Abstract
Background
There is a continuous demanding for tightly regulated prokaryotic expression systems, which allow functional synthesis of toxic proteins in Escherichia coli for bioscience or biotechnology application. However, most of the current promoter options either are tightly repressed only with low protein production levels, or produce substantial protein but lacking of the necessary repression to avoid mutations initiated by leaky expression in the absence of inducer. The aim of this study was to develop a tightly regulated, relatively high-efficient expression vector in E. coli based on the principle of iron uptake system.
Results
By using GFP as reporter, PfhuA with the highest relative fluorescence units, but leaky expression, was screened from 23 iron-regulated promoter candidates. PfhuA was repressed by ferric uptake regulator (Fur)-Fe2+ complex binding to Fur box locating at the promoter sequence. Otherwise, PfhuA was activated without Fur-Fe2+ binding in the absence of iron. In order to improve the tightness of PfhuA regulation for toxic gene expression, Fur box in promoter sequence and fur expression were refined through five different approaches. Eventually, through substituting E. coli consensus Fur box for original one of PfhuA, the induction ratio of modified PfhuA (named PfhuA1) was improved from 3 to 101. Under the control of PfhuA1, strong toxic gene E was successfully expressed in high, middle, low copy-number vectors, and other two toxic proteins, Gef and MazF were functionally synthesized without E. coli death before induction.
Conclusions
The features of easy control, tight regulation and relatively high efficiency were combined in the newly engineered PfhuA1. Under this promoter, the toxic genes E, gef and mazF were functionally expressed in E. coli induced by iron chelator in a tightly controllable way. This study provides a tightly regulated expression system that might enable the stable cloning, and functional synthesis of toxic proteins for their function study, bacterial programmed cell death in biological containment system and bacterial vector vaccine development.
Background
With the advent of the post-genomic era coming, the need is boosting to express a growing number of genes originating from different organisms [1]. Unfortunately, many of these foreign genes severely interfere with the survival of Escherichia coli cells, which could lead to bacteria death or cause significant defects in bacteria growth. What’s more, following the development of biological technology, many genetically engineered E. coli have been constructed and developed for different purposes, such as bioremediation, biomedicine and bioenergy [2]. However, their practical applications in the field are still restricted because engineered bacteria may cause new environmental contaminations. To minimize the potential risks, biological containment system was designed to monitor and restrict the distribution of engineered bacteria. Generally, containment systems are based on a ‘killer gene’ and a tight ‘regulatory circuit’ that controls expression of the killer gene in response to the presence or absence of environmental signals [3,4]. In order to activate the killer gene expression only at expected condition, usually a specific environment, the promoter that controls bacteria programmed cell death needs to be easily controllable, tightly regulated and environment-responsive.
In general, there are five solutions to manage killer or highly toxic genes expression: manipulation of transcriptional and translational control elements of highly toxic genes, manipulation of the coding sequence, manipulation of the copy number, addition of stabilizing sequences, and empirical selection of E. coli strains [5]. Among these solutions, manipulation of transcriptional control elements aims at blocking the leaky expression from the common used promoters, such as Plac, Ptrc, Ptac, PT7, PpL, PtetA or PlacUV5, which is critical in toxic gene expression [6,7]. As a result, the manipulation strategies of transcriptional control elements have been described to reduce the possibility of a unexpected toxic event. The strategies include suppression of basal expression from leaky inducible promoters, suppression of read-through transcription from cryptic promoters, tight control of plasmid copy numbers and proteins production as inactive (but reversible) forms [5,8-10].
Since the incomplete repression of promoter presents a major problem when cloning genes that encode lethal product to the bacterial host [11], it is obviously critical to tighten the control of gene expression by utilizing a tightly controllable promoter that permits E. coli normal growth until the very moment of highly toxic gene induction. Starting from this point, several tightly regulated expression circuits have been developed, such as Prha, hybrid Plac/ara-1, and PBAD-based vectors [8]. The widely used PBAD derivative expression systems have been verified as useful solutions for their stringentness in toxic protein production in E. coli. Expression is induced to high levels on media containing L-arabinose, and tightly shuts off on media with glucose but without L-arabinose, which shows more stringent regulation of target gene expression than other expression systems. However, some problems still exist in this system, such as few vectors available and catabolism repression by glucose [6].
Usually, environment-responsive promoters are commonly engineered in inducible expression system, such as promoters sensing pH, temperature, oxygen concentration or iron availability [12]. From iron-uptake systems that are evolved by bacteria growing in iron-limiting environment, many iron-related promoters have been reported [13-15]. The promoter from iron-uptake regulon is strongly repressed in iron rich conditions by Fur, but fully derepressed in absence of iron [16]. Usually, Fur protein complexing with ferrous irons binds with high affinity to the 19-bp inverted repeat consensus sequence known as the Fur box (GATAATGAT [A/T] ATCATTATC) in the relevant promoter area, which controls transcription of iron-responsive genes in microorganisms [17]. Fur inhibits transcription initiation by blocking the entry of RNA polymerase (RNAP) to the promoter. This ability is tightly dependent on the relative affinities of RNAP and Fur to binding sites in the DNA [17].
In this study, a novel tightly inducible expression system was developed as an alternative of the current ones. This system, designated pYPfhuA1, was capable of extremely tight regulation and allowed cloning of genes encoding highly toxic products within various copy-number plasmids. In detail, using E. coli Top10 as bacterial host, the strong iron-regulated promoter PfhuA was selected as the primary candidate. By modifying Fur box in promoter sequences and Fur repressor synthesis, the tightness of PfhuA was decreased to varying degrees. Thereinto, PfhuA1, which could be strict repressed by excess iron and efficiently induced by iron chelators, was the ideal promoter candidate for toxic gene expression in E. coli host.
Results
Preliminary screening for iron-regulated promoters
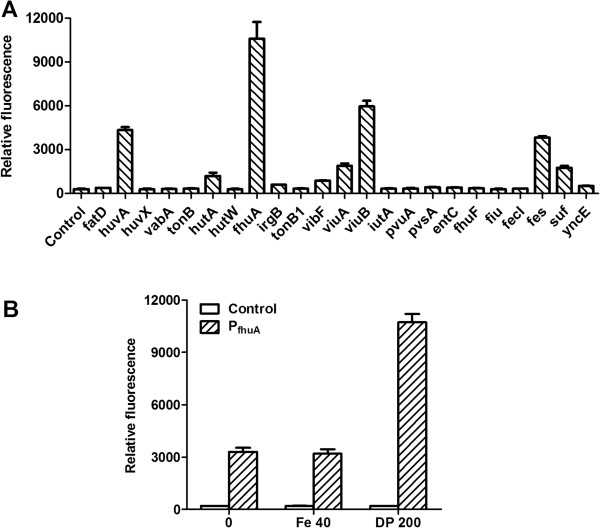
As described in the previous study [18], 23 promoter candidates were selected from Vibrio parahaemolyticus, Vibrio cholerae, E. coli and Vibrio anguillarum, and their transcription abilities were investigated with pUTtG as screening vector and iron chelator 2,2’-dipyridyl as inducer. Samples were adjusted to OD600 = 1 to measure the green fluorescence emitted by GFP, so the promoter strength and regulation could be simply correlated with the GFP fluorescence. As seen in Figure 1A, the promoters Psuf, Pfes, PhuvA, PfhuA, PviuB, and PviuA showed relatively high transcription activities with the relative fluorescence (RF) units of over 1,500 under iron-limiting medium. Among them, PfhuA owned the highest RF units of 10595, and was chosen as the primary candidate for further study.
Figure 1.
GFP expressions under different promoters in iron-limitation (A), under PfhuA in variant iron concentrations (B). A.E. coli Top10 transformants harboring ptPXG were cultured in LB medium to OD600 = 0.8 and induced by 200 μM 2,2’-dipyridyl. Samples were taken at 20 h after induction for GFP assay; B. LB medium with 40 μM FeSO4 as iron-rich condition, LB medium with 200 μM 2,2’-dipyridyl as iron-limiting condition. The error bars represent the standard deviation (SD) for one independent experiment, performed in triplicate, the experiment was repeated for three times.
To evaluate the regulation performance of PfhuA, the GFP synthesis was detected when Top10/ptPfhuAG growing in LB medium supplemented with repressor (40 μM FeSO4) or inducer (200 μM 2,2’-dipyridyl). As shown in Figure 1B, even with the repressor addition, an obvious leaky expression was detected under PfhuA transcription. Based on the data, the induction ratio that showed the tightness of promoter was calculated as 3.4, indicating that the tightness of PfhuA had to be improved for the toxic gene expression.
Modifications of PfhuA to improve its tightness
PfhuA was from V. cholerae ferrichrome outer membrane receptor encoded gene fhuA[19]. Its characteristic regions were demonstrated as the same result by three online promoter prediction tools-Softberry, BDGP and SCOPE. The Fur box [20] spans across −10 region (Figure 2). To improve PfhuA tightness, the Fur box sequences were remolded according to E. coli consensus Fur box sequence GATAATGAT[A/T]ATCATTATC [16]. As shown in Figure 2, Strategy 0 represented the original PfhuA sequence. In Strategy 1, the original Fur box of PfhuA was changed into E. coli consensus Fur box sequence by base substitution. In Strategy 2, Fur box was substituted by enhanced E. coli Fur box as described by Escolar et al. [17]. The enhanced Fur box contained five repeats of GATAAT motif to strengthen the Fur protein binding. In Strategy 3, two Fur boxes were used. One was the original one spanning −10 region, and the other one was E. coli consensus Fur box sequence across −35 region without changing the spacer distance between −35 and −10 regions. In Strategy 4, the fur fragment without Pfur was inserted into the upstream of PfhuAgfp TT cassette for more Fur protein synthesis. In Strategy 5, with the same aim, the fur fragment including Pfur was inserted. In theory, more Fur proteins would be synthesized in Strategy 5 than in Strategy 4. All the above-mentioned strategies aimed to enhance the affinity of Fur to the promoter and increase the transcription tightness sequentially.
Figure 2.
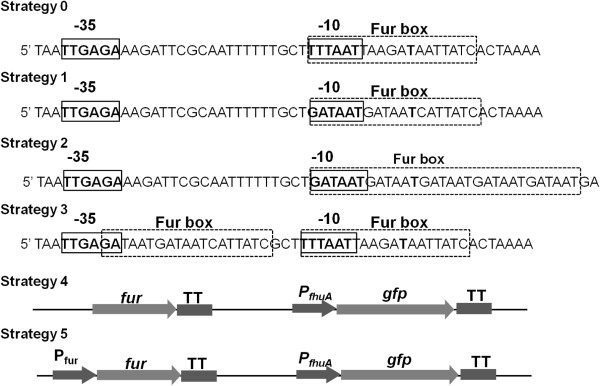
PfhuA modified strategies used in this study. Continuous line frame, -35 or −10 region; dash line frame, Fur binding box; PfhuA, fhuA promoter; TT, transcriptional terminator rrnBT1T2 derived from the rrnB rRNA operon of E. coli.
All the remolded recombinant plasmids were transformed into E. coli Top10, resulting in Top10/ptPfhuAG (Strategy 0), Top10/ptPfhuA1G (Strategy 1), Top10/ptPfhuA2G (Strategy 2), Top10/ptPfhuA3G (Strategy 3), Top10/ptPfhuAGSDfur (Strategy 4), Top10/ptPfhuAGPfurSDfur (Strategy 5) to check the promoter regulation behaviors. As shown in Table 1, the relative fluorescence units under repression and induction were used to calculate the induction ratio and relative induction fold. The highest relative induction fold of 51 and the lowest induction ratio of 3 were obtained with PfhuA. By replacing the original Fur box with E. coli consensus Fur box in Strategy 1, the repression of PfhuA was increased greatly from 3 to 101, but the induction was decreased by almost half of original value. The similar performances were also obtained in other strategies. Especially Strategy 2, 3, 5 displayed the high tightness under detection limit, but their relative induction folds were as low as 9, 3 and 12, respectively, which meant very limited transcription efficiency. As in Strategy 4, more GFPs were induced than in Strategy 5 following by less Fur protein synthesis, and showed the induction ratio of 108, and relative induction fold of 17. Under the same condition, Strategy 1 was comparable with the well-known tight regulated promoter PBAD with the induction ratio of 126 and relative induction fold of 30. Taken together, there was a balance between the tightness and transcription efficiency, and the improvement of tightness would be accompanied by the decrease of transcription efficiency. Taking these two criteria into consideration, PfhuA1 in Strategy 1, performed similarly as PBAD, was designated as the final promoter candidate for the next experiments.
Table 1.
Comparison of different promoter modified strategies
| Strategy | Modified strategies and descriptions | Repression RF | Induction RF | Induction ratio | Relative induction fold |
|---|---|---|---|---|---|
| NC |
E. coli Top10 |
200 ± 10 |
200 ± 9 |
0 |
1 |
| PC |
PBAD |
260 ± 11 |
7812 ± 31 |
126 |
30 |
| 0 |
Original fhuA promoter |
3292 ± 20 |
10116 ± 37 |
3 |
51 |
| 1 |
Changed the Fur box in −10 region into a conserved Fur box from E. coli[17] |
250 ± 12 |
5466 ± 26 |
101 |
27 |
| 2 |
Modified the Fur box in −10 region into an enhanced Fur box [17] |
196 ± 5 |
1885 ± 17 |
BDL |
9 |
| 3 |
Integrated a designed Fur box in −35 region |
200 ± 9 |
662 ± 19 |
BDL |
3 |
| 4 |
Inserted the SD-fur-TT circuit in the vector |
230 ± 9 |
3464 ± 24 |
108 |
17 |
| 5 | Inserted the Pfur-SDfur-TT circuit in the vector | 195 ± 7 | 2389 ± 26 | BDL | 12 |
Repression RF = Relative fluorescence units under repression condition (LB + 40 μM FeSO4);
Induction RF = Relative fluorescence units under induction condition (LB + 200 μM 2,2’-dipyridyl);
Induction ratio = (Induction RF – NC’s Induction RF)/ (Repression RF- NC’s Repression RF), indicating the tightness of promoter;
Relative induction fold = Induction RF/NC’s Induction RF, indicating the transcription efficiency of promoter;
BDL, beyond detection limit; NC, Negative control; PC, Positive control.
Performance evaluation of pPfhuA1 as an inducible tightly regulated expression system
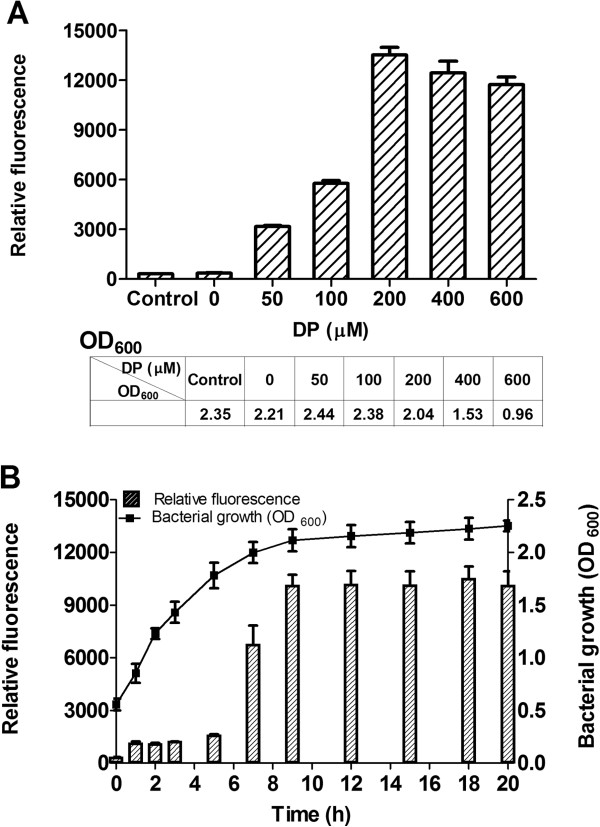
The ability to modulate the target gene expression by partial induction of the promoter activity by applying different concentrations of inducer is a desirable feature for a controllable expression system. To investigate the possibility to modulate the PfhuA1-initiated target gene expression in an inducer concentration-dependent manner, the specific GFP yields were measured from the cells of transformed with ptPfhuA1G induced with various concentrations of 2,2’-dipyridyl (0, 50, 100, 200, 400, and 600 μM). In Figure 3A, an exponential GFP yields response was observed with 2,2’-dipyridyl in the range of 0 to 200 μM, which culminated in a wide range of GFP synthesis with the relative fluorescence units from 350 to 13520. The maximal specific GFP yield was obtained in 200 μM 2,2’-dipyridyl. However, as 2,2’-dipyridyl concentration increased to 400 and 600 μM, GFP yields decreased gradually. This could be attributed to the harsh iron-limiting condition created by high concentration of 2,2’-dipyridyl for chelating divalent cations, which impaired the E. coli growth for the difficulty to uptake enough iron from the medium. It could be found that 2,2’-dipyridyl less than 200 μM exerted little influence on E. coli growth, and the OD600 values at the end of induction were between 2.0 and 2.5. As 2,2’-dipyridyl concentration continued to increase to 400 μM and 600 μM, OD600 value drastically decreased to 1.5 and 1.0, respectively. Therefore, 0 ~ 200 μM 2,2’-dipyridyl could partially induce and 200 μM was the preferable inducer concentration to fully switch on ptPfhuA1 expression system and to minimize the harmful effect of the inducer on E. coli growth.
Figure 3.
PfhuA1-controlled GFP expression induced by 2,2’-dipyridyl. A. GFP expression levels of E. coli Top10/ptPfhuA1 under the induction of different 2,2’-dipyridyl concentrations. OD600 in the table represented the OD600 value of cultures at 20 h post-induction. B. Time course analysis of GFP expression in E. coli Top10/ptPfhuA1 after induction with 200 μM 2,2’-dipyridyl. 2,2’-dipyridyl was added at 0 h. The error bars represent the standard deviation (SD) for one independent experiment, performed in triplicate, the experiment was repeated for three times.
In order to see the time response of ptPfhuA1 by which it is effectively turned on, GFP yields along with E. coli growth were detected. GFP expression in the ptPfhuA1-bearing culture was continuously increased until 9 h post induction (Figure 3B). During the first 1 h induction, the relative fluorescence value increased by 3.4 folds, which meant the expression circuit had been switched on. Afterwards, the GFP expression increased very slowly from 1 to 5 h, and underwent a jump from 5 to 9 h. At last, the GFP expression achieved its peak at 9 h post induction, corresponding to the beginning of stationary phase in E. coli growth. Thus, the whole expression circle for PfhuA1 lasted for 9 h. In other side, the GFP synthesis always lagged behind the E. coli growth. This is desirable in toxic gene expression to reduce the damage of protein products to cells.
Toxic protein expression under the control of modified PfhuA1
Based on the tight regulatory control in GFP expression, the potential of pYPfhuA1 in clone and expression of highly toxic gene products need to be verified. First, in order to check whether the basal expressions from PfhuA1 in different copy numbered vectors would allow the highly toxic protein synthesis in E. coli, pUT (pUC ori., high copy), pUTb (pBBR1 ori., middle copy), and pUTa (p15A ori., low copy) [21] were used as the back plasmids in toxic gene expression. Experimentally, toxic gene was inserted into the back plasmids with high-, middle- and low- copy number origins. Here the lysis gene of E. coli phage φX174, E, was selected as toxic gene, and its trace expression could induce lysis by formation of a transmembrane tunnel structure in the cell envelope of E. coli[22]. 200 μM 2,2’-dipyridyl as the optimal dose was added into LB medium for induction. As shown in Figure 4A, the similar growths of the strains bearing ptPfhuA1E, pbPfhuA1E and paPfhuA1E were observed as the control strain in medium with repressor. Because of extreme sensibility of E. coli to E protein [22], the normal growth of these strains indicated the tight control of PfhuA1 activity in different copy-numbered vectors under the repression condition. However, under the induction conditions, their behaviors were diverse from each other. After Top10/ptPfhuA1E, Top10/pbPfhuA1E and Top10/paPfhuA1E were induced at 0.48, 0.49 and 0.52 OD600, the highest OD600 (0.66, 0.95, and 1.15) was achieved 1, 2, and 2.5 h later, and the lowest OD600 (0.15, 0.21, and 0.23) appeared at 5, 6, and 7 h post induction, respectively. The differences of lysis performances caused by E protein production reflected the copy-number variant of vectors bearing PfhuA1. Sorting from fast to slow, although three strains with ptPfhuA1E, pbPfhuA1E, and paPfhuA1E lysed in 5, 6, and 7 h, respectively, their lysis kinetics were in similar mode. This kind of typical lysis curves indicated that E expression was under a controllable promoter. As a result, PfhuA1 is tight enough to regulate high toxic gene expression in different copy-number vectors.
Figure 4.
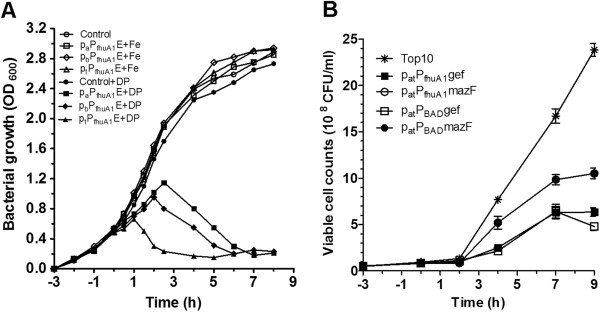
E. coli growth with toxic gene expression under the control of PfhuA1. A. Growth curves to demonstrate the tightly controllable expression of lysis gene E in ptPfhuA1E, pbPfhuA1E and paPfhuA1E under repression (+Fe, 40 μM FeSO4), or induction (+DP, 200 μM 2,2’dipyridyl). At −3 h, 40 μM FeSO4 was added; at 0 h, 200 μM 2,2’dipyridyl was added. Cultures were in duplicate. Bacterial growth was measured at OD600 nm. B. Colony forming units of Top10/patPfhuA1gef and Top10/patPfhuA1mazF after induction. At 0 h, 200 μM 2,2’dipyridyl was added. The error bars represent the standard deviation (SD) for three independent experiments, performed in triplicate.
With the successful application of pYPfhuA1 in the regulation of E gene expression, its broad availability was the next concern. Then, other two toxic proteins were synthesized using pYPfhuA1 in E. coli host. The gef encoding Gef of E. coli belongs to the hok killer gene family in Gram-negative bacteria and its expression kills the cell from the inside by interfering with a vital function in the cell membrane [23]. The mazF from a toxin-antitoxin module mazEF specifies a stable toxin that cleaves mRNA at a specific site(s) which is responsible for programmed cell death in E. coli[24]. Here patPfhuA1 was used, which is a middle-low-copy number vector with ~30 copies per cell [21]. Colony forming units (CFUs) of the transformants were detected in the presence of inducer (Figure 4B). Comparing with the negative control Top10, all the recombinant strains were grown similarly in the presence of iron (data are not shown), and no leaky expression phenotype was appeared. However, the growth of Top10/patPfhuA1gef and Top10/patPfhuA1mazF was repressed when cultured with 2,2’-dipyridyl as the result of gef and mazF expression. At 0 h, 2,2’-dipyridyl was added. During the first 2 h after induction, all three strains performed to adapt the iron-limiting condition and maintained in a similar density of 109 CFU/ml. However, after 2 h, their growth behaviors were obviously different. Top10 strain grew fast to 2.4×109 CFU/ml in exponential way until 9 h post induction. However, the strains Top10/patPfhuA1gef and Top10/patPfhuA1mazF grew exponentially until 7 h post induction and then kept at 6×108 CFU/ml and 1×109 CFU/ml, respectively. Therefore, the growth repression shown by recombinant strains revealed that toxic genes gef and mazF were functionally expressed after induction under the control of PfhuA1. Furthermore, pYPfhuA1 could be applied in different toxic gene expressions.
Discussion
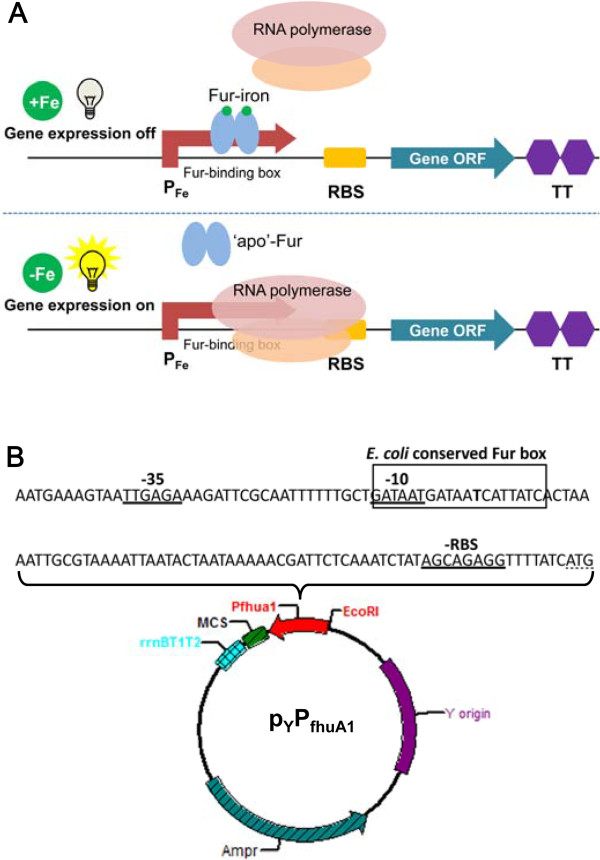
The wide variety of applications of recombinant proteins and genetically engineered E. coli signifies the increasing demand for varied regulatory circuits. The challenges of regulatory expression technology are multifaceted to meet the growing need, in terms of quantities, qualities, cost-effectiveness and some particular cases. When even a low level of gene expression is detrimental to bacterial growth, the expression system has to be tightly regulated and efficiently shut off in the absence of inducer. In this work, a modified Fur-dependent and iron-limiting inducible expression system was constructed, which owns several prominent features and benefits that confers it an attractive and versatile expression system for highly toxic foreign protein synthesis. In this iron regulation system (Figure 5A), abundant iron complexing with Fur repressor binds to PfhuA1 sequence to prevent RNA polymerase contacting with the promoter region and to shut down the transcription. In the other side, without enough iron, Fur is disassociated from the Fur box of PfhuA1 sequence to open a place for RNA polymerase and to switch on the expression of downstream gene. This system is inducible by iron chelator addition. Most of all, the induction ratio was improved via enhancing the binding capacity between the repressor Fur and PfhuA1, which made the system was tight enough to express different kinds of lethal genes in E. coli. As shown in Figure 5B, the ultimate plasmid was named as pYPfhuA1. According to different purposes of more or less recombinant protein needed, Y site could be replaced by high or low copy-numbered origin. Although the increased copy number of those vectors might cause higher levels of leaky expression, it was proved that high-copied ptPfhuA1 could successfully express highly toxic gene, such as E in E. coli. Besides, the exact PfhuA1 sequence and key regions of the plasmid were also shown. A strong rrnBT1T2 terminator derived from the rrnB rRNA operon of E. coli located at downstream of multi-cloning sites. Taken together, when designing these vectors for toxic gene expression, the following properties had to be taken into account: a strong promoter capable of tightly regulation, strong terminators against read-through transcription from other plasmid-borne promoters, and different copy numbered origins of replication for gene expression with different degrees of toxicity. In all, this system is easily inducible, tightly controllable and potentially versatile for biotechnological application.
Figure 5.
A. A schematic diagram of the mechanism of tight Fur-dependent iron-limitation regulation switch developed for E. coli stains. The mechanism is described in Results and Discussion. B. Physical map of the pYPfhuA1 expression vector. Y, different replicate origins; MCS, multi-cloning sites; rrnBT1T2, ribosomal terminators T1 and T2.
One of the most notable merits for this system is its tight regulation by iron-limitation signal. Escolar et al. once pointed out that the actual sequences recognized by Fur consisted of a minimal array of three conserved 6 bp (GATAAT) units which could be extended laterally by discrete additions of repeats of the same unit, thereby giving rise to new sites to which the repressor binds with a range of affinities [17]. It was assumed that the affinity would vary depending on both the number of repeats present on each operator and the conservation of their sequences, which would allow an entire range and hierarchy of transcription responses depending on small changes in the iron status in the cell [17]. Based on this hypothesis, we designed three types of Fur boxes differing in sequence conservation and 6-bp unit repeat number. It was found that the more repeats of conserved 6-bp units or the more conserved Fur box, the stronger affinity between Fur and the promoter, which was following by the tighter promoter. This verified the hypothesis mentioned above. In another side, this strategy also provides a new way for modifying other inducible promoters. However, a problem in this strategy is that the transcription efficiency decreases along with the tightness improvement. This is in common, because most of the regulatory systems have an intrinsic limitation in the range of induction, and attempts to mutate promoters to reduce basal expression usually result in concomitant reduction of induced levels. To overcome this drawback, Royo et al. used the nasF attenuator and the NasR-dependent anti-termination system from Klebsiella oxytoca to construct a novel expression circuit which could conditionally prevent undesired transcription from any transcriptional initiation signal, while keeping induced levels intact [10,25]. This point of view gives us a new reference for further optimization of expression vector constructed here.
Although iron is one of the most abundant elements on Earth, in aerobic environments it is predominantly found as ferric (hydro)oxides that are relatively insoluble at neutral pH, and thus, ionic ferric (Fe3+) concentrations are exceedingly low [26]. Therefore, as an iron limitation responsive vector, pYPfhuA1 has great potential in developing ‘suicidal’ containment system for environmentally relevant application. Furthermore, in our previous research, iron-limiting condition was verified as an in vivo stimulating signal, and promoters derived from iron uptake system could be induced in vivo[18]. Therefore, this means PfhuA1 could be used in the construction of in vivo inducible antigen synthesis system, which could alleviate the toxicity or metabolic burden of the host strain and improve the immunogenicity for bacterial vector vaccine [27-32]. Moreover, in vivo regulated and delayed attenuation strategy has been developed for maintaining the invasive abilities of bacterial vector to the greatest extent, such that the recombinant vaccine has the ability as virulent wild-type strain to reach effector lymphoid tissues before display of attenuation to preclude onset of any disease symptoms [33]. pYPfhuA1 could also be applied in construction of in vivo delayed attenuation vaccine. In all, pYPfhuA1 could be induced by iron chelators in medium, in vitro aerobic environments and in vivo animal host, and has extensive application in biotechnology area.
Conclusions
In all, as the expression vector particularly developed for toxic protein synthesis, pYPfhuA1 owns its notable advantages, which include easy control, tight regulation, relatively high efficiency and multi-environments response potential. Consequently, pYPfhuA1 has great potential in functional study of toxic genes, construction of biological containment system, or bacterial vector vaccine development.
Methods
Bacterial strains and growth conditions
The common clone vector E. coli Top10F’ (F’[lacIq Tn10 (TetR)] mcrA Δ(mrr-hsdRMS-mcrBC) Φ80 lacZ ΔM15 ΔlacΧ74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG, Invitrogen) was chosen as bacterial host. The plasmids used in this study are listed in Table 2. E. coli strains were grown at 37°C in lysogeny broth (LB) medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl) [34]. When required, ampicillin (Amp, 100 μg/ml), FeSO4 (40 μM) and/or different concentrations of 2,2’-dipyridyl were added.
Table 2.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pUT |
pUC18 derivative with rrnBT1T2 terminator, Apr |
[18] |
| pUTtG |
pUT derivative, with a reporter gene gfp, Apr |
[18] |
| pUTb |
pUC18 derivative with rrnBT1T2 terminator, pBBR1 ori., Apr |
[21] |
| pUTa |
pUC18 derivative with rrnBT1T2 terminator, p15A ori., Apr |
[21] |
| pUTat |
pUC18 derivative with rrnBT1T2 terminator, pAT153 ori., Apr |
[21] |
| ptPfhuAG |
pUT derivative containing PfhuAgfp TT, Apr |
This study |
| ptPfhuA1G |
pUT derivative containing PfhuA1gfp TT, Apr |
This study |
| ptPfhuA2G |
pUT derivative containing PfhuA2gfp TT, Apr |
This study |
| ptPfhuA3G |
pUT derivative containing PfhuA3gfp TT, Apr |
This study |
| ptPfhuAGSDfur |
pUT derivative containing PfhuAgfp TT and SD-fur TT from E. coli, Apr |
This study |
| ptPfhuAGPfurSDfur |
pUT derivative containing PfhuAgfp TT and Pfurfur TT from E. coli, Apr |
This study |
| ptPfhuA1E |
pUT derivative containing PfhuA1E TT, Apr |
This study |
| pbPfhuA1E |
pUTb derivative containing PfhuA1E TT, Apr |
This study |
| paPfhuA1E |
pUTa derivative containing PfhuA1E TT, Apr |
This study |
| patPfhuA1E |
pUTat derivative containing PfhuA1E TT, Apr |
This study |
| patPfhuA1gef |
pUTat derivative containing PfhuA1gef TT, Apr |
This study |
| patPfhuA1mazF | pUTat derivative containing PfhuA1mazF TT, Apr | This study |
Plasmid construction
A reporter plasmid pUTtG constructed previously was used as the promoter screening vector [18]. The primers from the former work were applied to amplify the candidate promoters from bacterium chromosomes or plasmids [18]. The amplified promoter products, possessing the native start codon, Shine-Dalgarno sequence, and −35 and −10 promoter elements plus additional upstream bases, were inserted into pUTtG, and the resultant plasmids were transformed into E. coli Top10 named Top10/ptPXG (X mean different promoters) for iron-regulated promoter screening.
The primers listed in Table 3 were used to modify the PfhuA promoter sequences and its regulation. PfhuA1 and PfhuA3 fragments were all amplified from the original PfhuA sequence in the previous work [18], and PfhuA2 (TAATGAAAGTAATTGAGAAAGATTCGCAATTTTTTGCTGATAATGATAATGATAATGATAATGATAATGAAAAATTAATACTAATAAAAACGATTCTCAA) was synthesized by Life Technologies (Shanghai, China). The three promoter fragments fused with their relevant GFPs by overlap polymerase chain reaction (PCR), respectively. The resultant fragment was ligated into PvuII/EcoRI digested plasmid ptPfhuAG. The 276 bp E gene was amplified from PhiX174 genome, and the amplification of 153 bp gef, 336 bp mazF fragments were used E. coli DH5α (F-,φ80dlacZΔM15, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rk-,mk+), phoA, supE44, λ-, thi-1, gyrA96, relA1, Invitrogen) chromosome as templates. The modified PfhuA1 was inserted into EcoRI/BamHI sites of pUT, pUTb, pUTa and pUTat, and then the resultant plasmids were ligated with BamHI/PstI digested toxic genes E, gef, mazF to get recombinant plasmids ptPfhuA1E, pbPfhuA1E, paPfhuA1E, patPfhuA1gef and patPfhuA1mazF (Table 2). At last, all the recombinant plasmids were transformed into E. coli Top10 for PfhuA regulation test.
Table 3.
PCR primers used in this study
| Name | Sequence (5’-3’)a |
|---|---|
| PFhuA1-for |
CGGAATTCCAGTACCAGTGCCGCCATCGTCCA |
| PFhuA1-rev |
TGATTATCATTATCAGCAAAAAATTGCGAATCTTTCTC |
| GFP1-for |
ATAATGATAATCATTATCACTAAAATTGCGTAAAATTAAT |
| GFP1-rev |
AAAACTGCAGTTATTTGTACAGTTCATCCATGCCA |
| PFhuA2-for |
CGGAATTCCCTTAATGAAAGTAATTGAGAAAGAT |
| GFP2-for |
AACGATTCTCAAATCTATAGCAGAGGTTTTATCAT |
| GFP2-rev |
AAAACTGCAGTTATTTGTACAGTTCATCCATGC |
| PFhuA3-for |
CGGAATTCACCAGTACCAGTGCCGCCATCGTC |
| PFhuA3-rev |
TAATGATTATCATTATCTCAATTACTTTCATTAAGGCACC |
| GFP3-for |
ATGATAATCATTATCGCTTTTAATTAAGATAATTATCACT |
| GFP3-rev |
AAAACTGCAGTTATTTGTACAGTTCATCCATGCCAT |
| Fur-for |
CAGCTGGAGCAAATTCTGTCACTTCTTCTA |
| Fur-rev |
CGGAATTCATAAGTGAGAGCTGTAACTCTCGC |
| PFur-for |
CAGCTGCTGGCTGATGATGACCACTTTGT |
| PFur-rev |
CGGAATTCATAAGTGAGAGCTGTAACTCTCGC |
| E-for |
CGCGGATCCATGGTACGCTGGACTTTGTGGGA |
| E-rev |
AAAACTGCAGTCACTCCTTCCGCACGTAATTTT |
| gef-for |
CGCGGATCCATGAAGCAGCATAAGGCGATGATTG |
| gef-rev |
AAAACTGCAGTTACTCGGATTCGTAAGCCGTGAAA |
| mazF-for |
CGCGGATCCATGGTAAGCCGATACGTACCCGATAT |
| mazF-rev | AAAACTGCAGCTACCCAATCAGTACGTTAATTTT |
a Restriction enzyme sites used for cloning of PCR products are underlined.
General DNA procedure and online analysis tools
General DNA operations were carried out following the standard protocols. Automated DNA sequencing and primer synthesis were completed by Life Technologies (Shanghai, China). PfhuA characteristic regions were predicted by online tools: BPROM-bacterial promoter prediction program from Softberry (http://linux1.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb), BDGP-Promoter (http://www.fruitfly.org/seq_tools/promoter.html) and SCOPE-Suite for Computational identification Of Promoter Elements [35].
GFP synthesis detection
Overnight cell cultures were inoculated (1:100, v/v) into fresh LB medium containing appropriate antibiotics and cultured in a shaker at 200 rpm and 37°C. At middle log phase typically with an optical density at OD600 = 0.8-1.0, 2, 2’-dipyridyl was added to induce the expression of GFP. After 20 hours or defined time points of iron-limiting induction, 1 ml cell culture sample was taken, centrifuged at 12,000 g for 3 min, washed and resuspended in PBS (pH 7.2) to the same OD600 value (OD600 = 1.0). For each sample, 100 μl of cell suspension was added into a 96-well flat-bottom polystyrene plate (Costar, USA) and measured with a fluorescence plate reader (TECAN, GENios Pro, Austria). Excitation wavelength was set at 485 nm and emission was detected at 535 nm.
Toxic gene expression detection
The E. coli strains Top10/ptPfhuA1E, Top10/pbPfhuA1E, Top10/paPfhuA1E, Top10/patPfhuA1gef and Top10/patPfhuA1mazF were overnight grown at 37°C in LB medium supplemented with 40 μM FeSO4 to ensure tight repression of toxic gene. To induce toxic gene expression, the culture was diluted to an OD600 of 0.1 and cultured in a shaker at 200 rpm and 37°C. At early log phase (OD600 = 0.3-0.4), 2,2’-dipyridyl was added into culture to create iron-limiting condition. Cell samples were taken at different time points after induction to measure both the OD600 and the colony formation units (CFUs) to determine the bacteria growth in iron-limiting medium.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LG participated in the design of the study, carried out the cloning studies, performed the statistical analysis and drafted the manuscript. QL conceived of the study, helped to draft the manuscript and coordinated the study. CL carried out the molecular genetics and expression studies. YZ helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Lingyu Guan, Email: lingyuguan@gmail.com.
Qin Liu, Email: qinliu@ecust.edu.cn.
Chao Li, Email: momo0519777@163.com.
Yuanxing Zhang, Email: yxzhang@ecust.edu.cn.
Acknowledgements
This work was supported by the joint project, National Natural Science Foundation of China- Austrian Science Fund (30811130545).
References
- Kimelman A, Levy A, Sberro H, Kidron S, Leavitt A, Amitai G, Yoder-Himes DR, Wurtzel O, Zhu Y, Rubin EM. A vast collection of microbial genes that are toxic to bacteria. Genome Res. 2012;22(9):1–8. doi: 10.1101/gr.133850.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh JS, Abhilash P, Singh H, Singh RP, Singh D. Genetically engineered bacteria: An emerging tool for environmental remediation and future research perspectives. Gene. 2011;480(1–2):1–9. doi: 10.1016/j.gene.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Atlas RM. Molecular methods for environmental monitoring and containment of genetically engineered microorganisms. Biodegradation. 1992;3(2–3):137–146. doi: 10.1007/BF00129079. [DOI] [PubMed] [Google Scholar]
- Molin S. Environmental potential of suicide genes. Curr Opin Biotechnol. 1993;4(3):299–305. doi: 10.1016/0958-1669(93)90099-I. [DOI] [PubMed] [Google Scholar]
- Saida F, Uzan M, Odaert B, Bontems F. Expression of highly toxic genes in E. coli: special strategies and genetic tools. Curr Protein Pept Sci. 2006;7(1):47–56. doi: 10.2174/138920306775474095. [DOI] [PubMed] [Google Scholar]
- Jana S, Deb J. Strategies for efficient production of heterologous proteins in Escherichia coli. Appl Microbiol Biotechnol. 2005;67(3):289–298. doi: 10.1007/s00253-004-1814-0. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Morel L, Le Francois T, Bourque D, Bourget L, Groleau D, Massie B, Miguez CB. Novel, versatile, and tightly regulated expression system for Escherichia coli strains. Appl Environ Microbiol. 2010;76(15):5058–5066. doi: 10.1128/AEM.00413-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacalone MJ, Gentile AM, Lovitt BT, Berkley NL, Gunderson CW, Surber MW. Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. Biotechniques. 2006;40(3):355–363. doi: 10.2144/000112112. [DOI] [PubMed] [Google Scholar]
- Glascock C. J Weickert M. Using chromosomal lacIQ1 to control expression of genes on high-copy-number plasmids in Escherichia coli. Gene. 1998;223(1):221–231. doi: 10.1016/S0378-1119(98)00240-6. [DOI] [PubMed] [Google Scholar]
- Royo JL, Manyani H, Cebolla A, Santero E. A new generation of vectors with increased induction ratios by overimposing a second regulatory level by attenuation. Nucleic Acids Res. 2005;33(19):e169–e169. doi: 10.1093/nar/gni168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony LC, Suzuki H, Filutowicz M. Tightly regulated vectors for the cloning and expression of toxic genes. J Microbiol Methods. 2004;58(2):243–250. doi: 10.1016/j.mimet.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Voigt CA. Genetic parts to program bacteria. Curr Opin Biotechnol. 2006;17(5):548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Allen CE, Schmitt MP. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J Bacteriol. 2009;191(8):2638–2648. doi: 10.1128/JB.01784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffelner H, Rieder G, Haas R. Helicobacter pylori vaccine development: Optimisation of strategies and importance of challenging strain and animal model. Int J Med Microbiol. 2008;298(1–2):151–159. doi: 10.1016/j.ijmm.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Lim JM, Hong M, Kim S, Oh D, Kang H, Kwon O. Iron chelator-inducible expression system for Escherichia coli. J Microbiol Biotechnol. 2008;18(8):1357–1363. [PubMed] [Google Scholar]
- Baichoo N, Helmann JD. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol. 2002;184(21):5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolar L, Pérez-Martín J, de Lorenzo V. Binding of the Fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J Mol Biol. 1998;283(3):537–547. doi: 10.1006/jmbi.1998.2119. [DOI] [PubMed] [Google Scholar]
- Guan LY, Mu W, Champeimont J, Wang QY, HZh W, Xiao JF, Lubitz W, Zhang YX, Liu Q. Iron-regulated lysis of recombinant Escherichia coli in host releases protective antigen and confers biological containment. Infect Immun. 2011;79(7):2608–2618. doi: 10.1128/IAI.01219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MB, Sexton JA, DeCastro GJ, Calderwood SB. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J Bacteriol. 2000;182(8):2350–2353. doi: 10.1128/JB.182.8.2350-2353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect Immun. 2005;73(12):8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Liu Q, Chen H, Zhang Y. A stable plasmid system for heterologous antigen expression in attenuated Vibrio anguillarum. Vaccine. 2011;29(40):6986–6993. doi: 10.1016/j.vaccine.2011.07.036. [DOI] [PubMed] [Google Scholar]
- Witte A, Wanner G, Sulzner M, Lubitz W. Dynamics of PhiX174 protein E-mediated lysis of Escherichia coli. Arch Microbiol. 1992;157(4):381–388. doi: 10.1007/BF00248685. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Poulsen L, Thisted T, Nielsen A, Martinussen J, Andreasen P. The hok killer gene family in gram-negative bacteria. New Biol. 1990;2(11):946–956. [PubMed] [Google Scholar]
- Amitai S, Yassin Y, Engelberg-Kulka H. MazF-mediated cell death in Escherichia coli: a point of no return. J Bacteriol. 2004;186(24):8295–8300. doi: 10.1128/JB.186.24.8295-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina C, Camacho EM, Flores A, Mesa-Pereira B, Santero E. Improved Expression Systems for Regulated Expression in Salmonella Infecting Eukaryotic Cells. PLoS One. 2011;6(8):e23055. doi: 10.1371/journal.pone.0023055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm W, Morgan JJ. Aquatic chemistry. 3. New York, USA: John Wiley & Sons; 1996. [Google Scholar]
- Mu W, Guan L, Yan Y, Liu Q, Zhang Y. A novel in vivo inducible expression system in Edwardsiella tarda for potential application in bacterial polyvalence vaccine. Fish Shellfish Immunol. 2011;31(6):1097–1105. doi: 10.1016/j.fsi.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Loessner H, Endmann A, Leschner S, Bauer H, Zelmer A, Westphal K, Weiss S. zur Lage S. Improving live attenuated bacterial carriers for vaccination and therapy. Int J Med Microbiol. 2008;298(1–2):21–26. doi: 10.1016/j.ijmm.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Wang S, Li Y, Shi H, Sun W, Roland KL, Curtiss R III. Comparison of a regulated delayed antigen synthesis system with in vivo-inducible promoters for antigen delivery by live attenuated Salmonella vaccines. Infect Immun. 2011;79(2):937–949. doi: 10.1128/IAI.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John M, Crean TI, Calderwood SB, Ryan ET. In vitro and in vivo analyses of constitutive and in vivo-induced promoters in attenuated vaccine and vector strains of Vibrio cholerae. Infect Immun. 2000;68(3):1171–1175. doi: 10.1128/IAI.68.3.1171-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin CE, Kaper JB. Use of stabilized luciferase‐expressing plasmids to examine in vivo‐induced promoters in the Vibrio cholerae vaccine strain CVD 103‐HgR. FEMS Immunol Med Mic. 2009;57(1):69–79. doi: 10.1111/j.1574-695X.2009.00580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husseiny MI, Hensel M. Evaluation of an intracellular-activated promoter for the generation of live Salmonella recombinant vaccines. Vaccine. 2005;23(20):2580–2590. doi: 10.1016/j.vaccine.2004.11.035. [DOI] [PubMed] [Google Scholar]
- Curtiss R III, Xin W, Li Y, Kong W, Wanda S, Gunn B, Wang S. New technologies in using recombinant attenuated Salmonella vaccine vectors. Crit Rev Immunol. 2010;30(3):255–270. doi: 10.1615/CritRevImmunol.v30.i3.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Carlson JM, Chakravarty A, DeZiel CE. SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res. 2007;35(suppl 2):W259–W264. doi: 10.1093/nar/gkm310. [DOI] [PMC free article] [PubMed] [Google Scholar]