An analogous approach for investigating functional networks would be to investigate the intersubject covariance of local RSFC parameters. This would provide information about underlying RSFC (e.g., spatial patterns, intersubject variability, and robustness) in a manner separate from and complementary to the usual time-series driven approaches. It is known that, in the resting-state BOLD signal, the amplitudes of the LFFs, as well as the shape of spectra in general, vary among subjects. We hypothesize that the spatial patterns of covariance of RSFC patterns in known networks will show that these amplitudes and the spectral shapes are due to influences of the networks as a whole, instead of due to local and independent features. This would suggest a common influence underlying the given property within a network.
Here, we use the intersubject variability of ALFF, fALFF, H, and σ to determine the presence of nonlocal patterns in RSFC parameters across the brain. Similar analysis is also performed with non-RSFC parameters, the mean of the BOLD signal and GMV (from MPRAGE) parameters, to compare results using quantities which are variously weighted for metabolic, hemodynamic, and anatomical content. Patterns of spatial covariance of the relevant RSFC parameters (not the time series) across subjects are here referred to as functional covariance networks (FCNs) and are shown to be related to known RSNs. Spatial ICA and seed-based correlation are used to analyze data sets formed by concatenating individual parameter maps to determine spatial maps based on intersubject variance. Two independent data sets are analyzed separately, testing the robustness of the method and of the results.
We hypothesize that meaningful RSNs will be elucidated by analysis of the covariance of RSFC parameters and not by non-RSFC parameters. We note that such patterns would not be observed if RSFC parameters (particularly the amplitudes of the low frequencies, ALFF) were determined without some form of global influence (per individual). We show that several of the components determined in this manner correlate strongly with known RSFC maps, calculated from over 1000 data sets as part of the 1000 Functional Connectomes Project (FCP) (Biswal et al., 2010). By examining several parameters in similar fashion, we are also able to compare ways in which each represents the RSFC on a group level, potentially influencing the parameter choice in future studies.
Materials and Methods
Data acquisition
In this study, two subsets of the publicly available FCP database were analyzed. Data from a total of 391 subjects having both a resting state fMRI (rs-fMRI) and a T1-weighted MPRAGE scan were downloaded from the www.NITRC.org website. The first group contained 198 subjects collected at Beijing Normal University (BNU) (76 M/122F; age 18–26; time of repetition (TR)=2 sec; volumes=235); functional resolution was 3.125×3.125×3 mm with 64×64×36 voxels, and anatomical (T1) resolution was 1.330×1.0×1.0 mm with 128×177×186 voxels. The second group contained 193 subjects collected at the Harvard University (HU) (75M/123F, age 18–26; TR=3 sec; volumes=119); functional resolution was 3×3×3 mm with 72×72×47 voxels, and anatomical resolution was 1.2×1.198×1.198 mm with 144×192×192 voxels. Additionally, results from analysis of the entire FCP database were utilized for comparison, in particular, a set of 20 resting-state ICA-derived spatial maps shown in Figure S3 of Biswal et al. (2010).
Preprocessing
All subject data were preprocessed using a standard scheme implemented with AFNI (Cox, 1996) and FSL software tools (Jenkinson and Smith, 2001; Smith et al., 2004), following the procedure described by Biswal et al. (2010). Data were motion corrected with respect to the mean image; spatial smoothing was performed with a 6-mm full-width at half-maximum (FWHM) Gaussian blur, and temporal filtering (when utilized) was done in the LFF range of 0.01–0.1 Hz. Linear trends up to second order polynomials were removed from the data. All the preprocessed data sets were also transformed in the 3-mm Montreal Neurological Institute (MNI) standard brain (Mazziotta et al., 2001). Five subjects were removed from the BNU group due to significant head motion, leaving a total of 193 subjects in that data set. Statistics of maximum subject motion parameters (mean, standard deviation, and maximum for linear translation and rotation) for each group are given in Table 1, with motion in all cases being approximately ≤1 mm.
Table 1.
Summary Statistics for Maximal Motion of Each Subject in Both Data Sets
| Roll | Pitch | Yaw | dS | dL | dP | |
|---|---|---|---|---|---|---|
| BNU max | 0.59 | 1.23 | 0.94 | 1.07 | 0.73 | 0.74 |
| BNU mean | 0.07 | 0.18 | 0.08 | 0.13 | 0.07 | 0.08 |
| BNU std | 0.08 | 0.20 | 0.12 | 0.15 | 0.09 | 0.13 |
| HU max | 0.63 | 1.16 | 0.56 | 0.85 | 0.49 | 0.51 |
| HU mean | 0.09 | 0.25 | 0.11 | 0.20 | 0.09 | 0.16 |
| HU std | 0.07 | 0.18 | 0.08 | 0.13 | 0.04 | 0.08 |
Rotations (roll, pitch, and yaw) are given in degrees, and translations (dS, dL, and dP, in the superior–inferior, left–right and posterior–anterior directions, respectively), in mm.
BNU, Beijing Normal University; HU, Harvard University.
For each subject, ALFF, fALFF, Hurst exponent [calculated using a publicly available, wavelet-based method of Abry and Veitch (1999)], and σ were estimated on a voxel-wise basis for the whole brain. Additionally, the mean (μ) of unfiltered resting-state BOLD signal was also calculated on a voxel-wise basis. (Analysis with σLFF, as calculated in the Appendix, was also performed, but, as expected, this yielded nearly identical results to those of σ, so that only those of the latter are presented here.) Finally, as a control representing anatomical structure, GMV values (from the MPRAGE/T1 weighted images) were computed as well. Similar to the approach implemented by Mechelli et al. (2005), segmentation of the anatomical image was performed using a unified segmentation algorithm (Ashburner and Friston, 2000, 2005) implemented in Statistical Parametric Mapping Software (SPM8) with Matlab (MathWorks, Natick, MA). For each of the subjects in the HU and BNU groups, the anatomical MPRAGE image was segmented in to gray matter and transformed into MNI 3-mm standard space. An additional modulation step was also performed on the segmented images as part of the voxel-based morphometry (VBM) processing scheme, to preserve the total amount of gray matter in each voxel before and after the transformation into MNI standard space (Good et al., 2001). The group-wise analysis of the six variables of interest is described below (the parameter formulations and relations are given in the Appendix).
Calculations of connectivity maps
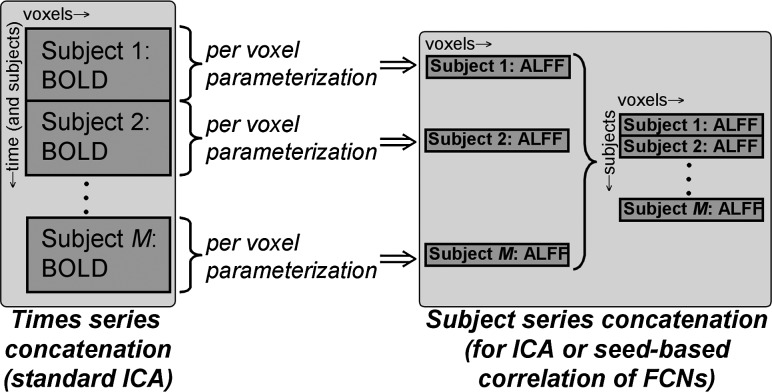
For each data set having a total of M subjects, a parameter of interest (for example, ALFF) was calculated for each of the voxels in the brain (see schematic diagram in Fig. 1). This process was repeated for each of the M subjects in the group, and the standard space parameter maps were concatenated to form a 4D data set of M discrete points; that is, the end result resembled a typical whole-brain set of time series, but in this case each time point was an individual subject and each voxel value was a parameter value (in this example, ALFF). To avoid confusion with the MR-acquired time series, we refer to this set as a “subject series.” All analyses conducted were independent of the arbitrary order of subjects, and randomly selected subsets of each group produced similar results. For both BNU and HU data sets, this process was repeated to create a subject series for the following types of parameters: resting state functional (ALFF, fALFF, H, and σ), μ, and GMV.
FIG. 1.
Schematic diagram for creating subject series matrix for ICA of FCNs. At the left is shown the standard matrix for group ICA, with each column containing a concatenation of BOLD time series from all M subjects in standard space. From this matrix, a parameter of interest (e.g., ALFF) is calculated from each subject's time series, and the results are concatenated as shown to produce a matrix with rows of ALFF values per subject. Each column then contains a subject series of parameter values for a given voxel, which provides the input to the ICA (as well as for seed-based correlation) for calculating FCNs. ICA, independent component analysis; FCN, functional covariance network; BOLD, blood oxygen level dependent; ALFF, amplitude of low-frequency fluctuation.
Spatial ICA was then implemented on each whole-brain, parametric subject series data set using probabilistic ICA implemented in MELODIC software (http://fmrib.ox.ac.uk/analysis/research/melodic/). The number of independent components (ICs) was selected to be 20, with this commonly chosen model order in resting-state analyses matching that of the FCP data analysis. Each of the ICA maps was transformed into an unthresholded Z-score map, which was directly compared to RSFC maps (also containing unthresholded Z-scores) calculated from the whole FCP data set (Biswal et al., 2010) to find corresponding or matching components. It should be noted that the FCP resting-state maps were calculated from standard ICA of concatenated time series (not from subject series analysis).
To derive matching components between the subject series, ICs with the known RSN components, a correlation matrix (weighted by Z-score) between the set of 20 FCP spatial maps, and a set of 20 subject series ICA maps was generated. For each FCP component, the IC with a maximum matrix value was selected (and visually verified); unweighted spatial cross-correlation coefficients for each matched component were recorded for comparison and significance testing (Smith et al., 2009). For both the HU and BNU data sets, standard group ICA (for 20 components) was performed on the BOLD time series, using the temporal concatenation approach available through FSL. Resulting components were matched to FCP ICs in similar manner as those of the subject series. For all cases, similarity of Z-score maps was quantified using correlation coefficients, from which a significance level can be roughly approximated. We note that taking into account the conservative estimate of ≈500 total degrees of freedom (≈37,000 GM and WM voxels, 6-mm FWHM smoothing over 3-mm- isotropic voxels, and left–right symmetry) and multiple comparisons (20 by 20) for the data sets, correlation values r>0.25 correspond to a corrected significance p<10−5 (Forman et al., 1995; Smith et al., 2009).
Additionally, seed-based correlations of the subject series (calculated as shown in Fig. 1) were calculated to investigate connectivity within the DMN. The seed voxel was selected from within the posterior cingulate cortex (PCC) with a coordinate (−5, −49, 40), as used by, for example (Fox et al., 2005; Biswal et al., 2010).
Results
ICA comparisons
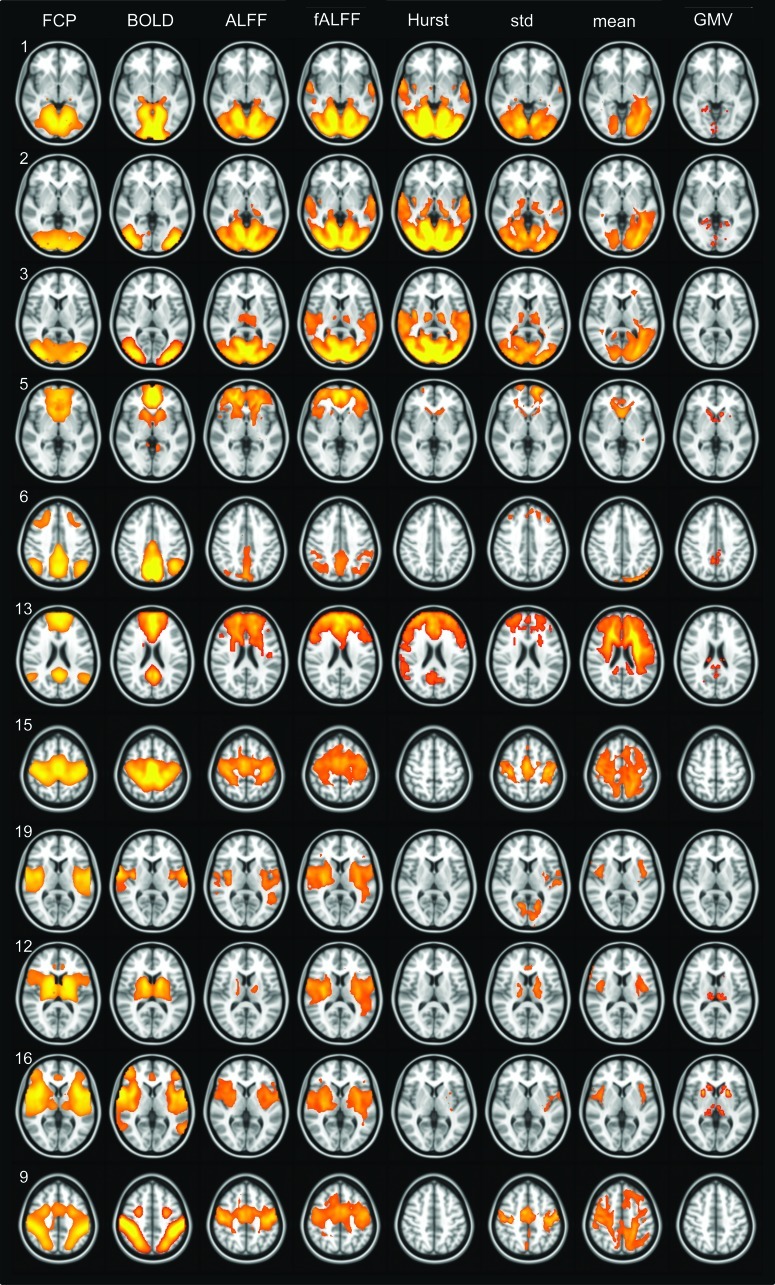
Figure 2 shows a comparison of FCP axial slices and matched BNU subject series ICs. After correlation, the BNU ICs were thresholded for visualization, using cluster-based thresholding with Z>2.3 and a cluster extent of p<0.01. The first column represents the FCP maps (with component number shown), followed by the ICs for the standard BOLD time series, ALFF, fALFF, H, σ, μ, and GMV. [Axial slice numbers were chosen to match those of the Biswal et al. (2010), except for IC 6.] Several known RSNs are represented by individual or groups of FCP components. Relevant ones for this study are shown, grouped appropriately, in the first columns of Figure 2 (and Fig. 3). The visual network is shown by FCP 1, 2, and 3, with striate and extrastriate portions being distinct. The DMN is represented by FCP 5, 6, and 13, with highest Z-scores, respectively, in the ventral–medial prefrontal, the posterior cingulate, and the medial prefrontal cortices. The sensorimotor network is given by FCP 15 and 19 (somatosensory), and FCP 12 shows the basal ganglia network. FCP 16 corresponds to the anterior insula, part of the salience network, and FCP 9, to the dorsal attention network. The associated (unweighted) spatial cross-correlation coefficient values for the subject series parameters are given in Table 2. A large overlap of high Z-score regions is observed for most resting-state parameters, with corresponding correlation values r >0.25; as estimated in the Methods, this correlation value corresponds to a corrected significance of p<10−5. In many cases, correlation between ICs was much larger than r>0.4.
FIG. 2.
Z-score maps from ICA for the FCP (first column) and BNU (columns 2–7), where the given parameter for the latter is given at the top of each column. The number of each FCP component corresponds to that of the original article (Biswal et al., 2010), with components grouped by network: FCP 1–3, visual (vis); FCP 5, 6, and 13, default mode (DM); FCP 15 and 19, sensorimotor (sm); FCP 12, basal ganglia (bas); FCP 16, salience (sal); and FCP 9, the dorsal attention (datt). Thresholding of each map is described in the text, with highest Z-scores in yellow. FCP, Functional Connectome Project; BNU, Beijing Normal University.
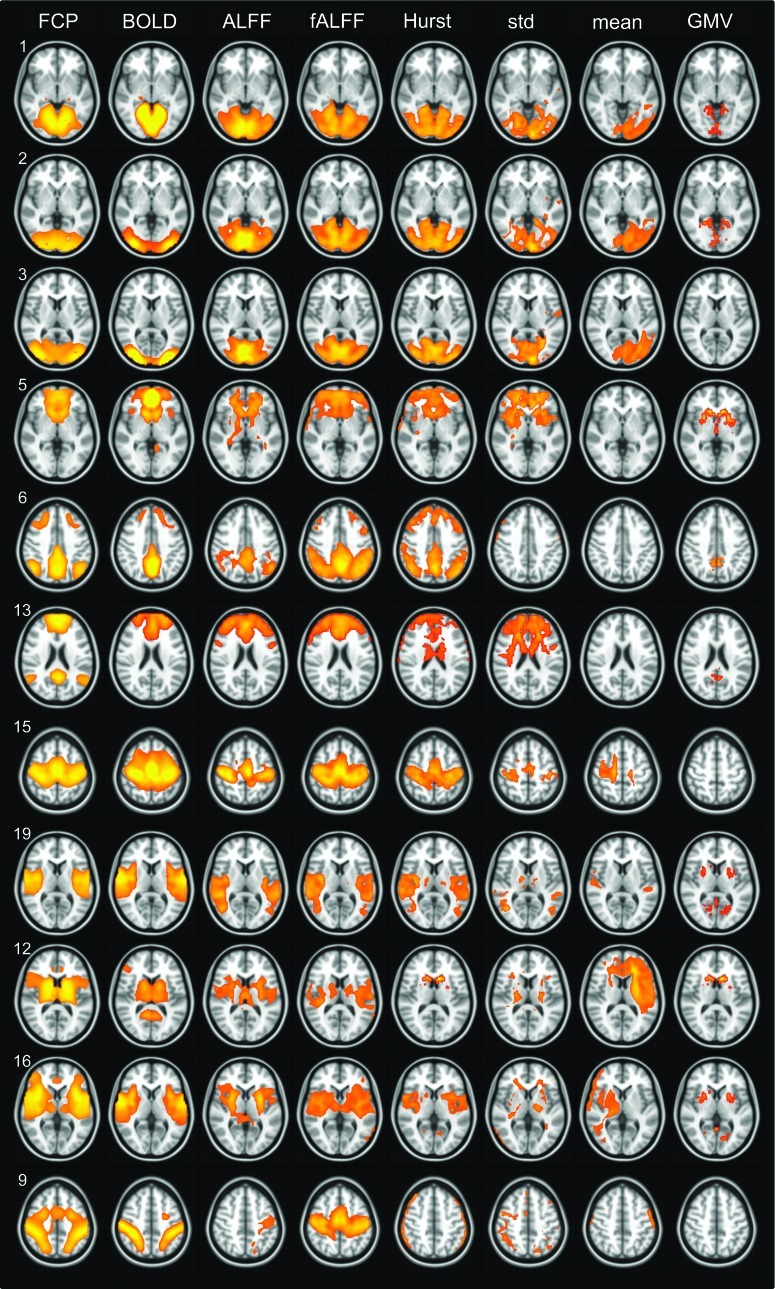
FIG. 3.
Analogous maps to Figure 2 for HU data set. HU, Harvard University.
Table 2.
Correlation Coefficients Between The Significant Functional Connectome Project Components and Each of the Associated Independent Component Analysis Results of the BNU and HU Data Sets are Shown, Grouped by Network
| |
Correlation coefficients |
||||||
|---|---|---|---|---|---|---|---|
|
FCP IC |
BOLD |
ALFF |
fALFF |
Hurst |
Std |
Mean |
GMV |
| (network) | BNU; HU | BNU; HU | BNU; HU | BNU; HU | BNU; HU | BNU; HU | BNU; HU |
| 1 (vis) | 0.63; 0.75 | 0.79; 0.72 | 0.79; 0.78 | 0.78; 0.76 | 0.70; 0.63 | 0.40; 0.43 | 0.14; 0.22 |
| 2 (vis) | 0.63; 0.69 | 0.78; 0.76 | 0.70; 0.73 | 0.64; 0.66 | 0.74; 0.62 | 0.40; 0.39 | 0.11; 0.18 |
| 3 (vis) | 0.75; 0.69 | 0.75; 0.66 | 0.76; 0.78 | 0.80; 0.81 | 0.76; 0.58 | 0.40; 0.40 | 0.06; 0.08 |
| 5 (DM) | 0.67; 0.61 | 0.52; 0.44 | 0.35; 0.50 | 0.44; 0.51 | 0.33; 0.40 | 0.28; 0.28 | 0.14; 0.13 |
| 6 (DM) | 0.64; 0.71 | 0.34; 0.45 | 0.40; 0.55 | 0.18; 0.46 | 0.11; 0.08 | 0.13; 0.07 | 0.08; 0.14 |
| 13 (DM) | 0.67; 0.44 | 0.53; 0.35 | 0.55; 0.58 | 0.37; 0.40 | 0.41; 0.33 | 0.29; 0.12 | 0.03; 0.06 |
| 15 (sm) | 0.72; 0.68 | 0.72; 0.65 | 0.64; 0.81 | 0.36; 0.75 | 0.53; 0.41 | 0.29; 0.17 | 0.02; 0.03 |
| 19 (sm) | 0.66; 0.57 | 0.69; 0.61 | 0.66; 0.72 | 0.34; 0.70 | 0.47; 0.36 | 0.22; 0.14 | 0.03; 0.05 |
| 12 (bas) | 0.66; 0.54 | 0.29; 0.47 | 0.28; 0.52 | 0.40; 0.39 | 0.41; 0.36 | 0.26; 0.27 | 0.28; 0.25 |
| 16 (sal) | 0.73; 0.63 | 0.42; 0.36 | 0.59; 0.63 | 0.43; 0.54 | 0.27; 0.23 | 0.27; 0.25 | 0.12; 0.11 |
| 9 (datt) | 0.61; 0.64 | 0.49; 0.21 | 0.50; 0.45 | 0.26; 0.26 | 0.25; 0.19 | 0.31; 0.15 | 0.04; 0.03 |
| AVE. | 0.67; 0.63 | 0.57; 0.52 | 0.57; 0.52 | 0.46; 0.57 | 0.45; 0.38 | 0.29; 0.24 | 0.10; 0.12 |
| ST. DEV. | 0.05; 0.09 | 0.18; 0.17 | 0.17; 0.13 | 0.20; 0.18 | 0.21; 0.18 | 0.08; 0.12 | 0.08; 0.07 |
Mean values of correlation for each column are also shown, for which fALFF shows a maximum for both data sets (tied with H in HU and with ALFF in BNU sets).
FCP, Functional Connectome Project; IC, independent component; BOLD, blood oxygen level dependent; LFF, low-frequency fluctuation; ALFF, amplitude of LFF; Falff, fractional ALFF; vis, visual; DM, default mode; sm, sensorimotor; datt, dorsal attention; sal, salience; bas, basal ganglia; GMV, gray matter volume.
An analogous set of images for the HU group is shown in Figure 3, with associated correlation values also given in Table 2. In several cases (and depending on the parameter), multiple FCP components were represented by the same IC from the subject series (see Table 3); this particularly occurred for the visual network, the sensorimotor network, and the prefrontal components of the DMN. In general, the dorsal attention network was poorly represented by each subject series in terms of both high Z-score overlap and spatial cross-correlation.
Table 3.
The Component Numbers Corresponding to the Spatial Maps in Figures 2 and 3 are Shown, Grouped by Network (Defined in Table 2)
| |
IC number |
||||||
|---|---|---|---|---|---|---|---|
|
FCP IC |
BOLD |
ALFF |
fALFF |
Hurst |
Std |
Mean |
GMV |
| (network) | BNU; HU | BNU; HU | BNU; HU | BNU; HU | BNU; HU | BNU; HU | BNU; HU |
| 1 (vis) | 10; 13 | 1; 2 | 1; 2 | 1; 4 | 1; 4 | 1; 6 | 7; 6 |
| 2 (vis) | 6; 19 | 1; 2 | 1; 2 | 1; 4 | 1; 4 | 1; 6 | 7; 6 |
| 3 (vis) | 6; 19 | 1; 2 | 1; 2 | 1; 4 | 1; 4 | 1; 6 | 6; 1 |
| 5 (DM) | 13; 4 | 3; 18 | 6; 6 | 16; 13 | 18; 1 | 5; 15 | 2; 7 |
| 6 (DM) | 4; 5 | 8; 14 | 14; 7 | 15; 6 | 20; 7 | 1; 6 | 15; 19 |
| 13 (DM) | 13; 12 | 3; 3 | 6; 6 | 7; 13 | 4; 1 | 2; 15 | 15; 19 |
| 15 (sm) | 16; 7 | 2; 1 | 4; 3 | 8; 2 | 2; 17 | 8; 4 | 14; 10 |
| 19 (sm) | 15; 20 | 2; 1 | 4; 3 | 8; 2 | 2; 17 | 20; 12 | 14; 16 |
| 12 (bas) | 7; 10 | 5; 13 | 19; 15 | 12; 11 | 20; 15 | 9; 16 | 4; 7 |
| 16 (sal) | 5; 20 | 10; 13 | 4; 15 | 19; 20 | 19; 15 | 20; 16 | 4; 16 |
| 9 (datt) | 20; 17 | 2; 1 | 4; 3 | 17; 2 | 2; 7 | 8; 12 | 6; 2 |
Relevant for each column is the repetition of a given component (IC numbers are unrelated across columns and data sets) across different networks (i.e., that the parameter did not separate the networks into separate components), such as for the repeated correlation of IC 5 in the BNU set for fALFF.
It should be noted that typically the same subset of FCP components had high correlation with ICA results of the BNU and HU data sets. For example, in both the HU and BNU subject series data sets, a single IC represented the visual cortex, while the visual network was split into three components for the FCP case (and two components for the BOLD time series). Moreover, the BNU and HU data sets typically yielded similar looking results for a given parameter, particularly for resting-state variables, as well as roughly similar correlation values, in most cases (Table 2). For both subject series, fALFF was the parameter which yielded highest overall correlation to FCP components (tied with ALFF for the BNU set; as seen in average values in Table 2) with GMV being the least.
Mean and standard deviation of intersubject variability of ICs
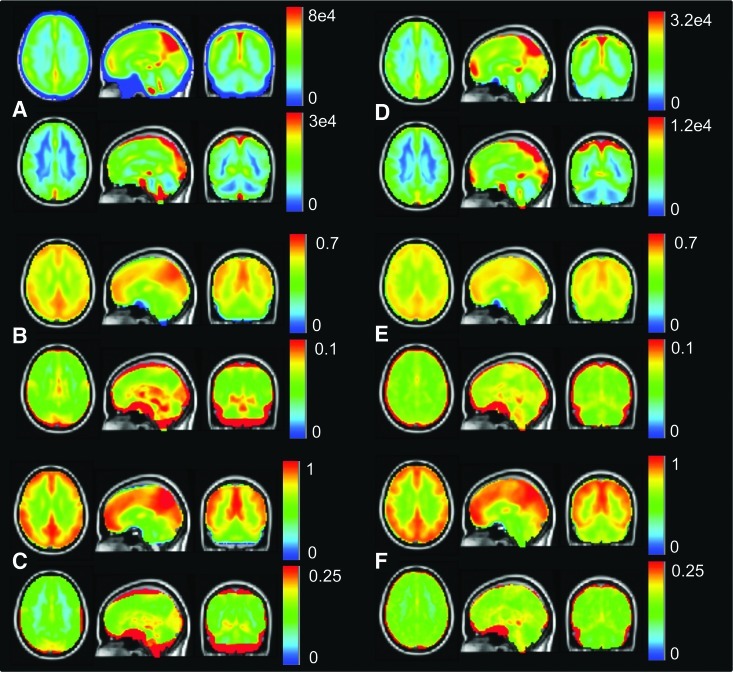
The mean and standard deviations of the HU and BNU subject series values of RSFC parameters are shown in left and right columns, respectively, of Figure 4. The BNU values of ALFF are approximately a factor of 2.5 times greater than those of HU, though the maps of each parameter show very similar locations of high mean and standard deviation (typically in the gray matter). Additionally, the analogous fALFF and Hurst maps of the two data sets are very similar, again showing largest standard deviation in the gray matter, with fairly uniform values throughout (with a maximum occurring around the superior parietal lobule and precuneus).
FIG. 4.
The mean and standard deviation (upper and lower portion of each panel, respectively), of each subject series are shown for the BNU (left column) and HU (right column) data sets for the RSFC parameters: ALFF (A, D), fALFF (B, E), and the Hurst exponent (C, F). Standard deviation is uniformly higher in gray matter than in white matter regions, with similar patterns of variation observed in both HU and BNU sets in all cases (with ALFF differing in a single brain-wide constant). RSFC, resting-state functional connectivity.
Correlating intersubject variability of ICs
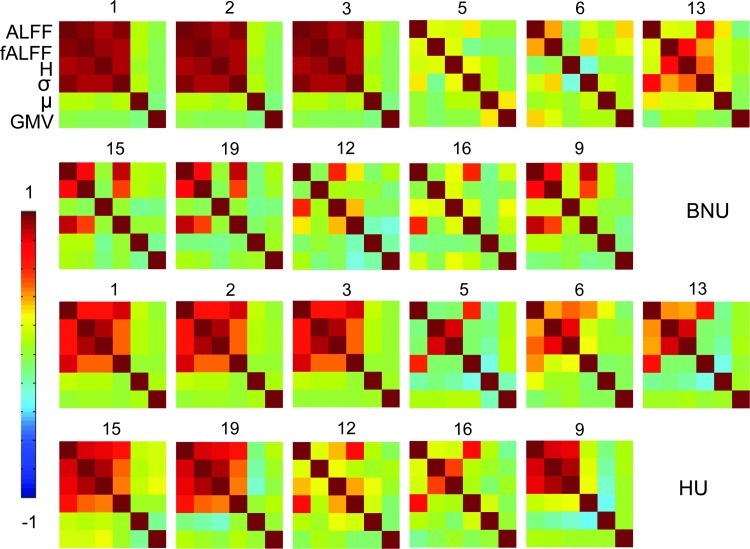
The properties of the subject series associated with each ICA spatial map from the subject series data sets were also investigated. These series represent the intersubject variability associated with each spatial IC map. To compare the intersubject variability of the parameters, Figure 5 shows correlation matrices of the ICA-produced series for a given associated FCP component (i.e., corresponding to rows in Figs. 2 and 3). The top two and bottom two rows, respectively, show values from the BNU and HU data sets. Within the visual (FCP 1–3) and HU sensorimotor (FCP 15 and 19) networks, ALFF, fALFF, H, and σ were positively correlated, as were subsets of these parameters within the DMN (FCP 5, 6, and 13). The mean and GMV series were typically uncorrelated with other parameters.
FIG. 5.
Per parameter correlation matrices for the time series associated with each component from spatial ICA (spatial ICs shown in Figs. 2 and 3) for the BNU (top two rows) and the HU data sets (bottom two rows), with each matrix labeled by the corresponding FCP component number. For each matrix, the parameters are in the following order (descending the y-axis and rightward along the x-axis): ALFF, fALFF, Hurst, standard deviation, and mean. The correlation coefficients are generally high among the first three components, as well as with the standard deviation in several cases; the series of means is often uncorrelated with the other parameters. fALFF, fractional ALFF.
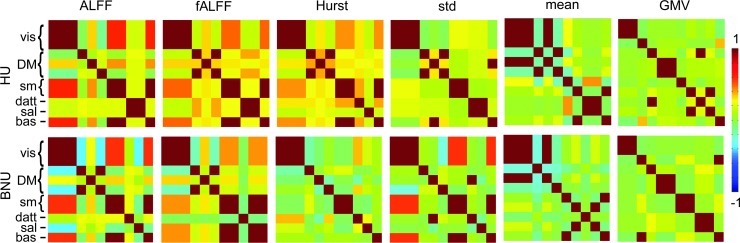
Also of interest is the correlation of the intersubject variability associated with different spatial ICs for a given parameter (i.e., corresponding to columns in Figs. 2 and 3). These are shown in Figure 6. The visual network was typically represented by a single component, and the DMN by two components. Variability with fALFF showed several cases of positive correlation among networks, particularly for HU, such as between vis and sm, and vis and bas; these networks also showed positive correlation with ALFF, and also for H in the HU dataset (though BNU showed much less correlation of variability between networks) and for σ in the BNU dataset. Separate networks were generally uncorrelated in terms of the mean and GMV parameters.
FIG. 6.
Per IC correlation matrices for the time series associated with each component from spatial ICA (spatial ICs shown in Figs. 2 and 3) for the HU (top) and the BNU data sets (bottom), with column labeled by parameter. High correlation corresponds to similarity of a parameter in intersubject variability between components. For resting-state parameters, several networks are represented by a single IC (e.g., the visual and sensorimotor for most parameters), or by highly correlated time series (e.g., default mode networks). IC, independent components.
Seed-based correlation
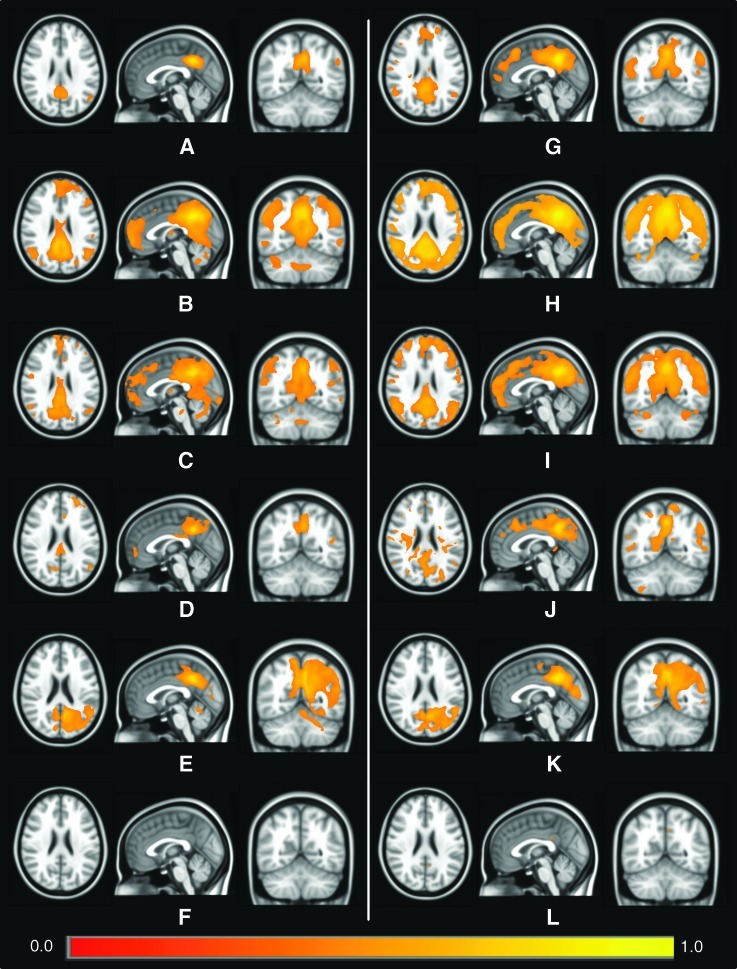
Figure 7 shows results of seed-based correlation for the parameter subject series within the DMN, for a seed voxel in the PCC with MNI coordinate (−5, −49, and 40) and images thresholded at r>0.4. The left and right columns show BNU and HU data, respectively, and rows contain results of correlation using (in descending order): ALFF, fALFF, the Hurst exponent, standard deviation, mean, and GMV. Varied amounts of the DMN are visible in each case; fALFF and H show correlated regions in the PCC, left and right inferior parietal cortices (IPC), and the ventral anterior cingulated cortex (vACC), as well as surrounding areas; ALFF shows small regions in the PCC, and for HU in the vACC, l/rIPC, and vACC. Activity appears in the PCC for standard deviation and mean, with only the rIPC showing any correlation in the rest of the DMN, though additional regions of activity appear. The GMV showed very few voxels of high correlation (and images with lower thresholds did not show coherent regions).
FIG. 7.
Spatial maps of significant correlation from seed-based analyses using a seed voxel in the posterior cingulate cortex with Montreal Neurological Institute coordinate (−5, −49, 40). All images are thresholded at r>0.4. The first column shows values from the BNU set for (A) ALFF, (B) fALFF, (C) Hurst exponent, (D) standard deviation, (E) mean, and (F) GMV, with respective parameters for the HU data set in (G–L).
Discussion
In this study, subject series of rs-fMRI parameters were analyzed using ICA and seed-based correlation to: (1) determine a spatial covariance structure of local LFF features across different subjects; (2) compare the organization of resting-state parameter covariance with known RSNs from a large set of data, the FCP; and (3) compare this covariance structure with that of control measures, the mean of the BOLD signal and GMV. We hypothesized that the presence of covariance structures would suggest a common influence underlying the given property within a network. Moreover, in the presence of such organization, comparisons with control parameters would test against null hypotheses of whether the influence was largely hemodynamic and anatomical.
To create these subject series, the resting parameters ALFF, fALFF, Hurst exponent, and σ (which has been used as a resting-state parameter) were calculated across each individual's whole brain, and the results were concatenated across the group. ICA of the RSFC parameters produced several spatial maps, which correlated highly with known resting-state components (defined by FCP data) in both the BNU and HU sets. Of these parameters, σ had lowest overall correlation to FCP RSNs (Table 2), with significantly lower values than fALFF and Hurst for the HU data set, but not for any other resting-state parameters for BNU (Table 4). Spatial maps derived from the mean of the BOLD signal and GMV tended to have lower correlation with FCP components, with the greatest overlap with in the visual network, but having significantly lower correlations to RSNs than any resting-state parameter (except for mean in BNU, for Hurst, and standard deviation; Table 4).
Table 4.
Pearson Correlation Coefficients from Columns in Table 2 were Fisher Transformed to Z-Scores and Compared Using Paired t-Tests, with the Resulting p-values Shown for the HU and BNU Data Set in the Upper and Lower Triangles, Respectively
| |
p-Values of paired t-tests |
||||||
|---|---|---|---|---|---|---|---|
| BOLD | ALFF | fALFF | Hurst | Std | Mean | GMV | |
| BOLD | — | 3.30e-02 | 5.92e-01 | 3.37e-01 | 3.42e-04 | 3.74e-07 | 5.11e-08 |
| ALFF | 1.95e-01 | — | 8.47e-04 | 9.41e-02 | 1.12e-03 | 1.28e-04 | 5.04e-05 |
| Falff | 8.80e-02 | 5.74e-01 | — | 4.85e-03 | 3.45e-05 | 5.24e-06 | 3.98e-06 |
| Hurst | 9.11e-03 | 3.27e-02 | 3.86e-02 | — | 1.06e-03 | 1.12e-04 | 8.01e-05 |
| Std | 7.95e-03 | 2.95e-03 | 1.93e-02 | 9.11e-01 | — | 8.77e-04 | 6.15e-04 |
| Mean | 5.31e-08 | 1.85e-04 | 1.27e-04 | 9.64e-03 | 9.08e-03 | — | 2.68e-03 |
| GMV | 3.17e-09 | 9.77e-05 | 6.79e-05 | 7.91e-04 | 9.59e-04 | 1.47e-04 | — |
Bold values highlight matrix elements below the p<0.05 threshold (corrected for 42 multiple comparisons), showing significant differences. For both data sets, typically mean and GMV values showed significant differences with other parameters, while ALFF, fALFF, and Hurst showed little difference among each other (excepting ALFF and fALFF for HU). Standard deviation showed significant differences with all parameters for HU, while differing significantly only with GMV for BNU.
Although in this study we have used rs-fMRI information from each subject to look at spatial patterns of connectivity, other investigators have previously used different measures in a similar manner (i.e., analyzing subject series). For example, He et al. (2007) have used region-wise cortical thickness values of subject series for graph theoretical analyses, and Xu et al. (2009) have used gray matter volume for ICA, and Zhang et al. (2011) have used ALFF for seed-based correlations. We note that the FCN approach performed in this work necessarily has a higher resolution than those approaches, as the RSFC parameters were calculated per voxel. As large data sets become more easily available, network analysis both within and between subjects to aid in understanding functional brain networks should be more feasible.
Networks from ICA with resting parameters
Spatial ICA of the resting-state parameters produced several known RSNs, as found through direct comparison with components of the FCP data set. Various components of the visual network (FCP 1–3) were typically represented as a single IC, with (left and right) regions of the superior temporal gyrus and insula included in the fALFF and Hurst sets (BNU). Features of the DMN were separated into distinct components, with extra activation appearing in the medial and inferior frontal gyri, though both Hurst and standard deviation maps were missing several regions. Bilateral components of the superior temporal gyrus (FCP 13) typically did not appear in any components. For the sensorimotor components (FCP 15 and 19), the left and right precentral gyri and the cingulate gyri were represented by all RSFC parameters, excepting Hurst for HU, and not by mean and GMV. Thalamic components of the basal component appeared in most parameter sets, with a generally smaller spatial extent than in FCP 12. The salience network in FCP 16 was best represented in ALFF and fALFF for both data sets. Both the inferior parietal and paracentral lobule features of the dorsal attention network (FCP 9) were missing for all parameters, and correlations were typically quite low (with the same component for the sensorimotor network showing highest correlation); possibly weak correlation is shown for the right inferior parietal component for HU ALFF, but most likely this component was not uniquely identified by the intersubject covariances.
The recapitulation of several known RSNs by ICA of the resting-state parametric subject series (Figs. 2 and 3), as well as by seed-based correlations (Fig. 7), provide a new group-based method of reconfirming the components of RSNs themselves and, importantly, their large-scale organization across the brain. In this case, parameters of resting-state properties vary across subjects independently, but they appear to have similar variance patterns within a given network; hence, we term these “functional covariance networks” (FCNs). That is, knowing how the fALFF value, for example, in voxel “x” varies across subjects would yield significant information about how the fALFF value of voxel “y” (within the same RSN component) would vary; however, there would be essentially no information about voxel “z” (outside the RSN).
While RSNs are often identified as independent spatial maps, some individual regions are located in multiple networks, and the networks themselves exhibit some interrelationships, such as in divisions of “task-positive” and “task-negative” networks (Fox et al., 2005; Mennes et al., 2010). The manner in which such regions participate in multiple networks is not well understood. Recent work by Smith et al. (2012) has undertaken to study these regions by examining temporal correlations using a combination of spatial and temporal ICA [a sequence that has been variously implemented in, e.g., Seifritz et al. (2002) and Alkan et al. (2011)]. The FCN approach discussed here offers another means for studying the interrelation of spatially independent networks on a group level, via intersubject covariance of resting-state properties. In some cases, the variability of parameters in FCNs, which corresponded to separate RSNs, was highly correlated, as shown in Figure 6 for the visual and sensorimotor networks, which may suggest higher level organization between these networks. Such possible relations between these and other networks will be investigated further.
RSFC properties
Numerous studies have shown that resting-state BOLD time series exhibit temporal correlations across spatially extended and disconnected networks across the brain. Parameters based on the spectral features of the signal, namely, the amplitudes (both relative and absolute) of LFFs and the spectral slope, have been shown to be useful in per voxel and region-wise comparisons. In this study, we have demonstrated the covariance of these RSFC spectral parameters across subjects in coherent patterns, which correspond to several known RSNs, and this suggests that the amplitudes and relative contributions of LFFs are mediated across the entire networks in each subject.
A remaining question in resting-state studies has been the relative contribution of neuronal signals of interest in the BOLD signals (and therefore, in the obtained RSNs themselves) and those of other physiological factors (confounds, such as cardiac rate, respiration, and vasculature). Previous analyses of the time series themselves have shown that cardiac and respiratory contributions to RSNs can be minimized by using appropriate filters. Importantly, the FCN results provide insight into the dependence of RSFC on the underlying structure and vasculature.
The spatial covariance of GMV, the anatomical parameter, showed no correspondence to any RSN. The parameter with greatest dependence on the underlying vasculature and hemodynamics, the mean of the BOLD signal, showed the lowest overall correlation with RSNs (averaged across ICs in Table 2) and showed only minor areas of high Z-score overlap (Figs. 2 and 3) with RSN components. These occurred in the visual network, also greatly lateralized for both data sets. The GMV components were typically uncorrelated to any known RSN.
Finally, the RSFC parameters (ALFF, fALFF, H, and σ) produced high correlation and large overlap of high Z-score regions with known RSNs. The standard deviation of the BOLD signal had generally lower correlations than the other RSFC parameters; while σ also has known dependence on vasculature and, as an unfiltered parameter, may be more affected by the scanner noise than other parameters, however, it has also been shown to have a strong correlation with the filtered RSFC parameter, σLFF (see the Appendix) and has been used as an RSFC parameter at times (e.g., Kannurpatti and Biswal, 2008). High Z-score overlap with FCP components was greatest in the visual networks and also in the somatosensory component (HU data set).
The visual network was well represented by covariance patterns of all RSFC parameters, though it is interesting to note that, whereas the network was subdivided into three FCP components, such differentiation was typically not found in the ICA of subject series (Table 3); the standard ICA of concatenated BOLD time series also did not differentiate all three components for either data set. Similar patterns were found across RSFC variables for the somatosensory (except with σ in HU and H). The majority of the extent of the DMN was represented by the parameters, though in several cases subcomponent regions were missing. The dorsal attention network appeared in standard group time series group analysis, but not for any RSFC (or other) parameters. These findings suggest that the majority of the networks arising from the use of the RSFC parameters are not primarily due to vascular origin, since then the voxels containing the vascular connections themselves would have been observed in the ICA components as well; such patterns were not observed, even for the case of the mean BOLD signal parameter, or for the anatomical GMV parameter. It is likely that these RSFC parameters and the consequent covariance networks arising may be due to neurophysiological mechanisms.
Physiological connection
In this study, we have obtained several independent networks from the intersubject covariance of rs-fMRI parameters. From the independent data sets examined here, these seem to have distinct patterns across healthy subject populations. It is possible that the predominant mechanism giving rise to this phenomenon is the underlying vasculature (blood flow and volume, vascular density), or even due to varied subject state (mood, caffeine content, time of day, etc.). However, in the case of simple vascular density, then we would expect to see RSN-like networks from the covariance of the BOLD signal mean, which was not observed. This would instead suggest the interesting possibility that the underlying mechanism relates to a neurophysiological basis for these covariance structures. Future studies are planned to further test this with positron emission tomography (PET) and arterial spin labeling (ASL) scans, whose measures relate directly to the metabolic rate (Vaishnavi et al., 2010). Additionally, studies of certain pathologies, such as epilepsy and schizophrenia, which lead to the alteration of the neurophysiological signal (and likewise of FCN structure) provide another avenue for investigating the underlying mechanism. Whereas previous aging studies linked the presence of structural covariance networks to various forms of maturation (Bergfield et al., 2010; Zielinski et al., 2010), the precise mechanism of FCN elucidation in healthy populations remains to be determined; further studies, including cognitive or behavioral measures, for example, would be useful in determining further interpretations of FCNs.
RSFC parameter comparisons
In both the HU and BNU groups, fALFF-derived results provided highest mean correlation between ICs and the FCP components, and generally great similarity was observed with the high Z-score maps. For BNU, ALFF provided nearly equivalent correlations, with Hurst substantially lower (Table 2). Paired t-tests of the correlation results showed that overall average results were not significantly different between RSFC parameters, except for σ with fALFF and Hurst for HU. However, it should be noted that for most components, visual inspection of the thresholded Z-score maps showed greater spatial overlap for the fALFF and ALFF results (Figs. 2 and 3). Lower H scores may be due in part to the duration of resting-state scans (235 time points for BNU and 119 for HU, relatively short for Hurst estimation), since it has been shown that estimates of the Hurst exponent depend strongly on time series length (DePetrillo et al., 1999). Lower σ scores and lower results for visual comparison were discussed in the previous section.
In the seed-based correlations (seeded in the PCC, with expectation of recapitulating the DMN), fALFF and Hurst provided high correlation coverage of most of the DMN, though both also included a large number of non-DMN regions as well. ALFF typically had incomplete spatial extent of the DMN, and the BOLD standard deviation showed only sparse coverage. Further studies are required to test the observed greater sensitivity, but lower specificity, of fALFF to intersubject covariance, which may be due to the dual aspects of increasing the information content by including amplitudes of the whole spectrum, while potentially adding more noise from the higher frequencies at the same time. As shown in Figure 5, the time series for a given IC associated with each of the resting-state parameters tended to have relatively high mutual correlation. Even so, spatial maps for the Hurst exponent occasionally showed large deviations from those of the (f)ALFF.
The robustness of the FCN results is shown by the strong similarity of significant networks shown by the independent data sets (HU and BNU) examined in parallel here. We note that these data sets have different TRs, and previous ICA studies of resting-state data have shown that cortical components show small changes with sampling rates (Kiviniemi et al., 2004). Additionally, ICA had been performed using 20 and 40 components, as well as determining the number to explain 90% of total variance, with similar results obtained in general. Thus, the spatial structure of RSFC appears robust even in the presence of significant intersubject variability, which has been used here to determine the global structure of amplitudes of BOLD signals. Further studies should include larger numbers of subjects to determine if the subdivisions of networks observed in the FCP may be obtained using FCN analysis.
Further applications
For large clinical data sets, analyses of intersubject covariance may help us to better understand group differences between clinical populations and healthy controls. One would expect that in clinical populations, the characteristic physiological covariance observed here would be altered in some regions so that the local LFF properties no longer reflected the same global influence as the rest of the network, potentially reflecting the disintegration of particular networks in either common or distinct manners. Such FCN-based analysis may provide additional information to that of the RSN-based techniques to better understand and differentiate the neural changes of the pathology. Such evidence would also be of importance for current attempts to classify individual brain states based on representative regions and their local LFF features. The characteristic disengagement of certain regions from the observed networks remains subject to future clinical studies, including appropriate clinical covariates.
Further analysis of this methodology for analyzing RSNs in groups will focus on the effects of group size, particularly with regard to robustness. It is possible that more networks or greater differentiation within networks would be observed by including more subjects. While in standard spatial and temporal ICA of individual and group BOLD time series, the origin of several artifactual ICs due to subject motion and vasculature are known, analogous interpretations for subject series artifactual ICs are not so clear. Certainly, some artifactual components will be due to intersubject motion and misalignment of mapped regions in standard space, but additional studies of these must be performed. The fact that the resting-state ICs in both groups were quite similar here suggests that artifacts due to head motion, misalignment, etc. were minimal. Certain populations in which known changes in brain structure exist (e.g., tumor, stroke, and traumatic brain injury) may require additional considerations (for example, in terms of alignment) for the analysis of FCNs as described here.
Variability due to the inclusion of multiple data sets (i.e., from separate institutions) also needs to be determined. However, data sets themselves are in general getting larger, making such analyses more practical. Also, the method appears to be robust, as separate analysis on two different data sets (from separate institutions) produced very similar results.
Conclusions
We have tested a data-driven, group-level approach for investigating RSNs. Using ICA on two large groups of subjects, the variance of resting-state parameters across subjects was shown to occur in spatial patterns, termed FCNs, several of which matched with known components of RSNs. These highly dynamic markers of resting-state behavior appear to change in a coordinated fashion in separate FCNs across the whole brain. The appearance of FCNs due to intersubject covariance suggests the presence of some common underlying influence for the RSFC properties within each network, rather than the LFFs being determined locally and independently. The correlation with known networks was much greater with resting-state parameters compared to BOLD mean and standard deviation; this suggests that the covarying properties of RSNs are significantly greater than those from a purely vascular/hemodynamic response or underlying anatomy (i.e., driven by GMV), and that the resting-state parameters themselves (σ and Hurst exponent, particularly ALFF and fALFF) contain significant information of neuronal/metabolic responses within the RSNs.
Appendix
Relations of resting-state BOLD parameters
In this study, several resting-state BOLD parameters are utilized during analysis, and their results in elucidating RSNs are compared. For reference, we briefly describe the mathematical relations of the resting-state BOLD parameters here.
For discrete time series, {xn}, with N time points, the following (complex) Fourier relations hold:
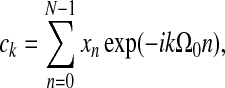 |
(1) |
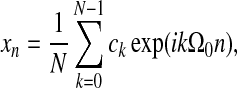 |
(2) |
with fundamental frequency Ω0=2π/N. The further interrelation of time variables (xn) and frequency variables (ck) is given by Parseval's Theorem, which states:
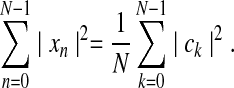 |
(3) |
The right hand side (RHS) describes the power spectrum of a time series, with |ck|2/N being the power spectral density (PSD) at a given frequency, kΩ0; the square root of each term, |ck|/N1/2, is called the amplitude. For a zero-mean time series (or one from which the mean, μ, has been subtracted) the LHS is simply proportional to the variance, σ2:
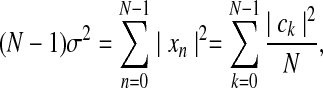 |
(4) |
where the RHS was rearranged to reflect the inclusion of the factor of N in the PSD. From this relation, the standard deviation is simply,
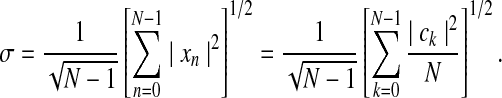 |
(5) |
Kannurpatti and Biswal (2008) showed that a related parameter, the standard deviation of the LFF-filtered time series, was highly correlated with σ. This new parameter, called the resting state BOLD fluctuation amplitude (RSFA) or σLFF, can be calculated directly from the frequency relations as:
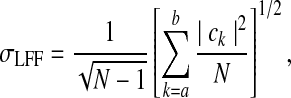 |
(6) |
where a and b correspond to 0.01 and 0.1 Hz, respectively.
Instead of PSD terms, Zang et al. (2007) used amplitude terms of filtered series to formulate a resting-state parameter. As its name denotes, ALFF is simply the sum of amplitudes in the LFF:
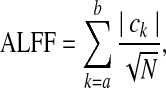 |
(7) |
though slight variations of ALFF (such as frequency average and global mean) exist as well. Comparing this term with the RHS expression for σLFF in Eq. (6), one would expect high correlation (and therefore high correlation generally with σ, as the terms are essentially the L1 and L2 norms of spectral amplitudes (with the latter scaled by [N-1]1/2). The two formulations encode qualitatively similar information. Mathematically, L1 is always greater than or equal to L2, but the latter measure exhibits greater percentage change when outliers are introduced to a signal. Therefore, the relative amplitudes of subdivisions of LFFs [which have been shown to be relevant by, e.g., Buzsáki and Draguhn (2004); Salvador et al. (2008)] may be represented differently by either L1- or L2-type parameters (i.e., ALFF or σ, respectively), and with different susceptibility to noise (greater in the L2 case) FALFF is given simply as the ratio of ALFF to the sum of all frequency amplitudes (up to Nyquist), so that, from Equation (7):
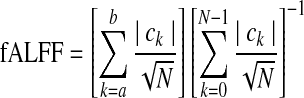 |
(8) |
It is possible to describe the Hurst exponent in terms of frequency properties of the resting-state time series, as well, though most estimation methods utilize other factors, such as wavelets (Bullmore et al., 2004). For small frequencies, one can show that
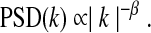 |
(9) |
The spectal slope, β, lies in the range [−1,1] and relates to the Hurst exponent as β=2H-1. Thus, the relative contributions of LFF, as can be observed in the spectral slope, affect H directly, whereas σ and (f) ALFF depend on summed values of LFFs.
Acknowledgments
The authors would like to thank Doug Ward for several useful comments on this article. This work was supported in part by NIH grants R01 AG032088 and R01 EB000215 (B.B.) and DFG-SFB 779 (M.W.).
Author Disclosure Statement
The authors declare no competing financial interests.
References
- Abry P. Veitch D. A wavelet based joint estimator of the parameters of long-range dependence. IEEE Trans Info Theory. 1999;45:878–897. [Google Scholar]
- Alkan Y. Biswal BB. Taylor PA. Alvarez TL. Segregation of frontoparietal and cerebellar components within saccade and vergence networks using hierarchical independent component analysis of fMRI. Vis Neurosci. 2011;28:247–261. doi: 10.1017/S0952523811000125. [DOI] [PubMed] [Google Scholar]
- Ashburner J. Friston KJ. Voxel-based morphometry: the methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner J. Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Beckmann CF. DeLuca M. Devlin JT. Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergfield KL. Hanson KD. Chen K. Teipel SJ. Hampel H. Rapoport SI. Moeller JR. Alexander GE. Age-related networks of regional covariance in MRI gray matter: reproducible multivariate patterns in healthy aging. Neuroimage. 2010;49:1750–1759. doi: 10.1016/j.neuroimage.2009.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM. Diamond JB. Smith MA. Bandettini PA. Separating respiratory-variation related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Biswal B. Yetkin FZ. Haughton VM. Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Biswal BB. Mennes M. Zuo XN. Gohel S. Kelly C. Smith SM. Beckmann CF. Adelstein JS. Buckner RL. Colcombe S. Dogonowski AM. Ernst M. Fair D. Hampson M. Hoptman MJ. Hyde JS. Kiviniemi VJ. Kötter R. Li SJ. Lin CP. Lowe MJ. Mackay C. Madden DJ. Madsen KH. Margulies DS. Mayberg HS. McMahon K. Monk CS. Mostofsky SH. Nagel BJ. Pekar JJ. Peltier SJ. Petersen SE. Riedl V. Rombouts SA. Rypma B. Schlaggar BL. Schmidt S. Seidler RD. Siegle GJ. Sorg C. Teng GJ. Veijola J. Villringer A. Walter M. Wang L. Weng XC. Whitfield-Gabrieli S. Williamson P. Windischberger C. Zang YF. Zhang HY. Castellanos FX. Milham MP. Toward discovery science of human brain function. Proc Natl Acad Sci U S A. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E. Fadili J. Maxim V. Sendur L. Whitcher B. Suckling J. Brammer M. Breakspear M. Wavelets and functional magnetic resonance imaging of the human brain. Neuroimage. 2004;23:S234–S249. doi: 10.1016/j.neuroimage.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Buzsáki G. Draguhn A. Neuronal oscillations in cortical networks. Science. 2004;304:1926–1929. doi: 10.1126/science.1099745. [DOI] [PubMed] [Google Scholar]
- Calhoun VD. Pekar JJ. McGinty VB. Adali T. Watson TD. Pearlson GD. Different activation dynamics in multiple neural systems during simulated driving. Hum Brain Mapp. 2002;16:158–167. doi: 10.1002/hbm.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. Cunningham JP. Glover GH. Influence of heart rate on the BOLD signal: the cardiac response function. Neuroimage. 2009;44:857–869. doi: 10.1016/j.neuroimage.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS. Rombouts SA. Barkhof F. Scheltens P. Stam CJ. Smith SM. Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M. Beckmann CF. De Stefano N. Matthews PM. Smith SM. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. Neuroimage. 2006;29:1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- DePetrillo PB. Speers D. Ruttimann UE. Determining the Hurst exponent of fractal time series and its application to electrocardiographic analysis. Comput Biol Med. 1999;29:393–406. doi: 10.1016/s0010-4825(99)00018-9. [DOI] [PubMed] [Google Scholar]
- Forman SD. Cohen JD. Fitzgerald M. Eddy WF. Mintun MA. Noll DC. Improved assessment of significant activation in functional magnetic resonance imaging (fMRI): use of a cluster-size threshold. Magn Reson Med. 1995;33:636–647. doi: 10.1002/mrm.1910330508. [DOI] [PubMed] [Google Scholar]
- Fox MD. Snyder AZ. Vincent JL. Corbetta M. Van Essen DC. Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good CD. Johnsrude I. Ashburner J. Henson RN. Friston KJ. Frackowiak RS. Cerebral asymmetry and the effects of sex and handedness on brain structure: a voxel-based morphometric analysis of 465 normal adult human brains. NeuroImage. 2001;14:685–700. doi: 10.1006/nimg.2001.0857. [DOI] [PubMed] [Google Scholar]
- Greicius MD. Krasnow B. Reiss AL. Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. Chen ZJ. Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 2007;17:2407–2419. doi: 10.1093/cercor/bhl149. [DOI] [PubMed] [Google Scholar]
- Jenkinson M. Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jones TB. Bandettini PA. Kenworthy L. Case LK. Milleville SC. Martin A. Birn RM. Sources of group differences in functional connectivity: an investigation applied to autism spectrum disorder. Neuroimage. 2010;49:401–414. doi: 10.1016/j.neuroimage.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannurpatti SS. Biswal BB. Detection and scaling of task induced fMRI-BOLD response using resting state fluctuations. Neuroimage. 2008;40:1567–1574. doi: 10.1016/j.neuroimage.2007.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi V. Kantola J-H. Jauhiainen J. Hyvarinen A. Tervonen O. Independent component analysis of nondeterministic fMRI signal sources. NeuroImage. 2003;19:253–260. doi: 10.1016/s1053-8119(03)00097-1. [DOI] [PubMed] [Google Scholar]
- Kiviniemi V. Kantola JH. Jauhiainen J. Tervonen O. Comparison of methods for detecting nondeterministic BOLD fluctuation in fMRI. Magn Reson Imaging. 2004;22:197–203. doi: 10.1016/j.mri.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Lowe MJ. Mock BJ. Sorenson JA. Functional connectivity in single and multislice echoplanar imaging using resting state fluctuations. Neuroimage. 1998;7:119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- Maxim VT. Sendur LS. Fadili J. Suckling J. Gould R. Howard R. Bullmore ET. Fractional Gaussian noise, functional MRI and Alzheimer's disease. Neuroimage. 2005;25:141–158. doi: 10.1016/j.neuroimage.2004.10.044. [DOI] [PubMed] [Google Scholar]
- Mazziotta J. Toga A. Evans A. Fox P. Lancaster J. Zilles K. Woods R. Paus T. Simpson G. Pike B. Holmes C. Collins L. Thompson P. MacDonald D. Iacoboni M. Schormann T. Amunts K. Palomero-Gallagher N. Geyer S. Parsons L. Narr K. Kabani N. Le Goualher G. Boomsma D. Cannon T. Kawashima R. Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MJ. Jung TP. Makeig S. Brown G. Kindermann SS. Lee TW. Sejnowski TJ. Spatially independent activity patterns in functional MRI data during the stroop color-naming task. Proc Natl Acad Sci U S A. 1998;95:803–810. doi: 10.1073/pnas.95.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechelli A. Friston KJ. Frackowiak RS. Price CJ. Structural covariance in the human cortex. J Neurosci. 2005;25:8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennes M. Kelly C. Zuo XN. Di Martino A. Biswal B. Xavier Castellanos F. Milham MP. Inter-individual differences in resting state functional connectivity predict task-induced BOLD activity. Neuroimage. 2010;50:1690–1701. doi: 10.1016/j.neuroimage.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME. MacLeod AM. Snyder AZ. Powers WJ. Gusnard DA. Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador R. Martinez A. Pomarol-Clotet E. Gomar J. Vila F. Sarro S. Capdevila A. Bullmore E. A simple view of the brain through a frequency-specific functional connectivity measure. Neuroimage. 2008;39:279–289. doi: 10.1016/j.neuroimage.2007.08.018. [DOI] [PubMed] [Google Scholar]
- Seeley WW. Menon V. Schatzberg AF. Keller J. Glover GH. Kenna H. Reiss AL. Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmueli K. van Gelderen P. de Zwart JA. Horovitz SG. Fukunaga M. Jansma JM. Duyn JH. Low-frequency fluctuations in the cardiac rate as a source of variance in the resting-state fMRI BOLD signal. Neuroimage. 2007;38:306–320. doi: 10.1016/j.neuroimage.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifritz E. Esposito F. Hennel F. Mustovic H. Neuhoff JG. Bilecen D. Tedeschi G. Scheffler K. Di Salle F. Spatiotemporal pattern of neural processing in the human auditory cortex. Science. 2002;297:1706–1708. doi: 10.1126/science.1074355. [DOI] [PubMed] [Google Scholar]
- Smith S. Jenkinson M. Woolrich M. Beckmann C. Behrens T. Johansen-Berg H. Bannister P. De Luca M. Drobnjak I. Flitney D. Niazy R. Saunders J. Vickers J. Zhang Y. De Stefano N. Brady J. Matthews P. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23:208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fox PT. Miller KL. Glahn DC. Fox PM. Mackay CE. Filippini N. Watkins KE. Toro R. Laird AR. Beckmann CF. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM. Miller KL. Moeller S. Xu J. Auerbach EJ. Woolrich MW. Beckmann CF. Jenkinson M. Andersson J. Glasser MF. Van Essen DC. Feinberg DA. Yacoub ES. Ugurbil K. Temporally-independent functional modes of spontaneous brain activity. PNAS. 2012 doi: 10.1073/pnas.1121329109. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnavi SN. Vlassenko AG. Rundle MM. Snyder AZ. Mintun MA. Raichle ME. Regional aerobic glycolysis in the human brain. Proc Natl Acad Sci U S A. 2010;107:17757–17762. doi: 10.1073/pnas.1010459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Buuren M. Gladwin TE. Zandbelt BB. van den Heuvel M. Ramsey NF. Kahn RS. Vink M. Cardiorespiratory effects on default-mode network activity as measured with fMRI. Hum Brain Mapp. 2009;30:3031–3042. doi: 10.1002/hbm.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RG. Ide K. Poulin MJ. Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21:1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Xiong J. Parsons LM. Gao JH. Fox PT. Interregional connectivity to primary motor cortex revealed using MRI resting state images. Hum Brain Mapp. 1999;8:151–156. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<151::AID-HBM13>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. Groth KM. Pearlson G. Schretlen DJ. Calhoun VD. Source-based morphometry: the use of independent component analysis to identify gray matter differences with application to schizophrenia. Hum Brain Mapp. 2009;30:711–724. doi: 10.1002/hbm.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang YF. He Y. Zhu CZ. Cao QJ. Sui MQ. Liang M. Tian LX. Jiang TZ. Wang YF. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev. 2007;29:83–91. doi: 10.1016/j.braindev.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Liao W. Zuo XN. Wang Z. Yuan C. Jiao Q. Chen H. Biswal BB. Lu G. Liu Y. Resting-state brain organization revealed by functional covariance networks. PLoS One. 2011;6:e28817. doi: 10.1371/journal.pone.0028817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski BA. Gennatas ED. Zhou J. Seeley WW. Network-level structural covariance in the developing brain. Proc Natl Acad Sci U S A. 2010;107:18191–18196. doi: 10.1073/pnas.1003109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou QH. Zhu CZ. Yang Y. Zuo XN. Long XY. Cao QJ. Wang YF. Zang YF. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: fractional ALFF. J Neurosci Methods. 2008;172:137–141. doi: 10.1016/j.jneumeth.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X.N. Di Martino A. Kelly C. Shehzad Z.E. Gee D.G. Klein D.F. Castellanos F.X. Biswal B.B. Milham M.P. The oscillating brain: complex and reliable. Neuroimage. 2010;49:1432–1445. doi: 10.1016/j.neuroimage.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]