Abstract
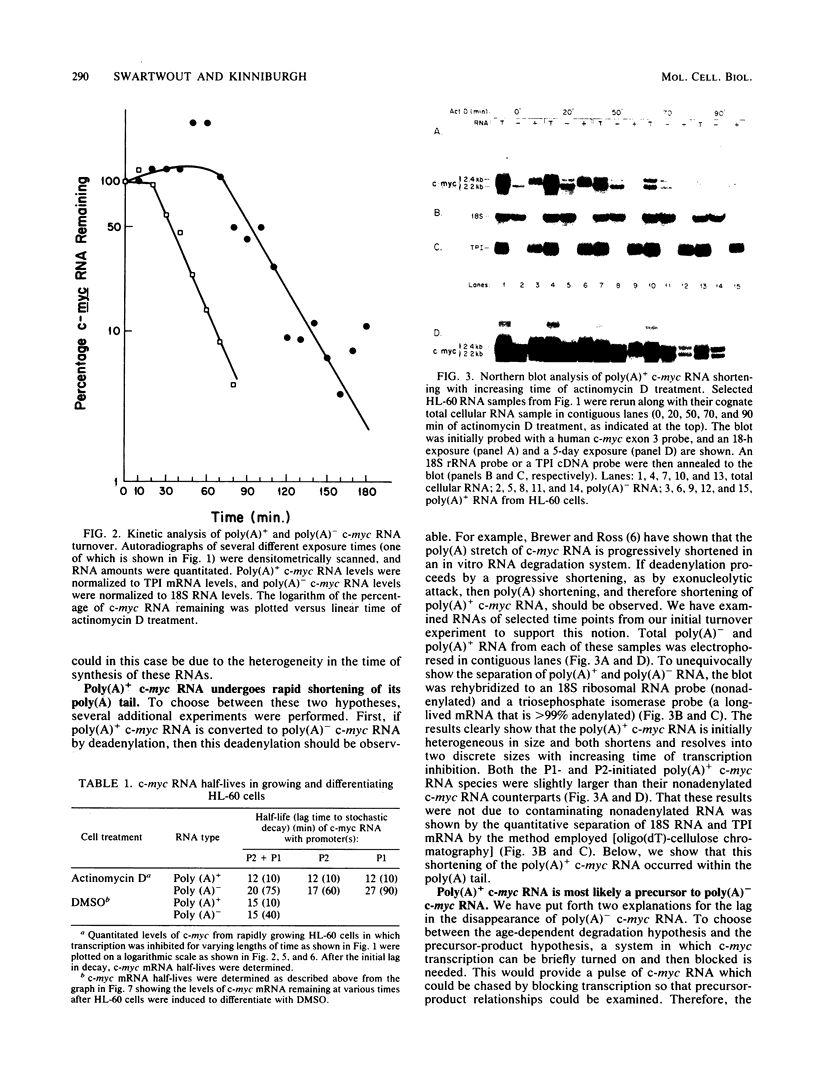
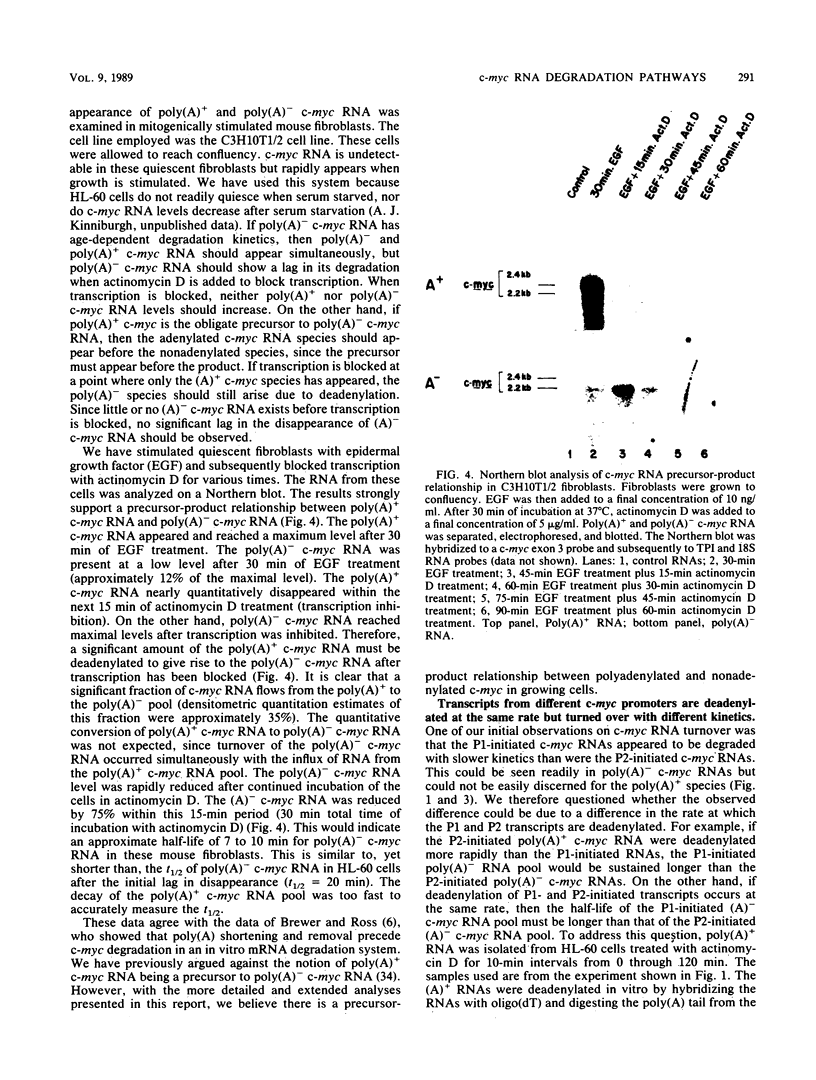
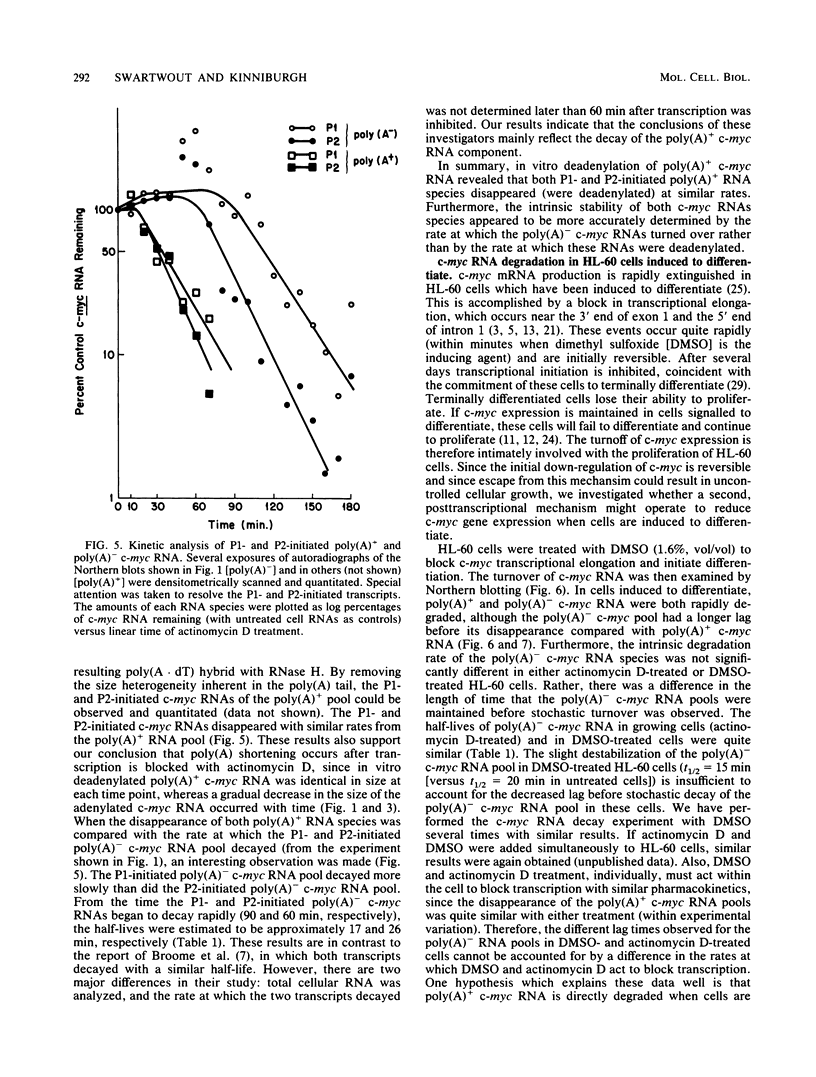
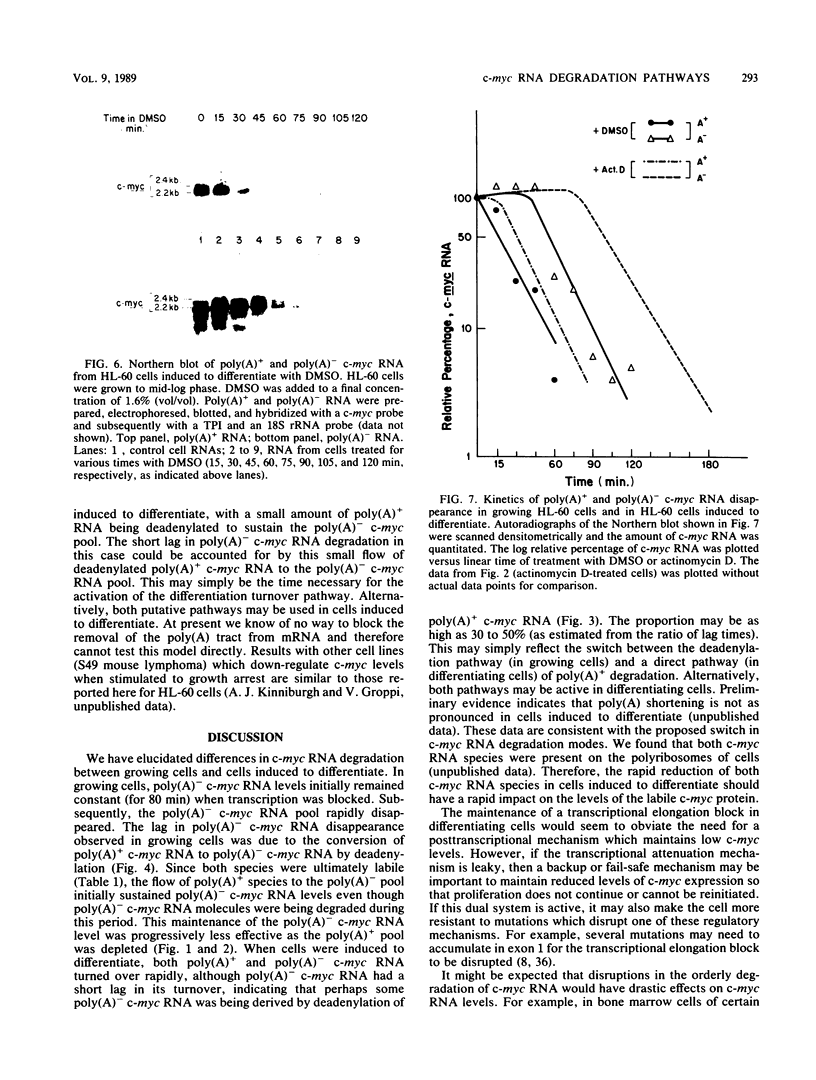
Transcripts of the proto-oncogene c-myc are composed of a rapidly degraded polyadenylated RNA species and an apparently much more stable, nonadenylated RNA species. In this report, the extended kinetics of c-myc RNA turnover have been examined in rapidly growing cells and in cells induced to differentiate. When transcription was blocked with actinomycin D in rapidly growing cells, poly(A)+ c-myc was rapidly degraded (t1/2 = 12 min). c-myc RNA lacking poly(A) initially remained at or near control levels; however, after 80 to 90 min it was degraded with kinetics similar to those of poly(A)+ c-myc RNA. These bizarre kinetics are due to the deadenylation of poly(A)+ c-myc RNA to form poly(A)- c-myc, thereby initially maintaining the poly(A)- c-myc RNA pool when transcription is blocked. In contrast to growing cells, cells induced to differentiate degraded both poly(A)+ and poly(A)- c-myc RNA rapidly. The rapid disappearance of both RNA species in differentiating cells suggests that a large proportion of the poly(A)+ c-myc RNA was directly degraded without first being converted to poly(A)- c-myc RNA. Others have shown that transcriptional elongation of the c-myc gene is rapidly blocked in differentiating cells. We therefore hypothesize that in differentiating cells a direct, rapid degradation of poly(A)+ c-myc RNA may act as a backup or fail-safe system to ensure that c-myc protein is not synthesized. This tandem system of c-myc turnoff may also make cells more refractory to mutations which activate constitutive c-myc expression.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985 Dec 12;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol Cell Biol. 1986 Oct;6(10):3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3' A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988 Apr;8(4):1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome H. E., Reed J. C., Godillot E. P., Hoover R. G. Differential promoter utilization by the c-myc gene in mitogen- and interleukin-2-stimulated human lymphocytes. Mol Cell Biol. 1987 Aug;7(8):2988–2993. doi: 10.1128/mcb.7.8.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E., Dalla-Favera R., Bentley D., Groudine M. Mutations in the first exon are associated with altered transcription of c-myc in Burkitt lymphoma. Science. 1987 Nov 27;238(4831):1272–1275. doi: 10.1126/science.3685977. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola J. A., Cole M. D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986 Apr 24;320(6064):760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- Dmitrovsky E., Kuehl W. M., Hollis G. F., Kirsch I. R., Bender T. P., Segal S. Expression of a transfected human c-myc oncogene inhibits differentiation of a mouse erythroleukaemia cell line. Nature. 1986 Aug 21;322(6081):748–750. doi: 10.1038/322748a0. [DOI] [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erikson J., Finger L., Sun L., ar-Rushdi A., Nishikura K., Minowada J., Finan J., Emanuel B. S., Nowell P. C., Croce C. M. Deregulation of c-myc by translocation of the alpha-locus of the T-cell receptor in T-cell leukemias. Science. 1986 May 16;232(4752):884–886. doi: 10.1126/science.3486470. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Holt J. T., Redner R. L., Nienhuis A. W. An oligomer complementary to c-myc mRNA inhibits proliferation of HL-60 promyelocytic cells and induces differentiation. Mol Cell Biol. 1988 Feb;8(2):963–973. doi: 10.1128/mcb.8.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M., Itani T., Yamaguchi M., Ariga H. c-myc protein can be substituted for SV40 T antigen in SV40 DNA replication. Nucleic Acids Res. 1987 Jun 25;15(12):4889–4899. doi: 10.1093/nar/15.12.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachman H. M., Skoultchi A. I. Expression of c-myc changes during differentiation of mouse erythroleukaemia cells. Nature. 1984 Aug 16;310(5978):592–594. doi: 10.1038/310592a0. [DOI] [PubMed] [Google Scholar]
- Langdon W. Y., Harris A. W., Cory S., Adams J. M. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell. 1986 Oct 10;47(1):11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- Morse B., Rotherg P. G., South V. J., Spandorfer J. M., Astrin S. M. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature. 1988 May 5;333(6168):87–90. doi: 10.1038/333087a0. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H., Leder P. Nuclear localization and DNA binding properties of a protein expressed by human c-myc oncogene. Science. 1984 Aug 17;225(4663):718–721. doi: 10.1126/science.6463648. [DOI] [PubMed] [Google Scholar]
- Preisler H. D., Kinniburgh A. J., Wei-Dong G., Khan S. Expression of the protooncogenes c-myc, c-fos, and c-fms in acute myelocytic leukemia at diagnosis and in remission. Cancer Res. 1987 Feb 1;47(3):874–880. [PubMed] [Google Scholar]
- Prochownik E. V., Kukowska J. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. 1986 Aug 28-Sep 3Nature. 322(6082):848–850. doi: 10.1038/322848a0. [DOI] [PubMed] [Google Scholar]
- Reitsma P. H., Rothberg P. G., Astrin S. M., Trial J., Bar-Shavit Z., Hall A., Teitelbaum S. L., Kahn A. J. Regulation of myc gene expression in HL-60 leukaemia cells by a vitamin D metabolite. Nature. 1983 Dec 1;306(5942):492–494. doi: 10.1038/306492a0. [DOI] [PubMed] [Google Scholar]
- Rothberg P. G., Erisman M. D., Diehl R. E., Rovigatti U. G., Astrin S. M. Structure and expression of the oncogene c-myc in fresh tumor material from patients with hematopoietic malignancies. Mol Cell Biol. 1984 Jun;4(6):1096–1103. doi: 10.1128/mcb.4.6.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Bressler P., Kelly K. Two distinct mechanisms of transcriptional control operate on c-myc during differentiation of HL60 cells. Mol Cell Biol. 1988 Feb;8(2):867–874. doi: 10.1128/mcb.8.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. L., Watt R. A., Sullivan N. F. The v- and c-myc oncogene proteins colocalize in situ with small nuclear ribonucleoprotein particles. Oncogene. 1987 Mar;1(1):5–12. [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Studzinski G. P., Brelvi Z. S., Feldman S. C., Watt R. A. Participation of c-myc protein in DNA synthesis of human cells. Science. 1986 Oct 24;234(4775):467–470. doi: 10.1126/science.3532322. [DOI] [PubMed] [Google Scholar]
- Swartwout S. G., Preisler H., Guan W. D., Kinniburgh A. J. Relatively stable population of c-myc RNA that lacks long poly(A). Mol Cell Biol. 1987 Jun;7(6):2052–2058. doi: 10.1128/mcb.7.6.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sümegi J., Spira J., Bazin H., Szpirer J., Levan G., Klein G. Rat c-myc oncogene is located on chromosome 7 and rearranges in immunocytomas with t(6:7) chromosomal translocation. Nature. 1983 Dec 1;306(5942):497–498. doi: 10.1038/306497a0. [DOI] [PubMed] [Google Scholar]
- Taub R., Kirsch I., Morton C., Lenoir G., Swan D., Tronick S., Aaronson S., Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac-Kaye M., Gelmann E. P., Levens D. A point mutation in the c-myc locus of a Burkitt lymphoma abolishes binding of a nuclear protein. Science. 1988 Jun 24;240(4860):1776–1780. doi: 10.1126/science.2454510. [DOI] [PubMed] [Google Scholar]