Abstract
MMP-9 (gelatinase B) participates in a variety of diverse physiologic and pathologic processes. We recently characterized a cyclooxygenase-2 (Cox-2)→PGE2→EP4 receptor axis that regulates macrophage MMP-9 expression. In the current studies, we determined whether MMPs, commonly found in inflamed and neoplastic tissues, regulate this prostanoid-EP receptor axis leading to enhanced MMP-9 expression. Results demonstrate that exposure of murine peritoneal macrophages and RAW264.7 macrophages to MMP-1 (collagenase-1) or MMP-3 (stromelysin-1) lead to a marked increase in Cox-2 expression, PGE2 secretion and subsequent induction of MMP-9 expression. Proteinase-induced MMP-9 expression was blocked in macrophages pre-incubated with the selective the Cox-2 inhibitor celecoxib or transfected with Cox-2 siRNA. Likewise, proteinase-induced MMP-9 was blocked in macrophages pre-incubated with the EP4 antagonist ONO-AE3-208 or transfected with EP4 siRNA. Exposure of macrophages to MMP-1 and MMP-3 triggered the rapid release of TNF-α, which was blocked by MMP-inhibitors. Furthermore, both Cox-2 and MMP-9 expression were inhibited in macrophages pre-incubated with anti-TNF-α IgG or transfected with TNF-α siRNA. Thus, proteinase-induced MMP-9 expression by macrophages is dependent on the release of TNF-α, induction of Cox-2 expression and PGE2 engagement of EP4. The ability of MMP-1 and -3 to regulate macrophage secretion of PGE2 and expression of MMP-9 defines a nexus between MMPs and prostanoids that is likely to play a role in the pathogenesis of chronic inflammatory diseases and cancer. These data also suggest that this nexus is targetable utilizing anti-TNF-α therapies and/or selective EP4 antagonists.
Introduction
MMP-9, a neutral endopeptidase, participates in diverse physiologic and pathologic processes (1–3). The degradation of extracellular matrix (ECM) components is commonly thought to be the principal role of MMP-9 in these diverse biological processes. However, in addition to degrading ECM, the biological actions of MMP-9 result from its ability to degrade and modify the activities of cytokines and chemokines, growth factors, and proteinase inhibitors (1,2).
MMP-9 expression is low or absent in most normal tissues, and markedly elevated during inflammation, wound healing, and neoplasia (1). Major inducers include inflammatory cytokines, growth factors, LPS, and ECM components (1). In this regard, we previously demonstrated that macrophage MMP-9 expression induced by TNF-α, LPS and ECM is dependent on enhanced Cox-2 expression, increased PGE2 synthesis and PGE2 binding to the EP4 prostanoid receptor (4,5). MMP-9 expression was markedly reduced in macrophages isolated from Cox-2 deficient mice, or in wild-type macrophages treated with selective Cox-2 inhibitors (4,5). Likewise, induction of macrophage MMP-9 expression was blocked by selective EP4 antagonists or EP4 silencing (5). Together these data point to the important role the Cox-2→PGE2→EP4 receptor axis plays in regulating macrophage MMP-9 expression.
MMP-9 is secreted as a zymogen (pro-MMP-9) and is activated through the proteolytic removal of its pro-domain by MMP-3 and indirectly by plasmin via the activation of pro-MMP-3 (6,7). However, the role of MMP-3 and other MMP family members in the regulation of macrophage MMP-9 expression has not been explored. In studies reported here, we determined whether MMPs commonly found in both inflamed and neoplastic tissues regulate macrophage MMP-9 expression by stimulating the Cox-2→PGE2→EP4 receptor axis. We found that macrophage exposure to MMP-1 or MMP-3 led to a marked increase in Cox-2 expression and PGE2 secretion, and subsequent induction of MMP-9. Proteinase-induced MMP-9 expression was blocked in macrophages pre-incubated with celecoxib or transfected with Cox-2 siRNA. Likewise, proteinase-induced MMP-9 was blocked in macrophages pre-incubated with the EP4 antagonist ONO-AE3-208 or transfected with EP4 siRNA. Exposure of macrophages to MMP-1 and MMP-3 triggered the rapid release of TNF-α, which was blocked by MMP-inhibitors. Furthermore, both Cox-2 and MMP-9 expression were effectively blocked in macrophages pre-incubated with anti-TNF-α IgG or transfected with TNF-α siRNA. Thus, proteinase-induced MMP-9 expression by macrophages is dependent on the release of TNF-α, induction of Cox-2 expression and PGE2 engagement of EP4. Together, these data identify a novel regulatory mechanism whereby MMP-1 and MMP-3 contribute to the inflammatory process by liberating TNF-α and stimulating PGE2 secretion via the induction of Cox-2, which leads, in turn, to increased MMP-9 expression. The ability of select MMPs to regulate macrophage secretion of PGE2 defines a nexus between MMPs and prostanoids that is likely to play a role in the pathogenesis of chronic inflammatory diseases and cancer.
Materials and Methods
Macrophages
Thioglycollate-elicited peritoneal macrophages were obtained from Swiss Webster mice by the method of Edelson and Cohn (8) as described previously (9). Mice were injected IP (3 ml/mouse) with 3% Brewer Thioglycollate Medium (DIFCO). Four days later, cells were harvested by lavage with cold Dulbecco’s PBS. Peritoneal cells were recovered by centrifugation and resuspended in DMEM supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 µg/ml) and 4 mM glutamine, and plated into tissue culture flasks or multi-well plates. Cells were allowed to adhere for 4 h and then washed free of nonadherent cells. All tissue culture supplies were obtained from Gibco/Life Technologies. The murine macrophage cell line RAW264.7 (10) was obtained from American Type Culture Collection, and maintained as adherent cultures in DMEM-10% FBS. All animal studies described in this report have been reviewed and approved by the Weill Cornell Medical College Institutional Animal Care and Use Committee.
MMPs
Human recombinant pro-MMP-1, active MMP-2, an active fragment of MMP-3 and active MMP-7 were obtained from Calbiochem. Human recombinant active MMP-9 was obtained from Oncogene. Prior to use, pro-MMP-1 was activated by incubation (2 h at 37°C) with 1 mM 4-amino-phenyl-mercuric acetate (APMA) in 50 mM Tris, pH 7.5, containing 0.05% Triton-X-100 and 5 mM CaCl2 (11). The conversion of pro-MMP-1 to its active form is associated with a loss of the pro-domain (~10 kDa), which was verified by SDS-PAGE and Western blot (data not shown).
Preparation of cell lysates
Cell lysates utilized in the analysis of Cox-2 were prepared by adding 1X SDS sample buffer directly to washed macrophage monolayers. In experiments designed to monitor levels of phosphorylated and total MAPKerk1/2, macrophages were lysed in TBS containing 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 1 mM sodium vanadate, 2 mM sodium pyrophosphate, 1 mM PMSF and 10 µg/ml aprotinin. Lysates were centrifuged (14,000 × g) for 20 min at 4°C. The supernatants were recovered, normalized for protein and mixed with SDS sample buffer with β-mercaptoethanol and boiled 5 min. Equal amounts of cell lysates were applied to gels based on protein content.
Western blots
Cell lysates were electrophoresed in 4–15% polyacrylamide gels and proteins were transferred to a PVDF membrane. The membrane was blocked in 5% defatted milk in TBST, washed in PBS and incubated 1 h in blocking buffer containing rabbit IgG raised against a peptide containing amino acids 584–598 of murine Cox-2 (0.5 µg/ml; Cayman Chemical) or rabbit anti-phosphospecific p44/p42 MAPK (i.e. MAPKerk1/2) IgG (75 ng/ml; Cell Signaling Technology). Membranes were washed 2X in TBST and incubated 1 h in blocking buffer containing goat anti-rabbit IgG conjugated to HRP (0.3 µg/ml; Transduction Laboratories). The membranes were washed 3X in TBST and bound HRP was visualized utilizing chemiluminescence (HyGlo). In the case of membranes probed with anti-phosphospecific MAPKerk1/2, following visualization of bound HRP, the membranes were stripped in 62.5 mM Tris buffer (pH 6.7) containing 100 mM β-mercaptoethanol and 2% SDS for 30 min at 50°C, washed and probed for total MAPKerk1/2 (Cell Signaling Technology).
Macrophage conditioned media were electrophoresed in gradient gels and proteins were transferred to a PVDF membrane. The membrane was placed in blocking buffer for 1 h, washed in PBS and incubated 2 h in blocking buffer containing rabbit anti-mouse MMP-9 IgG (0.2 µg/ml; Chemicon) or incubated at 4°C overnight in blocking buffer containing rabbit anti-murine TNF-α (2 µg/ml); Thermo Scientific). Membranes were washed 2X in TBST and incubated 1 h in blocking buffer containing goat anti-rabbit IgG conjugated to HRP or goat anti-mouse IgG conjugated to HRP.
Cox-2 and TNF-α siRNA transfection
RAW264.7 macrophages were transfected with either Cox-2, TNF-α or nonspecific siRNA (Santa Cruz) according to the manufacturer’s protocol, which was modified as follows. Macrophages (T-75 flask) were washed 3X with DPBS to remove serum, and medium replaced with antibiotic free DMEM. The cells were mechanically harvested, recovered by centrifugation, and resuspended in antibiotic free DMEM supplemented with 10% FBS. The cells (2 × 106/well) were aliquoted into a 6 well plate, and incubated overnight. The next morning, 9.1 µl of 10 µM siRNA was added to 152 µl transfection medium (Santa Cruz), and incubated 5 min at RT. In another tube 9.1 µl of the transfection reagent (Santa Cruz) was added to the transfection medium and incubated 5 min. The two tubes were combined and incubated 20 min to form the siRNA transfection reagent complex. Immediately before transfection, macrophage medium was removed and replaced with 1.5 ml antibiotic free DMEM supplemented with 10% FBS. The plate was placed on a rocker plate in the laminar flow hood, and the transfection reagent complex was added drop wise. Cells were incubated 72 h at 37° C.
EP4 siRNA transfection
RAW264.7 macrophages were transfected with EP4 or nonspecific siRNA (Thermo Scientific Dharmacon®) utilizing the Accell™ siRNA delivery protocol. Macrophages were aliquoted into a 6-well plate (0.5 × 105/well) in DMEM-10% FBS. The next day media was replaced with 1.5 ml Accell delivery medium containing 1 µM siRNA, and cells were incubated for 72 h at 37°C.
RT-PCR
RNA was prepared using Trizol reagent kits (Invitrogen). RNA (2 μg) was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (Roche Applied Science) and oligo d(T)16 primer. The resulting cDNA was then used for amplification. The primers and conditions for PCR analysis of murine Cox-2, MMP-9, EP4 and actin have been previously described (5). Primers for murine TNF-α were obtained from R&D Systems. PCR products were electrophoresed in a 1% agarose gel with 0.5 µg/ml ethidium bromide and photographed under UV light. PCR conditions for the target genes were optimized to ensure that experimental samples, excluding exuberant positive controls, are in the linear range when scanned. Densitometric analysis of the scanned target mRNAs were determined utilizing Scion Image (Scion Corporation) and normalized for levels of actin mRNA, which served as a loading control.
Determination of PGE2 and TNF-α levels in macrophage conditioned media
The concentrations of PGE2 in conditioned media were determined utilizing the STAT-PGE2 ELISA kit (Cayman Chemical). Concentrations of TNF-α were determined utilizing the Legend Max™ ELISA kit for mouse TNF-α (BioLegend).
Cytokine antibody array
The presence of 18 cytokines released into conditioned media from macrophages incubated 1 h with 50 nM MMP-3 was monitored utilizing an antibody array according to the manufactures’ instructions (Panomics). The assay is based on the sandwich ELISA method.
Evaluation of cellular toxicity
To rule out toxic effects of the inhibitors utilized in these studies, cellular morphology and/or protein content following incubation are regularly monitored. In addition, we test for potential cytotoxicity by monitoring mitochondrial dehydrogenase activity utilizing a MTT assay kit (Sigma-Aldrich). Levels of mitochondrial dehydrogenase activity were not significantly reduced following exposure to any of the inhibitors over the time periods described in the manuscript.
Cytokines and chemokines
Recombinant murine TNF-α, MIG (CXCL9), MIP-α (CCL3), INF-γ and RANTES (CCL5) were obtained from R&D Systems.
Results
MMP-1 and MMP-3 induce macrophage Cox-2 expression
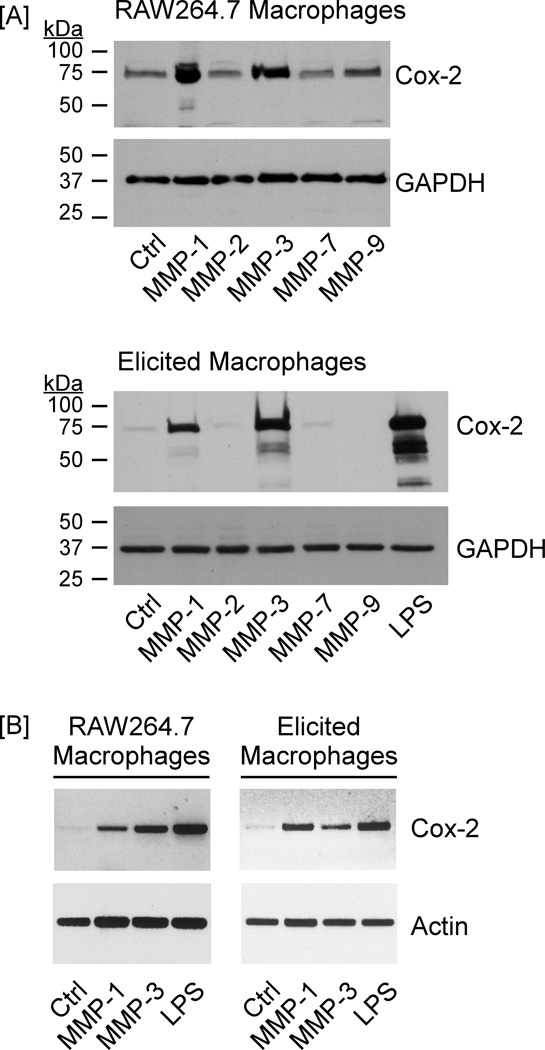
In previous studies, we characterized a Cox-2→PGE2→EP4 receptor axis that regulates macrophage MMP-9 expression (5). In the current study, we have determined whether MMP family members commonly found in inflamed and neoplastic tissues regulate expression of this prostanoid-EP receptor axis. For this purpose, RAW264.7 macrophages (10) and thioglycollate-elicited macrophages were incubated overnight with 50 nM active MMP-1, MMP-2 (gelatinase A), MMP-3, MMP-7 (matrilysin) or MMP-9. Incubation with 10 ng/ml LPS, a potent inducer of Cox-2 expression by macrophages, served as a positive control (12,13). Levels of Cox-2 in cell lysates were determined via Western blot. Overnight incubation with MMP-1 or MMP-3 strongly stimulated Cox-2 expression in RAW264.7 macrophages and elicited macrophages; in contrast, MMP-2, MMP-7 and MMP-9 had little or no effect (Figure 1A). MMP-induced changes in Cox-2 gene expression were monitored utilizing semi-quantitative RT-PCR. When normalized for actin mRNA levels (i.e. loading controls), levels of Cox-2 mRNA in MMP-1 and MMP-3 treated macrophages were elevated 17- and 28-fold, respectively in RAW264.7 macrophages, and 12- and 8-fold in elicited macrophages (Figure 1B).
Figure 1. MMP-1 and MMP-3 induce macrophage expression of Cox-2.
[A] RAW264.7 macrophages or thioglycollate-elicted peritoneal macrophages were suspended in DMEM containing 10% FCS and dispersed into a 48-well plate (2×105 cells/well). Following adherence, cells were washed 3X with DPBS and media were replaced with DMEM-0.1% low endotoxin (LE) BSA alone (Ctrl) or DMEM-0.1% LE-BSA containing 50 nM MMP-1, MMP-2, MMP-3, MMP-7, MMP-9 or 10 ng/ml LPS. The next day cell lysates were prepared, and levels of Cox-2 and GAPDH were determined utilizing Western blot. [B] Macrophages were dispersed into a 6-well plate (2×106 cells/well). Following adherence, cells were washed. Media were replaced with DMEM-0.1% LE-BSA alone (Ctrl) or DMEM-0.1% LE-BSA containing 50 nM MMP-1, MMP-3 or 10 ng/ml LPS. After overnight incubation, total RNA was isolated, and mRNA levels for Cox-2 and actin were determined utilizing RT-PCR. The data are representative of three separate experiments.
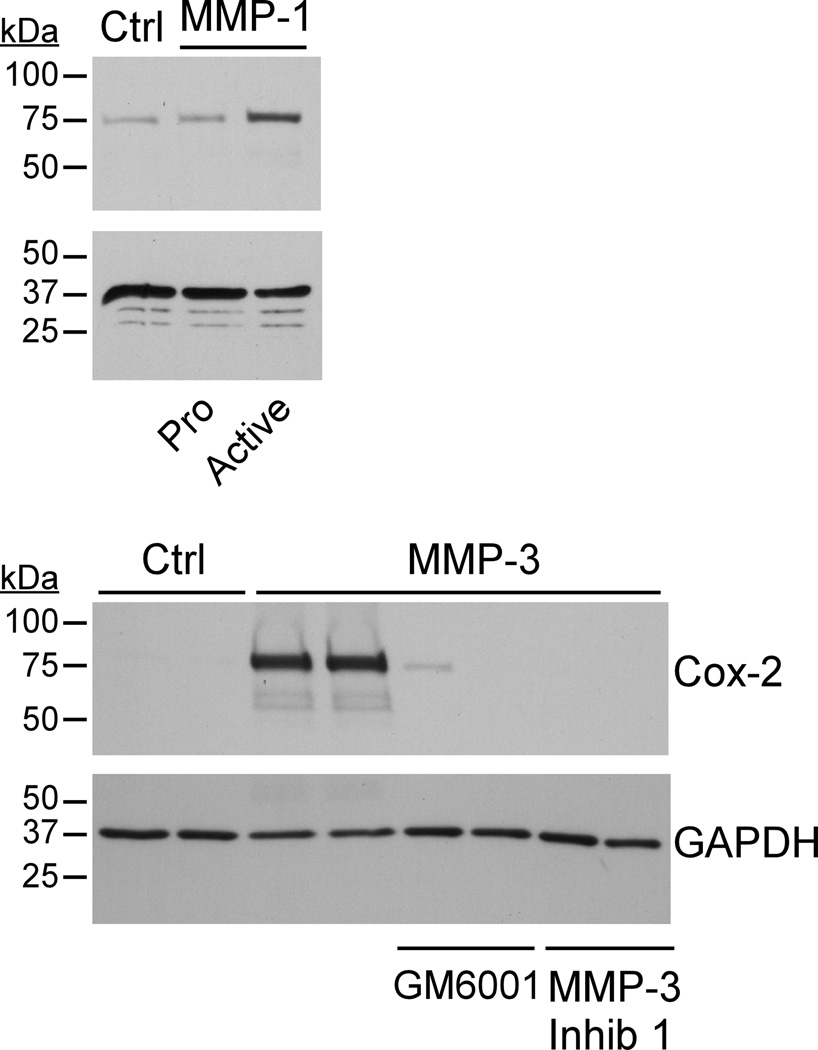
To determine whether MMP-induced Cox-2 expression was dependent on their enzymatic activity, we compared the abilities of pro-MMP-1 and APMA activated MMP-1 to induce Cox-2 expression in RAW264.7 macrophages. As seen in Figure 2 (top panel), in contrast to active MMP-1, incubation with pro-MMP-1 failed to induce Cox-2 expression. Next, we determined whether two unrelated MMP inhibitors could block MMP-3-induced Cox-2 expression. For this purpose, RAW264.7 macrophages were incubated with either a hydroxamic acid based broad spectrum MMP inhibitor (GM6001) or an inhibitory peptide corresponding to the pro-domain of MMP-3 (MMP-3 Inhibitor 1). As seen in Figure 2 (bottom panel), MMP-3-induced Cox-2 expression was completely blocked by either GM6001 or MMP-3 Inhibitor 1. Taken together, these data demonstrate that the induction of Cox-2 expression by MMP-1 and MMP-3 is dependent on their enzymatic activities.
Figure 2. MMP catalytic activity is required for Cox-2 induction.
RAW264.7 macrophages (48–well plate; 2×105 cells/well) were incubated in DMEM-0.1% LE-BSA containing 50 nM pro-MMP-1 or APMA activated MMP-1. In other wells, macrophages were pre-incubated for 15 min with the broad spectrum MMP inhibitor GM6001 (50 nM) or MMP-3 Inhibitor I (20 µM) prior to the addition of 50 nM MMP-3. The next day lysates were prepared, and levels of Cox-2 and GAPDH were monitored by Western blot. The data are representative of four separate experiments.
MMP-1 & MMP-3 induce macrophage MMP-9 expression
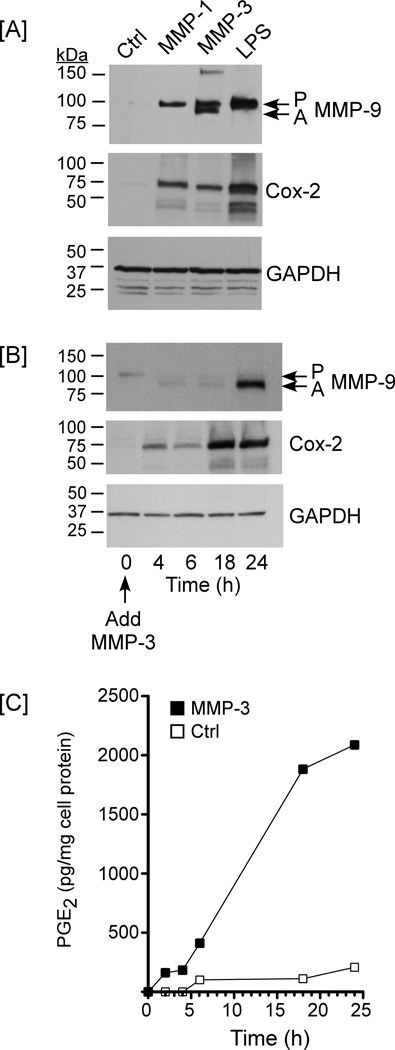
Macrophage MMP-9 expression is regulated by Cox-2-dependent synthesis of PGE2 (4,5). Therefore, we next determined whether MMP-1 and MMP-3 induced macrophage MMP-9 expression in a Cox-2-dependent manner. As seen in Figure 3A, the levels of Cox-2 and MMP-9 antigens in RAW264.7 lysates and conditioned media, respectively, were elevated following an overnight incubation with MMP-1, MMP-3 or LPS (positive control). The MMP-9 in conditioned media from MMP-1 and LPS treated macrophages was exclusively in the pro-form (105 kDa), which is slightly larger than the human pro-form (92 kDa). In contrast, both pro (P; 105 kDa) and active (A; 95 kDa) MMP-9 were identified in cultures incubated with MMP-3. MMP-3-dependent activation of pro-MMP-9 was confirmed utilizing zymography (not shown) as previously described (4), and is consistent with earlier reports indicating that pro-MMP-9 is activated extracellularly by MMP-3 (6,7,14,15).
Figure 3. MMP-1 and MMP-3 stimulate macrophage expression of PGE2 and MMP-9.
[A] RAW264.7 macrophages (48–well plate; 2×105 cells/well) were incubated in DMEM-0.1% LE-BSA alone (Ctrl) or DMEM-0.1% LE-BSA containing 50 nM active MMP-1, MMP-3 or 10 ng/ml LPS. Cells were incubated overnight, conditioned media collected, and lysates prepared. Levels of MMP-9 in conditioned media, and Cox-2 and GAPDH in lysates were determined by Western blot. [B] RAW264.7 macrophages were incubated with 50 nM MMP-3 for 0 – 24 h following which levels of MMP-9 in conditioned media, and Cox-2 and GAPDH in lysates were determined. [C] RAW264.7 macrophages were incubated in DMEM-0.1% LE-BSA alone or media containing 50 nM MMP-3 for 0 – 24 h. Levels of PGE2 in conditioned media were determined utilizing ELISA. The data in each panel are representative of three separate experiments.
In a subsequent experiment, we examined the temporal relationship between MMP-3-induced Cox-2, PGE2 and MMP-9 expression (Figure 3B & C). Levels of Cox-2 antigen were increased 4–6 h following the addition of MMP-3, and were strongly elevated at 18 and 24 h. Levels of PGE2 in media recovered from MMP-3 treated macrophages were markedly elevated at 18 and 24 h, as compared to PGE2 levels in media recovered from control cells. Following the increase in PGE2 levels, MMP-9 antigen in conditioned media of MMP-3 treated cells was strongly elevated at 24 h. In a separate experiment, levels of PGE2 release by RAW264.7 macrophages incubated 18 h with media alone or media containing 50 nM MMP-1 or MMP-3 were compared. Media derived from control cells contained 107 pg/mg cell protein (average of duplicate wells). Following incubation with MMP-1 or MMP-3, PGE2 levels increased to 661 and 905 pg/mg cell protein, respectively.
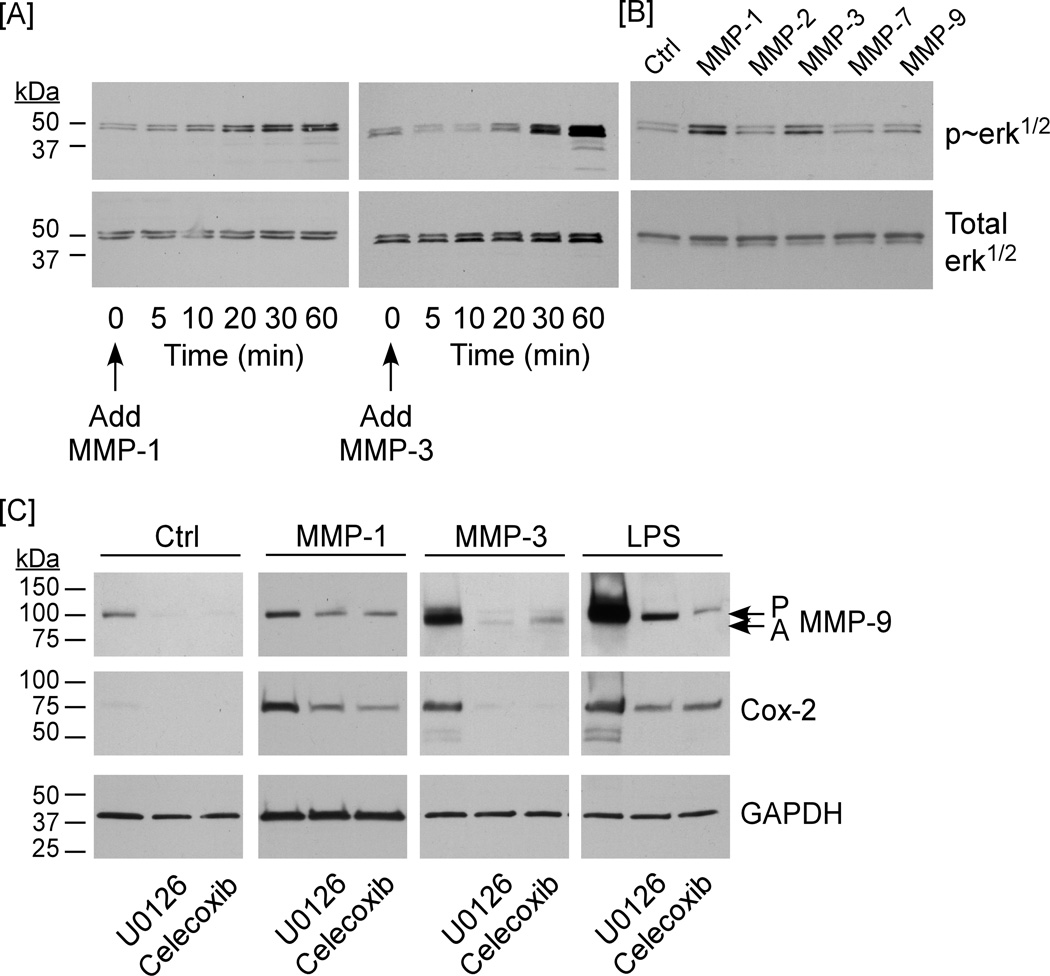
Activation of MAPKerk1/2 plays a causal role in MMP-induced Cox-2 expression and MMP-9 synthesis
We previously have shown that the activation of MAPKerk1/2 is a determinant of ECM-induced Cox-2 expression and MMP-9 synthesis (4,5). Therefore, we determined whether exposure of cells to MMP-1 or MMP-3 led to the activation of MAPKerk1/2. As seen in Figure 4A, the levels of phosphorylated MAPKerk1/2 in macrophages were elevated 20 – 30 min following the addition of active MMP-1 or MMP-3. In contrast, incubation with MMP-2, MMP-7 and MMP-9 did not result in the activation of MAPKerk1/2 (Figure 4B). Furthermore, pre-incubation of cells with the MEK-1 inhibitor U0126 (10 µM) blocked MMP-1 and MMP-3-induced phosphorylation/activation of MAPKerk1/2 (data not shown), and MMP-induced expression of Cox-2 and MMP-9 (Figure 4C). Thus, the activation of MAPKerk1/2 plays a causal role in MMP-induced Cox-2 expression and MMP-9 synthesis.
Figure 4. MMP-induced macrophage expression of MMP-9 is MAPKerk1/2 and Cox-2-dependent.
[A] RAW264.7 macrophages (48–well plate; 2.5×105/well) were cultured in DMEM-0.1% LE-BSA for 24 h. Cells were then incubated with 50 nM MMP-1 or MMP-3 for 5 – 60 min, and lysates were prepared. Levels of phosphorylated and total MAPKerk1/2 were determined utilizing Western blot. [B] RAW264.7 macrophages (48–well plate; 2.5×105/well) were cultured in DMEM-0.1% LE-BSA for 24 h. Cells were then incubated with media alone (Ctrl) or media containing 50 nM MMP-1, MMP-2, MMP-3, MMP-7 or MMP-9 for 30 min, and lysates were probed for phosphorylated and total MAPKerk1/2. [C] RAW264.7 macrophages (48 well plate; 2.5×105/well) were pre-incubated with U0126 (10 µM; MEK-1 inhibitor) or celecoxib (5 µM; Cox-2 inhibitor). Following an overnight incubation with 50 nM MMP-1, MMP-3 or 10 ng/ml LPS, levels of MMP-9 in conditioned media, and Cox-2 and GAPDH in lysates were determined by Western blot. The entire experiment was repeated twice with similar results.
MMP-induced MMP-9 synthesis is dependent on Cox-2
To examine the role of Cox-2 in proteinase-induced expression of MMP-9, macrophages were pre-incubated with the selective Cox-2 inhibitor celecoxib. Pre-treatment with 5 µM celecoxib blocked MMP-induced MMP-9 expression (Figure 4C), which was restored with the addition of PGE2 (not shown). The observation that celecoxib also attenuates Cox-2 expression is consistent with the previously reported positive feedback loop whereby PGE2 stimulates Cox-2 expression by engaging the EP4 receptor and triggering a Ras-MAPK signaling cascade (5,16).
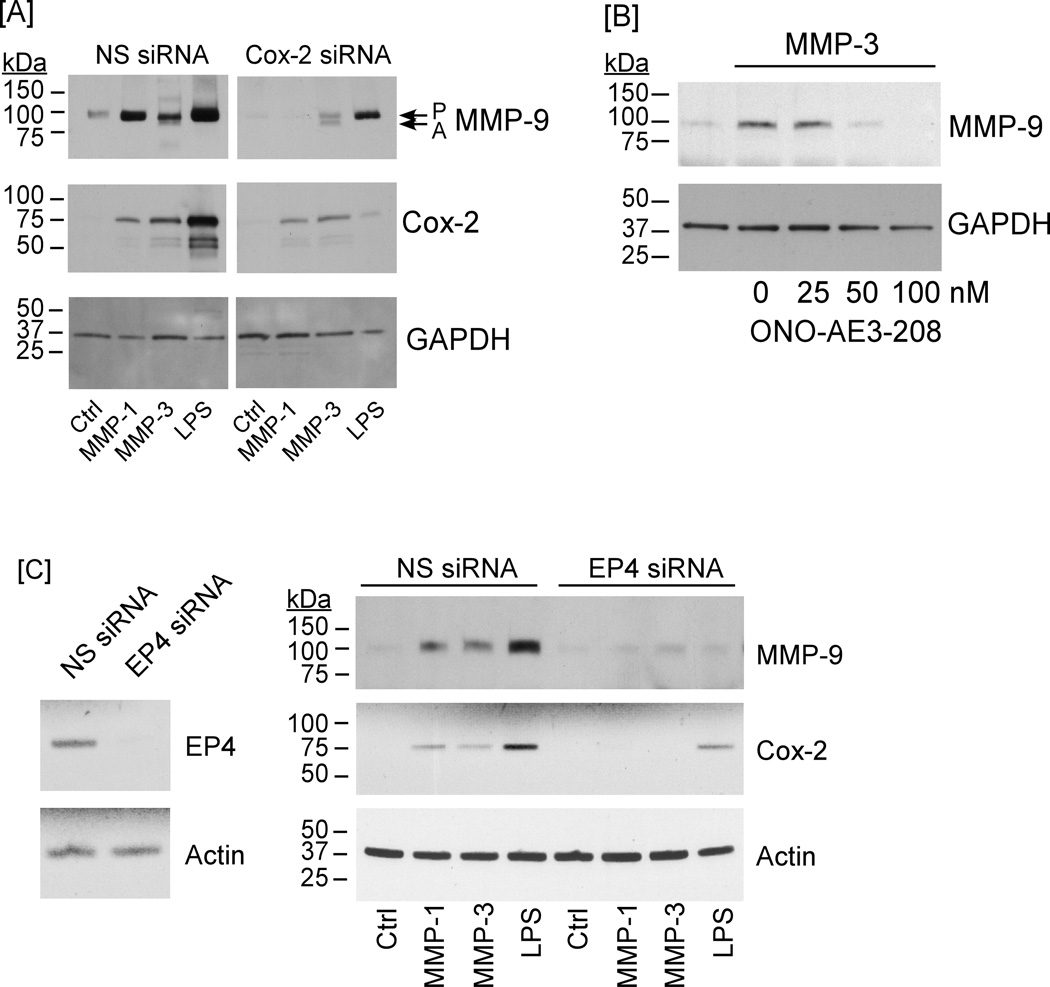
As an alternative strategy to examine the role of Cox-2 in MMP-induced expression of MMP-9, RAW264.7 macrophages were transfected with Cox-2 siRNA. As observed in Figure 5A, levels of Cox-2 antigen in cells transfected with nonspecific siRNA were increased following incubation with MMP-1, MMP-3 or LPS; whereas, reduced levels of Cox-2 were observed in MMP- or LPS-treated macrophages transfected with Cox-2 siRNA. When conditioned media were examined for MMP-9 antigen, MMP-1, MMP-3 and LPS treatment led to a large increase in MMP-9 expression, which was markedly attenuated in macrophages in which Cox-2 expression was knocked down.
Figure 5. Cox-2 silencing, EP4 antagonist or EP4 silencing block MMP-induced MMP-9 expression.
[A] RAW264.7 macrophages transfected with nonspecific (NS) or Cox-2 siRNA were suspended in DMEM-0.1% LE-BSA, and aliquoted into a 24-well plate. Following adherence, cells received media alone (Ctrl) or media containing 50 nM MMP-1, MMP-3 or LPS (10 ng/ml) and were incubated overnight. Levels of MMP-9 in conditioned media, and Cox-2 and GAPDH in lysates were determined by Western blot. [B] RAW264.7 macrophages were incubated with 50 nM MMP-3 alone or in the presence of 25 – 100 nM EP4 antagonist ONO-AE3-208 overnight. Levels of MMP-9 in conditioned media and GAPDH in lysates were determined by Western blot. [C] Macrophages transfected with NS or EP4 siRNA were incubated overnight in media containing 50 nM MMP-1, MMP-3 or LPS (10 ng/ml). Levels of MMP-9 in conditioned media, and Cox-2 and GAPDH in lysates were determined by Western blot. Levels of EP4 mRNA in transfected cells were determined utilizing RT-PCR. The entire experiment was repeated twice with similar results.
Finally, the diverse biologic actions of PGE2 are mediated by the EP1-4 family of prostanoid receptors (17,18). Earlier studies have demonstrated that PGE2-dependent MMP-9 expression was mediated via engagement of EP4 (5,19). Thus, we examined the ability of a selective EP4 antagonist ONO-AE3-208 to block MMP-induced MMP-9 expression. Pre-incubation of macrophages with 50 nM ONO-AE3-208 blocked MMP-3-induced MMP-9 expression (Figure 5B). In order to corroborate these findings, RAW264.7 macrophages were transfected with EP4 siRNA. The level of EP4 mRNA in cells transfected with EP4 siRNA was reduced ~95% when compared to cells transfected with nonspecific siRNA (Figure 5C; left panel). When transfected cells were exposed to proteinases, MMP-1, MMP-3 and LPS (positive control) failed to significantly induce Cox-2 or MMP-9 expression by macrophages transfected with EP4 siRNA (Figure 5C; right panel). Thus, the induction of MMP-9 expression by selected MMPs is causally linked to MAPKerk1/2-dependent stimulation in Cox-2 expression, and subsequent PGE2 engagement of EP4.
MMP-dependent release of TNF-α stimulates macrophage Cox-2 expression
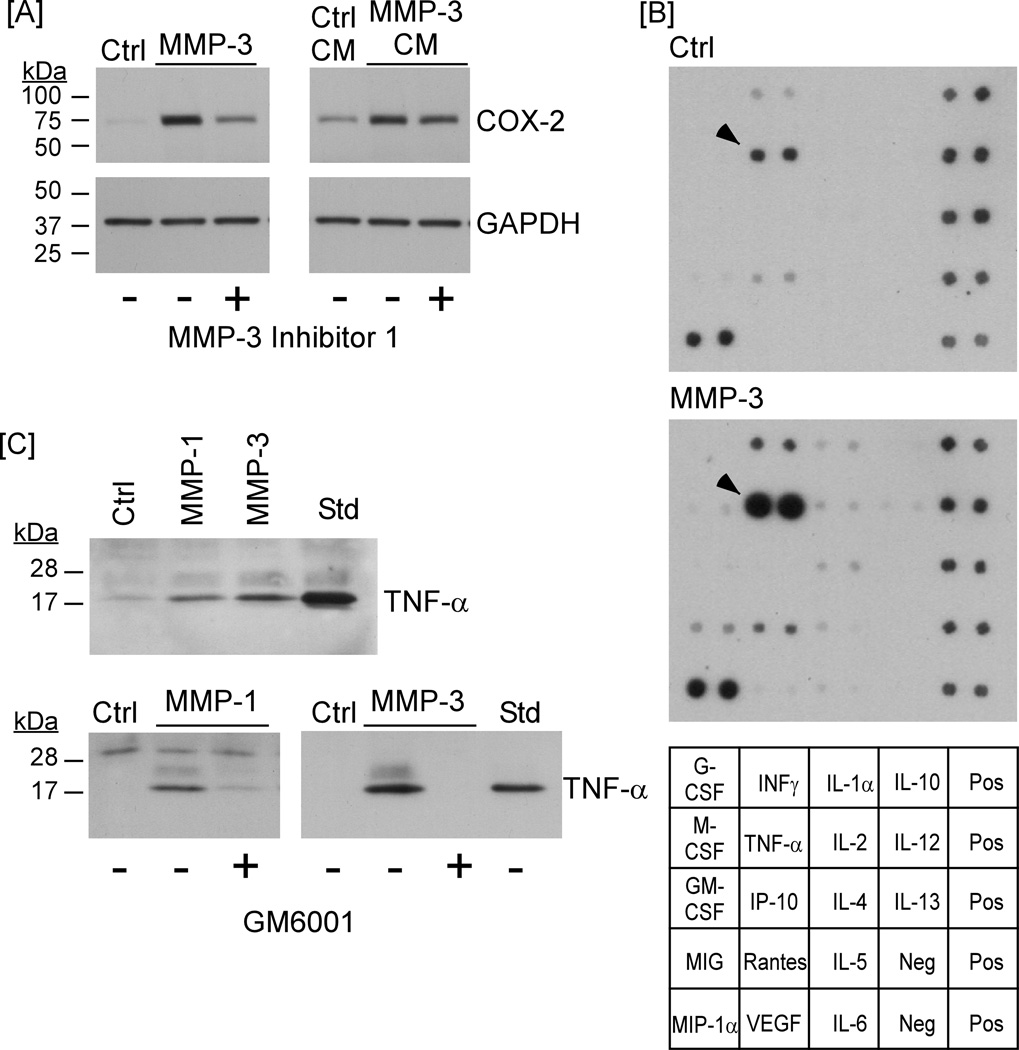
Proteinases can affect cellular function by releasing tethered growth factors (20–22), cytokines (23,24), fragments of extracellular matrix (25–27) or activating protease activated receptor family members (28,29). To determine whether MMP-induced Cox-2 expression resulted from the release or activation of a soluble factor, conditioned media from macrophages exposed to MMP-3 overnight were recovered, treated with MMP-3 Inhibitor 1 and transferred to naïve cells. As seen in Figure 6A (left panel), levels of Cox-2 expression were elevated in macrophages incubated overnight with MMP-3. The ability of MMP-3 to induce Cox-2 expression in macrophages co-incubated with MMP-3 Inhibitor 1 was noticeably reduced. Similar results are seen in Figure 2A. Likewise, conditioned media recovered from cells following an overnight incubation with MMP-3 stimulated Cox-2 expression by naïve macrophages (Figure 6A; right panel). The ability of conditioned media to up-regulate Cox-2 expression in naïve cells was relatively unaffected by the addition of MMP-3 Inhibitor 1. These data indicate that exposure of cells to MMP-3 released or activated a soluble factor that was responsible for MMP-induced Cox-2 expression.
Figure 6. MMP-1 and MMP-3 release TNF-α from macrophages.
[A] RAW264.7 macrophages (24–well plate; 2.5 × 105/well) were incubated in DMEM-0.1% LE-BSA containing 50 nM MMP-3 or MMP-3 and 20 µM MMP-3 Inhibitor 1 overnight. The next day conditioned media from MMP-3 treated macrophages were recovered and lysates were prepared. Conditioned media alone or media to which MMP-3 Inhibitor 1 was added (to block residual MMP-3 activity) were added to naïve macrophages and incubated overnight. Levels of Cox-2 and GAPDH were monitored by Western blot. The data are representative of three separate experiments. [B] Conditioned media recovered from RAW264.7 macrophages (6–well plate; 1.15 × 106/well), incubated with MMP-3 for 1 h, were collected and analyzed for the presence of the indicated cytokines and growth factors utilizing antibody array. [C] RAW264.7 macrophages (24–well plate; 2.5 × 105/well) were incubated in DMEM-0.1% LE-BSA containing 50 nM of the indicated MMPs alone or in the presence of the broad spectrum MMP inhibitor, GM6001 (50 nM) overnight. The next day conditioned media were recovered and levels of TNF-α were determined by Western blot. The data are representative of two separate experiments.
In an effort to identify the factor(s) released by macrophages following exposure to MMP-3, the presence of 18 cytokines was monitored utilizing antibody array. As seen in Figure 6B, 1 h treatment with MMP-3 resulted in a substantial increase in levels of TNF-α previously reported to be a potent stimulator of Cox-2 (5,30) and MMP-9 expression by macrophages (5,31). To corroborate and extend the antibody array data, conditioned media were recovered from RAW264.7 macrophages treated with MMP-1 and MMP-3 for 18 h, and probed for levels of TNF-α utilizing Western blot. Recombinant murine TNF-α (25 ng/lane) was utilized as a standard. When compared to the standard TNF-α, media samples recovered from MMP-treated macrophages contained relatively high levels of TNF-α (Figure 6C; upper panel). In contrast, TNF-α was not detected in media recovered from cells incubated overnight with MMP-1 or MMP-3 in the presence of the MMP inhibitor GM6001 (Figure 6C; lower panel). Finally, to obtain a quantitative measure of MMP-induced TNF-α release, media were recovered from RAW264.7 macrophages (48 well plate; 2.5×105/well) treated with 50 nM MMP-1, 50 nM MMP-3 or 10 ng/ml LPS (positive control) for 18 h, and probed for levels of TNF-α utilizing ELISA. Media from control cells contained 698 ± 109 pg/ml (mean ± SEM; n=3). Following exposure to MMP-1 or MMP-3, levels increased 25-fold (17,555 ± 6544) and 34-fold (23,767 ± 1574), respectively. TNF-α levels in media from cells incubated with LPS increased 160-fold (114,333 ± 1828).
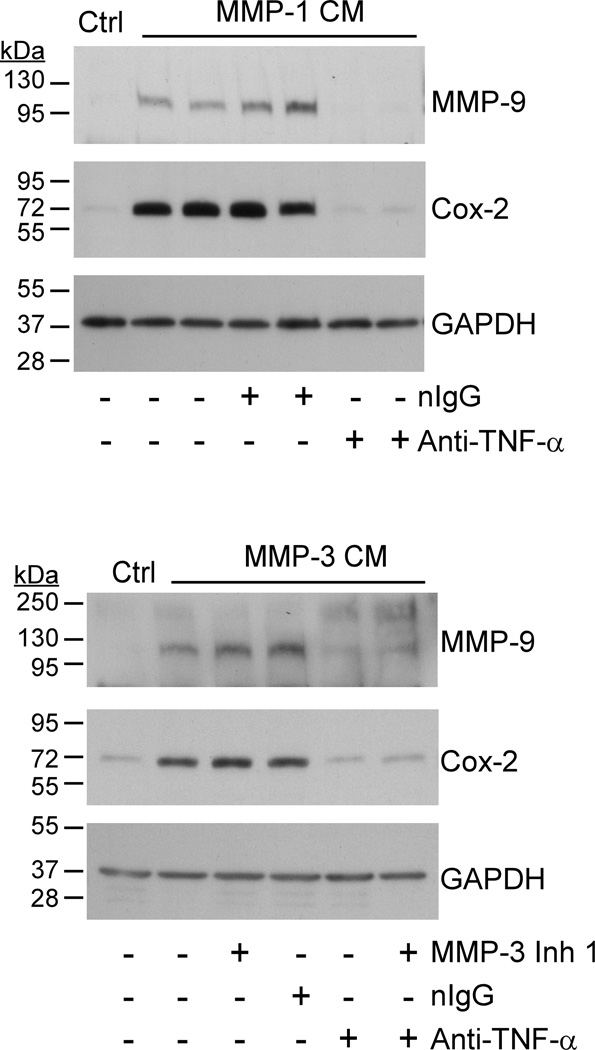
Next, we utilized a neutralizing rat monoclonal anti-murine TNF-α IgG to determine whether MMP-induced release of TNF-α was responsible for inducing Cox-2 and MMP-9 expression in RAW264.7 macrophages. For this purpose, conditioned media recovered from cells incubated with MMP-1 or MMP-3 were pre-incubated 2 h with 20 µg/ml anti-TNF-α IgG or normal IgG before transferring to naïve cells. As shown in Figure 7, conditioned media derived from MMP-1 treated cells stimulated Cox-2 and MMP-9 expression in naïve macrophages. Pre-incubation of the conditioned media with anti-TNF-α IgG blocked its ability to induce Cox-2 and MMP-9 expression; in contrast, normal IgG had no effect. Likewise, the addition of MMP-3 conditioned media to naïve macrophages stimulated their Cox-2 and MMP-9 expression. The addition of MMP-3 Inhibitor I or normal IgG to the conditioned media recovered from MMP-3 treated macrophages had no effect on its ability to induce Cox-2 or MMP-9 in naïve cells; in contrast, pre-incubation with anti-TNF-α IgG suppressed its ability to stimulate Cox-2 and MMP-9 by naïve macrophages.
Figure 7. Neutralizing anti-TNF-α IgG blocks MMP-induced Cox-2 expression.
Conditioned media recovered from RAW264.7 macrophages (24–well plate; 2.5 × 105/well) exposed (1 h) to 50 nM MMP-1 were subsequently incubated 2 h with non-immune IgG1 (20 µg/ml) or rat monoclonal anti-mouse TNF-α IgG1 (20 µg/ml). Conditioned media and antibody treated conditioned media were then added to naïve macrophages in duplicate wells. In a similar experiment, conditioned media recovered from cells incubated with MMP-3 were treated with MMP-3 Inhibitor 1 (20 µM) and/or anti-TNFα (20 µg/ml) prior to adding to naïve macrophages. Macrophages were incubated with conditioned media overnight and lysates prepared. Levels of Cox-2 and GAPDH in lysates were determined by Western blot. The data are representative of three separate experiments.
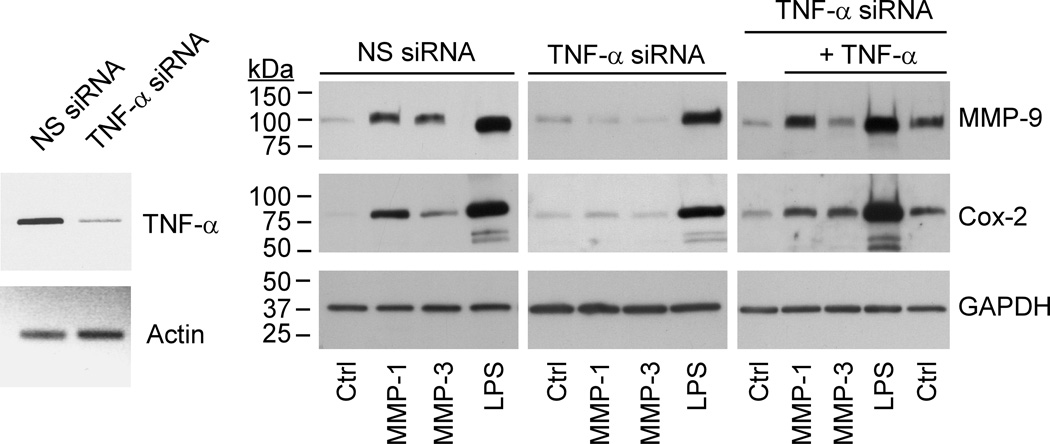
To directly test the role of TNF-α in proteinase-induced MMP-9 expression, RAW264.7 macrophages were transfected with either TNF-α siRNA or nonspecific siRNA. The level of TNF-α mRNA in cells transfected with TNF-α siRNA was reduced ~87% when compared to cells transfected with nonspecific siRNA (Figure 8; left panel). When TNF-α siRNA transfected cells were exposed to proteinases, MMP-1, MMP-3 failed to significantly induce Cox-2 or MMP-9 expression. Importantly, Cox-2 and MMP-9 expression were restored in these TNF-α siRNA transfected cells by the addition of exogenous TNF-α (Figure 8; right panel).
Figure 8. TNF-α silencing blocks MMP-induced MMP-9 expression.
RAW264.7 macrophages transfected with nonspecific (NS) or TNF-α siRNA were suspended in DMEM-0.1% LE-BSA, and aliquoted into a 24-well plate. Following adherence, cells received media alone (Ctrl) or media containing 50 nM MMP-1, MMP-3 or LPS (10 ng/ml) and were incubated overnight. In addition, cells transfected with TNF-α siRNA were incubated with MMP-1, MMP-3 or LPS in the presence of exogenous TNF-α (50 ng/ml). Levels of MMP-9 in conditioned media, and Cox-2 and GAPDH in lysates were determined by Western blot. Levels of TNF-α mRNA were determined utilizing RT-PCR. The data are representative of two separate experiments.
In these knockdown studies, LPS was included as a control to monitor non-specific effects of transfection with TNF-α siRNA. Although LPS potently stimulates macrophage expression of the inflammatory cytokines TNF-α and IL-1β, LPS-induced Cox-2 expression in human macrophages is independent of either inflammatory cytokine (32). Consistent with these observations, TNF-α knockdown had relatively little effect on LPS-induced Cox-2 and MMP-9 expression by murine RAW264.7 macrophages (Figure 8, right panel). Moreover, the presence of neutralizing anti-TNF-α IgG (20 µg/ml) failed to block LPS-induced Cox-2 and MMP-9 expression in murine RAW264.7 macrophages (data not show).
Finally, the antibody array in Figure 6 revealed that in addition to TNF-α several other cytokines and chemokines were induced by stimulation with MMP-3, although to a minor degree. These data suggest that other factors may contribute to the observed proteinase induction of Cox-2 and MMP-9 expression. However, an overnight incubation of RAW264.7 macrophages with 0.1 –1.0 ug/ml MIG (CXCL9), 0.5 – 50 ng/ml MIP-α (CCL3), 0.1 – 10 ng/ml INF-γ or 1–100ng/ml RANTES (CCL5) failed to induce Cox-2 or MMP-9 (data not shown). Taken together, these data indicate that MMP-1 and MMP-3 stimulate the release of TNF-α, which induces macrophage Cox-2-dependent MMP-9 expression.
Discussion
Genetic evidence indicates that MMP-9 participates in the recruitment of stem cells and tissue revascularization, skeletal growth plate angiogenesis and ossification, choroidal neovascularization following injury, tumor angiogenesis, dermal-epidermal separation in bullous pemphigoid, resolution of allergic inflammation, degradation of fibrin, and ventricular enlargement and collagen accumulation following myocardial infarction, and the pathogenesis of occlusive and aneurysmal vascular diseases (2,33,34). The activity of this pleiotropic proteinase is regulated at three levels: gene expression, pro-enzyme activation and inhibition (1,2). The proteolytic activation of pro-MMP-9 is currently understood to take place in the fluid-phase, and is catalyzed by MMP-3 (6,7,35). However, the ability of MMP-3 and/or other MMP family members to regulate MMP-9 expression has not been explored. Results of experiments reported here define a complex mechanism by which extracellular MMP-1 and MMP-3 stimulate macrophage MMP-9 expression via the liberation of TNF-α and subsequent induction of Cox-2, and PGE2 engagement of EP4 receptor.
PGE2, a member of the prostanoid family of autacoids, is an important mediator of the inflammatory response (36). PGE2 can stimulate vasodilation, increase permeability, pain, fever (37,38) and tissue destruction/remodeling via enhanced proteinase expression (4,39–42). Synthesis of PGE2 requires the liberation of arachidonic acid from membrane phospholipids, Cox-1 and/or Cox-2-dependent generation of PGH2, and its isomerization to PGE2 by members of the PGE synthase family (36). In most cells, Cox-1 expression is constitutive, whereas Cox-2 expression is induced in response to a variety of inflammatory mediators (43,44). As reported here, levels of Cox-2 mRNA and antigen in macrophages were selectively increased following exposure to catalytically active MMP-1 and MMP-3. Two mechanisms proposed for regulating Cox-2 expression may be relevant in MMP-induced Cox-2 expression: activation of MAPK superfamily pathways (45–48), and a PGE2-dependent positive feedback loop (5,16,49). Results of experiments reported here suggest both pathways are important. A causal role of activated MAPKerk1/2 in MMP-induced Cox-2 expression is supported by the observation that MMP-1 and MMP-3 trigger MAPKerk1/2 activation in macrophages, and pre-incubation of cells with a MEK-1 inhibitor blocks MMP-induced Cox-2 expression. Likewise, the failure of MMPs to induce Cox-2 expression in macrophages pre-treated with celecoxib or transfected with EP4 siRNA is consistent with the hypothesis that PGE2-dependent positive feedback loops regulate/amplify Cox-2 expression.
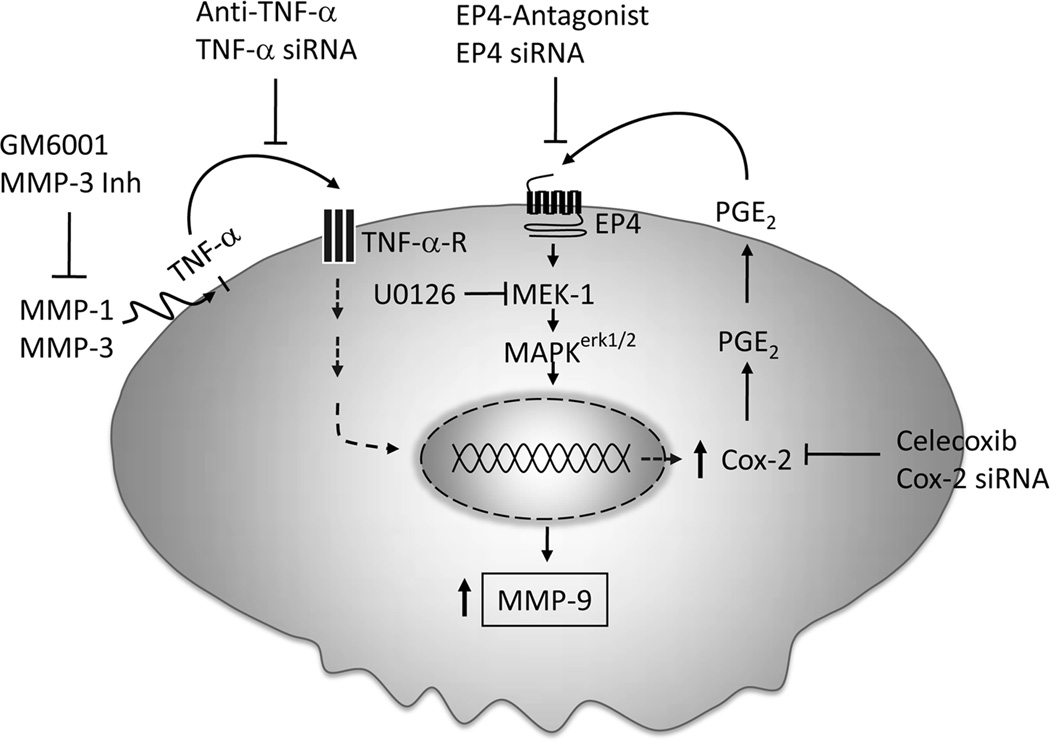
The release of TNF-α is a well defined mechanism by which proteinases regulate cell function. Pro-TNF-α, tethered to the cell membrane by its NH2-terminal domain, is efficiently processed to its mature form and released by membrane associated TNF-α converting enzyme (i.e. TACE) (50,51), which is a member of the ADAMs family of metalloproteinases (i.e. ADAM17) (52). In addition to TACE/ADAM17, several members of the MMP family are reported to process pro-TNF-α, although less efficiently (23,24). Studies reported here confirm the observation that MMP family members can process pro-TNF-α. We demonstrate that the levels of TNF-α in media recovered from macrophages incubated with active MMP-1 or MMP-3 were elevated as compared to control cells. The observed release of TNF-α provides a mechanism whereby these proteinases induce Cox-2-dependent MMP-9 expression. We and others have reported that macrophage Cox-2 (5,30) and MMP-9 expression (5,31) are stimulated by TNF-α, and knockdown of Cox-2 expression blocks TNF-α-induced MMP-9 expression (5). Moreover, as reported here, the ability of conditioned media recovered from macrophages incubated with MMP-1 or MMP-3 to induce Cox-2 and MMP-9 was blocked by neutralizing anti-TNF-α IgG. Furthermore, proteinases failed to induce either Cox-2 or MMP-9 in macrophages in which TNF-α expression was knocked down utilizing siRNA. Thus, MMP-1 and MMP-3-induced Cox-2 and MMP-9 expression by macrophages is dependent on the release of TNF-α (Figure 9).
Figure 9. MMP-1 and MMP-3 induce macrophage MMP-9 expression: Role of TNF-α and Cox-2.
Our data suggest that active MMP-1 and MMP-3 trigger the release TNF-α, which induces Cox-2 expression and PGE2 secretion. PGE2 binding to EP4 stimulates MAPKerk1/2-dependent increase in MMP-9 expression.
MMP family members are potent pro-inflammatory and immunoregulatory molecules that are implicated in the pathogenesis of chronic inflammatory diseases and cancer (2,53,54). Results of experiments reported here define a complex mechanism by which collagenase-1 and stromelysin-1, commonly found in both inflamed and neoplastic tissues, stimulate macrophage MMP-9 expression via the liberation of TNF-α and subsequent modulation the Cox-2→PGE2→EP4 receptor axis (Figure 9). The ability of MMPs to induce Cox-2-dependent MMP-9 expression by macrophages defines a nexus between MMPs and prostanoids that is likely to play a role in the pathogenesis of chronic inflammatory diseases and cancer. These data also suggest that this nexus is targetable utilizing anti-TNF-α therapies and/or selective EP4 antagonists.
Acknowledgements
These studies were supported by research grant HL073375 (to D.J. Falcone) from the National Institutes of Health, and grants from the New York Crohn’s Foundation (to A.J. Dannenberg) and Center for Cancer Prevention Research (to D.J. Falcone). The authors wish to thank Dr. Takayuki Maruyama (ONO Pharmaceutical Company, Osaka, Japan) for supplying the EP4 antagonist ONO-AE3-208.
Abbreviations used in this paper
- MMP
matrix metalloproteinase
- Cox-2
cyclooxygenase-2
- ECM
extracellular matrix
- APMA
4-amino-phenyl-mercuric acetate
- ADAM
a disintegrin and metalloproteinase
Reference List
- 1.Opdenakker G, Van den Steen PE, Van Damme J. Gelatinase B: a tuner and amplifier of immune functions. Trends in Immunol. 2001;22:571–579. doi: 10.1016/s1471-4906(01)02023-3. [DOI] [PubMed] [Google Scholar]
- 2.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nature Rev. Immunol. 2004;4:617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 3.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 4.Khan KMF, Howe LR, Falcone DJ. Extracellular matrix-induced cyclooxygenase-2 regulates macrophage proteinase expression. J. Biol. Chem. 2004;279:22039–22046. doi: 10.1074/jbc.M312735200. [DOI] [PubMed] [Google Scholar]
- 5.Pavlovic S, Du B, Sakamoto K, Khan KMF, Natarajan C, Breyer RM, Dannenberg AJ, Falcone DJ. Targeting prostaglandin E-2 receptors as an alternative strategy to block cyclooxygenase-2-dependent extracellular matrix-induced matrix metalloproteinase-9 expression by macrophages. J. Biol. Chem. 2006;281:3321–3328. doi: 10.1074/jbc.M506846200. [DOI] [PubMed] [Google Scholar]
- 6.Dreier R, Wallace S, Fuchs S, Bruckner P, Grassel S. Paracrine interactions of chondrocytes and macrophages in cartilage degradation: articular chondrocytes provide factors that activate macrophage-derived pro-gelatinase B (pro-MMP-9) J. Cell Sci. 2001;114:3812–3822. doi: 10.1242/jcs.114.21.3813. [DOI] [PubMed] [Google Scholar]
- 7.Ramos-DeSimone N, Hahn-Dantona E, Sipley J, Nagase H, French DL, Quigley JP. Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. J. Biol. Chem. 1999;274:13066–13076. doi: 10.1074/jbc.274.19.13066. [DOI] [PubMed] [Google Scholar]
- 8.Edelson PJ, Cohn ZA. Purification and cultivation of monocytes and macrophages. In: Bloom BR, David JR, editors. In vitro methods in cell mediated tumor immunity. New York: Academic Press; 1976. pp. 333–340. [Google Scholar]
- 9.Falcone DJ, Ferenc MJ. Acetyl-LDL stimulates macrophage-dependent plasminogen activation and degradation of extracellular matrix. J. Cell. Physiol. 1988;135:387–396. doi: 10.1002/jcp.1041350305. [DOI] [PubMed] [Google Scholar]
- 10.Raschke WC, Baird S, Ralph P, Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 1978;15:261–267. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- 11.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases - Structure, function, and biochemistry. Circ. Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto H, Naraba H, Murakami M, Kudo I, Yamaki K, Ueno A, Ohishi S. Concordant induction of prostaglandin E(2) synthase with cyclooxygenase-2 leads to preferred production of prostaglandin E(2) over thromboxane and prostaglandin D-2 in lipopolysaccharide-stimulated rat peritoneal macrophages. Biochem. Biophys. Res. Commun. 1997;230:110–114. doi: 10.1006/bbrc.1996.5894. [DOI] [PubMed] [Google Scholar]
- 13.Vogel S, Hirschfeld MJ, Perera PY. Signal integration in lipopolysaccharide (LPS)-stimulated murine macrophages. J. Endotoxin Res. 2001;7:237–241. [PubMed] [Google Scholar]
- 14.Ogata Y, Itoh Y, Nagase H. Steps involved in activation of the pro-matrix metalloproteinase 9 (progelatinase B)-tissue inhibitor of metalloproteinases- 1 complex by 4-aminophenylmercuric acetate and proteinases. J. Biol. Chem. 1995;270:18506–18511. doi: 10.1074/jbc.270.31.18506. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg GI, Strongin A, Collier IE, Genrich LT, Marmer BL. Interaction of 92-Kda Type-Iv Collagenase with the Tissue Inhibitor of Metalloproteinases Prevents Dimerization, Complex-Formation with Interstitial Collagenase, and Activation of the Proenzyme with Stromelysin. J. Biol. Chem. 1992;267:4583–4591. [PubMed] [Google Scholar]
- 16.Wang D, Buchanan FG, Wang H, Dey SK, DuBois RN. Prostaglandin E2 enhances intestinal adenoma growth via activation of the Ras-mitogen-activated protein kinase cascade. Cancer Res. 2005;65:1822–1829. doi: 10.1158/0008-5472.CAN-04-3671. [DOI] [PubMed] [Google Scholar]
- 17.Breyer RM, Bagdassarian CK, Myers SA, Breyer MD. Prostanoid receptors: subtypes and signaling. Annu. Rev. Pharmacol. Toxicol. 2001;41:661–690. doi: 10.1146/annurev.pharmtox.41.1.661. [DOI] [PubMed] [Google Scholar]
- 18.Hata AN, Breyer RM. Pharmacology and signaling of prostaglandin receptors: multiple roles in inflammation and immune modulation. Pharmacol. Ther. 2004;103:147–166. doi: 10.1016/j.pharmthera.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Cipollone F, Fazia ML, Iezzi A, Cuccurullo C, De Cesare D, Ucchino S, Spigonardo F, Marchetti A, Buttitta F, Paloscia L, Mascellanti M, Cuccurullo F, Mezzetti A. Association between prostaglandin E receptor subtype EP4 overexpression and unstable phenotype in atherosclerotic plaques in human. Arterioscler. Thromb. Vasc. Biol. 2005;25:1925–1931. doi: 10.1161/01.ATV.0000177814.41505.41. [DOI] [PubMed] [Google Scholar]
- 20.Saksela O, Rifkin DB. Release of basic fibroblasssst growth factor-Heparan sulfate complexes from endothelial cells by plasminogen activator- mediated proteolytic activity. J. Cell Biol. 1990;110:767–775. doi: 10.1083/jcb.110.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 2004;164:769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falcone DJ, McCaffrey TA, Haimovitz-Friedman A, Vergilio J-A, Nicholson AC. Macrophage and foam cell release of matrix bound growth factors: Role of plasminogen activation. J. Biol. Chem. 1993;268:11951–11958. [PubMed] [Google Scholar]
- 23.Gearing AJ, Beckett P, Christodoulou M, Churchill M, Clements J, Davidson AH, Drummond AH, Galloway WA, Gilbert R, Gordon JL. Processing of tumour necrosis factor-alpha precursor by metalloproteinases. Nature. 1994;370:555–557. doi: 10.1038/370555a0. [DOI] [PubMed] [Google Scholar]
- 24.Mohan MJ, Seaton T, Mitchell J, Howe A, Blackburn K, Burkhart W, Moyer M, Patel I, Waitt GM, Becherer JD, Moss ML, Milla ME. The tumor necrosis factor-alpha converting enzyme (TACE): a unique metalloproteinase with highly defined substrate selectivity. Biochem. 2002;41:9462–9469. doi: 10.1021/bi0260132. [DOI] [PubMed] [Google Scholar]
- 25.Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;277:225–228. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- 26.Khan KMF, Laurie GW, McCaffrey TA, Falcone DJ. Exposure of cryptic domains in the alpha1-chain of laminin-1 by elastase stimulates macrophages urokinase and matrix metalloproteinase-9 expression. J. Biol. Chem. 2002;277:13778–13786. doi: 10.1074/jbc.M111290200. [DOI] [PubMed] [Google Scholar]
- 27.Adair-Kirk TL, Atkinson JJ, Kelley DG, Arch RH, Miner JH, Senior RM. A chemotactic peptide from laminin alpha 5 functions as a regulator of inflammatory immune responses via TNF alpha-mediated signaling. J. Immunol. 2005;174:1621–1629. doi: 10.4049/jimmunol.174.3.1621. [DOI] [PubMed] [Google Scholar]
- 28.Coughlin SR. Protease-activated receptors in hemostasis, thrombosis and vascular biology. J. Throm. Haemostasis. 2005;3:1800–1814. doi: 10.1111/j.1538-7836.2005.01377.x. [DOI] [PubMed] [Google Scholar]
- 29.Boire A, Covic L, Agarwal A, Jacques S, Sherifl S, Kuliopulos A. PAR1 is a matrix metalloprotease-1 receptor that promotes invasion and tumorigenesis of breast cancer cells. Cell. 2005;120:303–313. doi: 10.1016/j.cell.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 30.Wu MH, Sun HS, Lin CC, Hsiao KY, Chuang PC, Pan HA, Tsai SJ. Distinct mechanisms regulate cyclooxygenase-1 and -2 in peritoneal macrophages of women with and without endometriosis. Mol. Hum. Reprod. 2002;8:1103–1110. doi: 10.1093/molehr/8.12.1103. [DOI] [PubMed] [Google Scholar]
- 31.Heidinger M, Kolb H, Krell HW, Jochum M, Ries C. Modulation of autocrine TNF-alpha-stimulated matrix metalloproteinase 9 (MMP-9) expression by mitogen-activated protein kinases in THP-1 monocytic cells. Biol. Chem. 2006;387:69–78. doi: 10.1515/BC.2006.010. [DOI] [PubMed] [Google Scholar]
- 32.Barrios-Rodiles M, Tiraloche G, Chadee K. Lipopolysaccharide modulates cyclooxygenase-2 transcriptionally and posttranscriptionally in human macrophages independently from endogenous IL-1 beta and TNF-alpha. J. Immunol. 1999;163:963–969. [PubMed] [Google Scholar]
- 33.Heissig B, Hattori K, Dias S, Friedrich M, Ferris B, Hackett NR, Crystal RG, Besmer P, Lyden D, Moore MAS, Werb Z, Rafii S. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of Kit-ligand. Cell. 2002;109:625–637. doi: 10.1016/s0092-8674(02)00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ducharme A, Frantz S, Aikawa M, Rabkin E, Lindsey M, Rohde LE, Schoen FJ, Kelly RA, Werb Z, Libby P, Lee RT. Targeted deletion of matrix metalloproteinase-9 attenuates left ventricular enlargement and collagen accumulation after experimental myocardial infarction. J. Clin. Invest. 2000;106:55–62. doi: 10.1172/JCI8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okada Y, Gonoji Y, Naka K, Tomita K, Nakanishi I, Iwata K, Yamashita K, Hayakawa T. Matrix metalloproteinase 9 (92-kDa gelatinase/type IV collagenase) from HT 1080 human fibrosarcoma cells. Purification and activation of the precursor and enzymic properties. J. Biol. Chem. 1992;267:21712–21719. [PubMed] [Google Scholar]
- 36.Park JY, Pillinger MH, Abramson SB. Prostaglandin E2 synthesis and secretion: the role of PGE2 synthases. Clin. Immunol. 2006;119:229–240. doi: 10.1016/j.clim.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Trebino CE, Stock JL, Gibbons CP, Naiman BM, Wachtmann TS, Umland JP, Pandher K, Lapointe JM, Saha S, Roach ML, Carter D, Thomas NA, Durtschi BA, McNeish JD, Hambor JE, Jakobsson PJ, Carty TJ, Perez JR, Audoly LP. Impaired inflammatory and pain responses in mice lacking an inducible prostaglandin E synthase. Proc. Natl. Acad. Sci. USA. 2003;100:9044–9049. doi: 10.1073/pnas.1332766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanov AI, Romanovsky AA. Prostaglandin E-2 as a mediator of fever: Synthesis and catabolism. Frontiers in Bioscience. 2004;9:1977–1993. doi: 10.2741/1383. [DOI] [PubMed] [Google Scholar]
- 39.Corcoran ML, Stetler-Stevenson WG, DeWitt DL, Wahl LM. Effect of cholera toxin and pertussis toxin on prostaglandin H synthase-2, prostaglandin E2, and matrix metalloproteinase production by human monocytes. Archives Biochem. Biophys. 1994;310:481–488. doi: 10.1006/abbi.1994.1196. [DOI] [PubMed] [Google Scholar]
- 40.Shankavaram UT, DeWitt DL, Funk SE, Sage EH, Wahl LM. Regulation of human monocyte matrix metalloproteinases by SPARC. J. Cell. Physiol. 1997;173:327–334. doi: 10.1002/(SICI)1097-4652(199712)173:3<327::AID-JCP4>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 41.Ottino P, Bazan HEP. Corneal stimulation of MMP-1, -9 and uPA by platelet-activating factor is mediated by cyclooxygenase-2 metabolites. Cur. Eye Res. 2001;23:77–85. doi: 10.1076/ceyr.23.2.77.5471. [DOI] [PubMed] [Google Scholar]
- 42.Dohadwala M, Batra RK, Luo J, Lin Y, Kostyantyn K, Pold M, Sharma S, Dubinett SM. Autocrine/paracrine prostaglandin E2 production by non-small cell lung cancer cells regulates matrix metalloproteinase-2 and CD44 in cyclooxygenase-2-dependent invasion. J. Biol. Chem. 2002;277:50828–50833. doi: 10.1074/jbc.M210707200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hla T, Bishop-Bailey D, Liu CH, Schaefers HL, Trifan OC. Cyclooxygenase-1 and -2 isoenzymes. Int. J. Biochem. Cell Biol. 1999;31:551–557. doi: 10.1016/s1357-2725(98)00152-6. [DOI] [PubMed] [Google Scholar]
- 44.Smith WL, Langenbach R. Why there are two cyclooxygenase isozymes. J. Clin. Invest. 2001;107:1491–1495. doi: 10.1172/JCI13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hendrickx N, Volanti C, Moens U, Seternes OM, De Witte PJ, Vandenheede R, Piette J, Agnostinis P. Up-regulation of cyclooxygenase-2 and apoptosis resistance by p38 MAPK in hypericin-mediated photodynamic therapy of human cancer cells. J. Biol. Chem. 2003;278:52231–52239. doi: 10.1074/jbc.M307591200. [DOI] [PubMed] [Google Scholar]
- 46.Pratt PF, Bokemeyer D, Foschi M, Sorokin A, Dunn MJ. Alterations in subcellular localization of p38 MAPK potentiate endothelin-stimulated COX-2 expression in glomerular mesangial cells. J. Biol. Chem. 2003;278:51928–51936. doi: 10.1074/jbc.M309256200. [DOI] [PubMed] [Google Scholar]
- 47.Mizumura K, Hashimoto S, Maruoka S, Gon Y, Kitamura N, Matsumoto K, Hayashi S, Shimizu K, Horie T. Role of mitogen-activated protein kinases in influenza virus induction of prostaglandin E-2 from arachidonic acid in bronchial epithelial cells. Clin. Exp. Allergy. 2003;33:1244–1251. doi: 10.1046/j.1365-2222.2003.01750.x. [DOI] [PubMed] [Google Scholar]
- 48.Subbaramaiah K, Marmo TP, Dixon DA, Dannenberg AJ. Regulation of cyclooxgenase-2 mRNA stability by taxanes - Evidence for involvement of p38, MAPKAPK-2, and HuR. J. Biol. Chem. 2003;278:37637–37647. doi: 10.1074/jbc.M301481200. [DOI] [PubMed] [Google Scholar]
- 49.Faour WH, He YL, He QW, de Ladurantaye M, Quintero M, Mancini A, Di Battista JA. Prostaglandin E-2 regulates the level and stability of cyclooxygenase-2 mRNA through activation of p38 mitogen-activated protein kinase in interleukin-1 beta-treated human synovial fibroblasts. J. Biol. Chem. 2001;276:31720–31731. doi: 10.1074/jbc.M104036200. [DOI] [PubMed] [Google Scholar]
- 50.Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, Hoffman CR, Kost TA, Lambert MH, Leesnitzer MA, McCauley P, McGeehan G, Mitchell J, Moyer M, Pahel G, Rocque W, Overton LK, Schoenen F, Seaton T, Su JL, Becherer JD. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature. 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 51.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 52.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol. Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Lint P, Libert C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J. Leukoc. Biol. 2007;82:1375–1381. doi: 10.1189/jlb.0607338. [DOI] [PubMed] [Google Scholar]
- 54.Fingleton B. Matrix metalloproteinases: roles in cancer and metastasis. Frontiers in Bioscience. 2006;11:479–491. doi: 10.2741/1811. [DOI] [PubMed] [Google Scholar]