Abstract
Processing of rRNA in mammalian cells includes a series of cleavages of the primary 47S transcript and results in producing three rRNAs: 18S, 28S and 5.8S. The sequence of the main processing events in human cells has been established, but little is yet known about the dynamics of this process, especially the dynamics of its early stages. In the present study, we used real-time PCR to measure levels of pre-rRNA after inhibition of transcription with actinomycin D. Thus we could estimate the half-life time of rRNA transcripts in two human-derived cell lines, HeLa and LEP (human embryonic fibroblasts), as well as in mouse NIH 3T3 cells. The primary transcripts seemed to be more stable in the human than in the murine cells. Remarkably, the graphs in all cases showed more or less pronounced lag phase, which may reflect preparatory events preceding the first cleavage of the pre-rRNA. Additionally, we followed the dynamics of the decay of the 5′ETS fragment which is degraded only after the formation of 41S rRNA. According to our estimates, the corresponding three (or four) steps of the processing in human cells take five to eight minutes.
Keywords: rRNA processing, cleavage, half-life time, primary transcript, human, mouse
Introduction
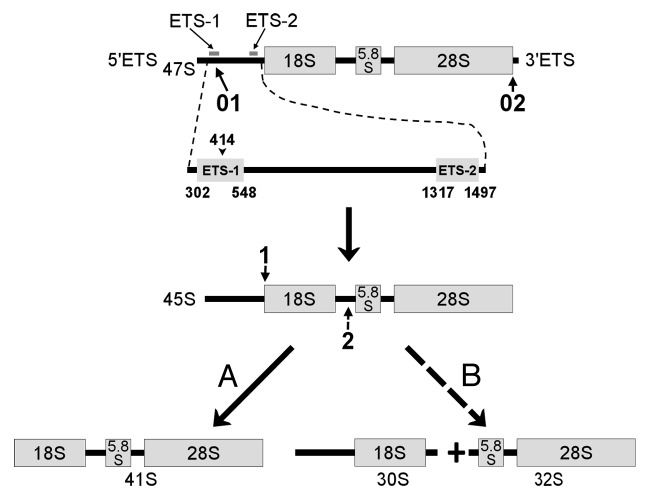
The RNA polymerase I (pol I) generates the largest fraction of all newly synthesized RNAs in growing eukaryotic cells by producing a single large rRNA (rRNA) precursor. This primary transcript (47S in mammalian species) undergoes processing into 18S, 5.8S and 28S rRNAs and assembly into mature ribosomal subunits1,2; reviewed in refs. 3–8. The rRNA processing consists in removing so-called external transcribed spacers (5′ETS and 3′ETS) and internal transcribed spacers (ITS), of which eukaryotic cells have at least two.3,9 The spacer sequences vary greatly in composition and length in different organisms, though the core elements in the mature rRNAs are highly conserved (reviewed in ref. 9). In human cells, the order of the processing events was chiefly determined in the early 1990s. The initial steps include cleavage of the primary transcript close to 5′ end (position +414) and removal of 3′ETS. The resulting 45S rRNA may be cleaved at the 5′ end of 18S RNA gene (the main pathway) or within the first internal transcribed spacer (relatively rare alternative pathway) (Fig. 1). In murine cells, the first processing cleavage of the primary transcript occurs in the position +650, then 3′ETS is removed.10-12 The resulting 45S rRNA may be cleaved either within ITS1 or at the additional cleavage site A0 which is situated within the 5′ETS and has an analog in human cells.10,13
Figure 1. First steps of human pre-RNA processing. (A) represents the main pathway. (B) is a relatively rare alternative. 01, Cleavage of 5′ETS at position +414; 02, elimination of 3′ETS; 1, cleavage at the 5′ end of 18S sequence, which is followed by degradation of 5′ETS remnant; 2, alternative cleavage within the first ITS. ETS-1 and ETS-2 are the fragments of 5′ETS amplified by real-time PCR in our work. Site A0 within 5′ETS is not shown.
Much less is known about the dynamics of the rRNA processing. Pulse-chase analysis with L-methyl 3H methionine so far allowed to investigate only later steps of the processing.14,15 In the work of Lazdins et al.,16 inhibition of transcription with actinomycin D (ActD) and S1 nuclease analysis were used to estimate the pre-rRNA half-life time in rodent cells. But for the human cells, even the life-span of pre-rRNA remains unknown.
In the present study, we employed real-time qPCR to measure levels of pre-rRNA and the processing products. This method is extremely powerful and can generate reliable, reproducible and biologically meaningful results; wherefore, in many cases qPCR is superseding other approaches, such as northern blotting and RNase protection essays.17,18 Using primers within 5′ETS region, we measured the levels of pre-rRNA in the human derived transformed HeLa cells and diploid LEP cells, as well as in murine NIH 3T3 cells, after inhibition of rDNA transcription with ActD. The obtained data allowed us to estimate the period of half-life of the human primary transcripts. Additionally, to estimate the rate of the few further steps of rRNA processing, we determined the expression of the part of 5′ETS fragment which is to be degraded only after the third (or, alternatively, the fourth) cleavage of the primary transcript (Fig. 1).
Results
Estimation of the half-life of the human and murine pre-rRNA
Since in human cells the primary transcript is cleaved inside 5′ETS at position +414,11,19 reviewed in ref. 10, we estimated the pre-rRNA half-life time in HeLa, LEP by amplification of the 246 bp long (from +302 to +548) 5′ETS fragment (ETS-1, Fig. 1), after inhibition of rDNA transcription with 20 μg/ml actinomycin D (ActD) for 1, 2, 5, 10, 15 and 20 min. ActD is known to inhibit pre-rRNA synthesis, but allows processing of previously transcribed rRNA intermediates.20,21 Similarly we treated murine 3T3 cells in which we amplified the fragment mETS (see Methods).
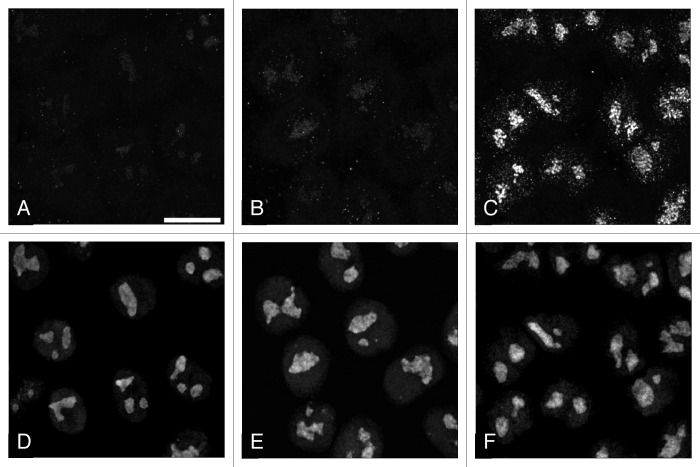
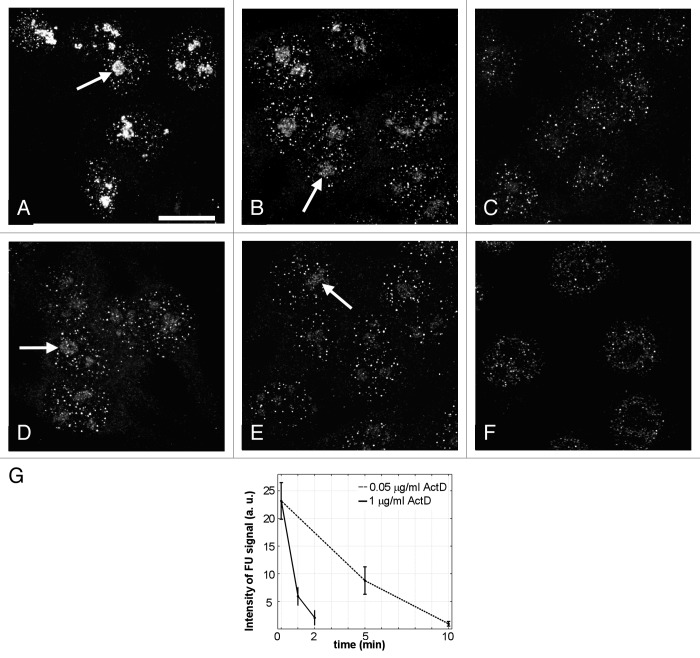
To assess the efficiency of the inhibition, the cells were incubated with 5-fluorouridine (FU), then fixed and processed for immunocytochemistry. An intensive fluorescent signal within nucleoli was observed in control after 1.5 min of incubation, but no signal after one minute of incubation with FU (Fig. 2). Thus we established that more than one minute is required for the effective incorporation of FU by the cells, but 1.5 min is sufficient for that. When, after incubation of the cells with FU for one minute, we added Act D in concentration 20 μg/ml for 0.5 min, without changing the cultivating medium, there was again no transcription signal in the nucleoli. From these results we inferred that the activity of pol I was efficiently arrested by ActD within half a minute. In other experiments we found that at lower concentrations of this inhibitor (0.05 μg/ml and 1 μg/ml) rRNA synthesis in the nucleoli continued for a long period (Fig. 3A–E). In such conditions, the levels of ETS-1 would not give us reliable data on the dynamic of processing. Therefore, for the estimation of the half-life time of pre-rRNA, we used the high concentration of ActD. The same concentration of the drug was used in the work of Lazdins et al.16
Figure 2. Visualization of the transcription signal and nucleolin in the nucleoli of HeLa cells. Incubation with FU for 60 sec (A, D), 75 sec (B, E), 90 sec (C, F). (A-C): FU; (D–F): nucleolin. Scale bar: 10 μm.
Figure 3. Effect of actinomycin D on rDNA transcription in HeLa cells. Transcription signal was visualized after incubation with FU for 2min. Arrows show the transcription signal in the nucleoli. (A) Control. Intensive transcription signal in the nucleoli and dot-like signals in the nucleoplasm of the cells. (B) 0.05 μg/ml ActD, 5 min. (C) 0.05 μg/ml ActD, 10 min. (D) 1 μg/ml ActD, one minute. (E) 1 μg/ml ActD, 2 min. (F) 20 μg/ml ActD, one minute. In contrast to F, a reduced FU signal is still observed within the nucleoli in B, D and E. Scale bar: 10 μm. (G) Integral intensities of the transcription (FU) signal in the nucleoli after ActD treatment.
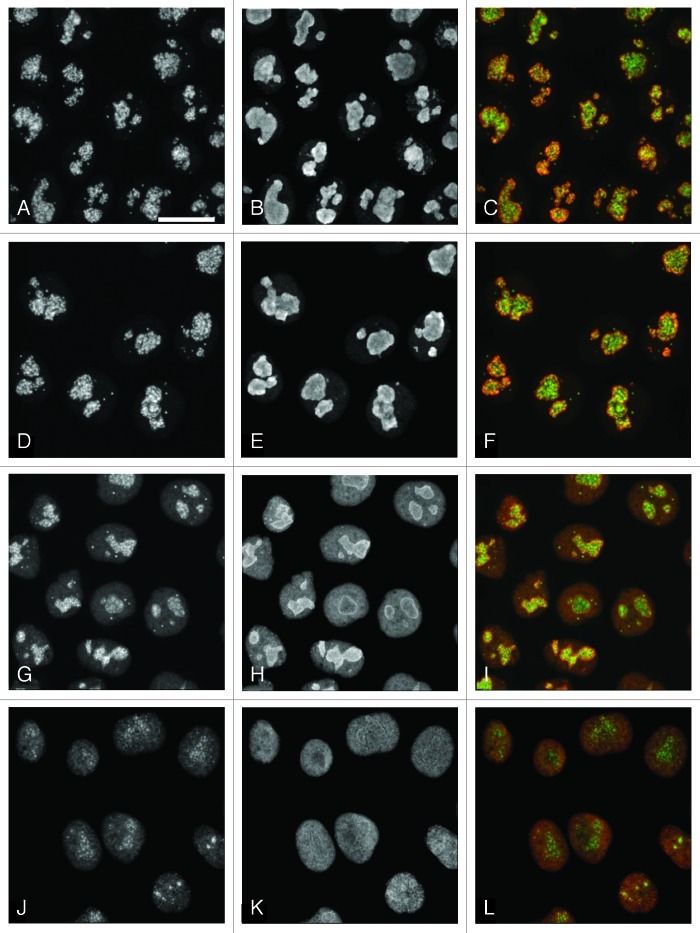
In such conditions, the possibility of side-effects, even after a short period of treatment, cannot be excluded. But we found no significant changes in the fibrillarin labeling pattern of the nucleoli after 10 min of the ActD treatment. After 20 min, the structure of nucleoli was altered; after 30 min, nucleoli were disrupted and their contents largely spread over the nucleoplasm (Fig. 4A–L). Since fibrillarin is a most important component of the nucleolar structure and, being a component of U3 snoRNP, plays a key role in rRNA processing,20,22-24 our data indicate that rRNA processing was not significantly affected during the short period of treatment.
Figure 4. Localization of fibrillarin (green) and nucleolin (red) after ActD treatment. (A–C): Control. (D–F): 20 μg/ml ActD, 10 min. Fibrillarin scaffold of the nucleoli appears to be intact. (G–I): 20 μg/ml ActD, 20 min. The size and shape of nucleoli are still preserved, but fibrillarin tends to form coarse clumps. (J–L): 20 μg/ml ActD, 30 min. Nucleoli are disrupted, fibrillarin is largely spread over the nucleoplasm. Scale bar: 7 μm.
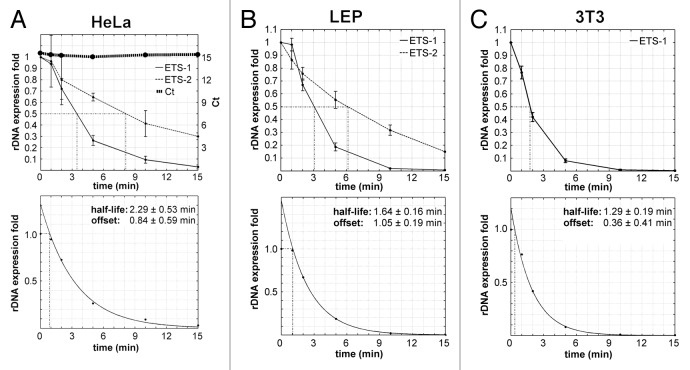
By means of real time PCR, we measured the fold changes of corresponding rRNA in relation to the control without ActD treatment in HeLa, LEP and 3T3 cells. As the internal control, we selected human, respectively murine, β actin gene. Expression of this gene did not change significantly during the incubation with ActD in the conditions of our experiments (data not shown). The results of the RT PCR measurements are presented in Figure 5A–C. After 11 min of ActD treatment in HeLa cells, after eight minutes in LEP cells and after seven minutes in 3T3 cells, the expression fell below 5% of the original level. In case of the LEP cell line, we observed no correlation of the results with the number of passages when the latter varied from 26 to 29.
Figure 5. Expression levels of ETS-1, ETS-2 and mETS measured by qPCR after incubation of cells with 20 μg/ml ActD. (A) HeLa cells. Experimental data for ETS-1 and ETS-2 (top) and exponential fitting for ETS-1 (bottom). The top line represents the values of cycle threshold (Ct) for the internal control (the product of human β actin gene). These values do not change significantly in the course of the experiment. (B) LEP cells. Experimental data for ETS-1 and ETS-2 (top) and exponential fitting for ETS-1 (bottom). (C) 3T3 cells. Experimental data for ETS-1 (top) and exponential fitting for ETS-1 (bottom). Three independent experiments were performed for each cell type. For each experiment, the exponential fitting was computed by constrained nonlinear least squares method32 in the Matlab environment (Mathworks). In the time interval from two to 15 min, we found optimal parameters a0 and t0 of the function y(t) = exp[a0 × (t − t0)], where a0 is the exponential decay and t0 is the time offset. The only constraint used was t0 > 0. Each curve for ETS-1 has a short initial segment (lag phase) and a long part well approximated by the exponential model.
After exponential fitting of the experimental curves, we obtained the following results for the half-life of the primary transcripts (mean value ± standard deviation): 2.29 ± 0.53 min in HeLa cells (Fig. 5A), 1.64 ± 0.16 min in LEP cells (Fig. 5B) and 1.29 ± 0.19 min in 3T3 cells (Fig. 5C). Additionally, we perceived that each experimental curve had an initial lag phase (Fig. 5). As a measure of the corresponding lag periods we used the offset values of the model exponents: 0.84 min for HeLa cells, 1.05 for LEP cells and 0.36 min for 3T3 cells (shown in the bottom graphs of Fig. 5). It should be observed that these values in the human cells exceeded the time required for the arrest of rDNA transcription with ActD. Moreover, the lag period for 3T3 cells (derived from the murine fibroblasts) was shorter than for LEP cells (derived from the human fibroblasts). These circumstances suggest that the observed lag phase is a regular feature of the rRNA processing dynamics, rather than a consequence of still continuing transcription. Apparently, these relatively short lag periods can be detected only if measurements of the ETS-1 begin as early as one minute after introduction of ActD.
Life time of ETS-2 in human cells
To estimate the rate of a few further processing steps in the human cells, we studied the life-time of ETS-2. This fragment located downstream of the primary processing site (+1317 to +1497) belongs to the part of 5′ETS that remains intact after the first processing cleavage, but decays after the formation of 41S rRNA, i.e., after the third processing cleavage (or the fourth in case of a relatively rare alternative pathway) (Fig. 1). Upon arresting the rRNA transcription with ActD, as in the above described experiments, we found that the level of ETS-2 expression was reduced to a half of its original value after 8.1 ± 1.9 min in HeLa cells (Fig. 5A) and after 6.1 ± 1.one minute in LEP cells (Fig. 5B). Comparing these data with the results for ETS-1, we may roughly estimate the period between the first and third (or fourth) cleavage events as five minutes in HeLa cells and three minutes in LEP cells.
Levels of ETS-1 and ETS-2 in human cells after treatment with low doses of ActD
In mammalian cells, ActD in concentration of 0.05 μg/ml inhibits pol I, but largely spares the activity of RNA polymerase II.25,26 Here we examined the effects of this low ActD concentration on the expression levels of ETS-1 and ETS-2. HeLa and LEP cells were incubated with 0.05 μg/ml ActD for 10, 30, 50, 70 90 and 120 min. In these experiments, the level of ETS-1 expression was decreased to one half of its original value in HeLa cells after 16 min (Fig. 6A) and in LEP cells after 8 min (Fig. 6B). Surprisingly, we observed the same dynamics in the expression of ETS-1 and ETS-2 (Fig. 6A and B), though the ETS-2 RNA persists longer than the ETS-1 RNA at the higher dose of ActD. To assess the efficiency of pol I inhibition in our experiments, we visualized the transcription signal in the nucleoli of HeLa cells after two minute incubation with FU. In the absence of ActD, all interphase cells showed intensive transcription signal in both nucleoli and nucleoplasm (Fig. 3A). After two minutes of the incubation with low dose ActD, the cells appeared as in the control; after five minutes of the treatment, the intensity of labeling was significantly decreased (Fig. 3B); after 10 min, the transcription signal disappeared from the nucleoli, though it was still intensive in the nucleoplasm (Fig. 3C). Thus the pol I was completely inactivated only after 10 min of the low dose ActD treatment. Accordingly, the peculiar results of our RT PCR measurements may be attributed to accumulation of aborted transcripts at low concentration of ActD. In these conditions, pol I activity is inhibited only partly,27,28 and the short products of transcription are likely subjected to a polyadenylation dependent decay.28 The remarkable coincidence of the expression curves for ETS-1 and ETS-2 at low doses of ActD (compare Figs. 5A, B and 6) suggests that these curves reflect the degradation of the incomplete transcripts rather than the processing itself. This may account for the absence of the lag phase on the graphs (Fig. 6).
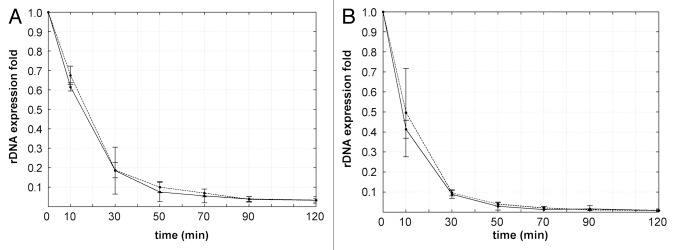
Figure 6. Expression levels of ETS-1 and ETS-2 in HeLa (A) and LEP (B) cells after incubation with 0.05 μg/ml ActD. In both human-derived cell lines the levels of ETS-1 and of ETS-2 showed a very similar dynamic (compare with Fig. 5A and B). Each curve presents the data of three independent experiments.
Discussion
The results of our work show that RT PCR may be used for the study of early stages of rRNA processing. One particular advantage of this approach, as compared with the hybridization methods,16,20,21 is the possibility of detecting regions with precisely defined 5′ and 3′ ends by selection of the proper couples of primers. Thus, we do not need to presume an immediate degradation of the removed spacers, but focus on the concrete cleavage events.
In this work we show that treatment of the human and murine cells with ActD in high concentration (20 μg/ml) efficiently inhibits rDNA transcription in less than half a minute. This finding allowed us to study the early dynamics of the rRNA processing in different cell lines by RT PCR.
The presence of a lag phase on all the curves depicting the expression of ETS-1 (Fig. 5) indicates that the early processing of the 47S rRNA may be divided into two periods. We suppose that the first of them, lasting for about one minute in the human-derived HeLa and LEP cells, is required for the preparatory events (such as recognition of the specific rRNA targets, base modification, formation of various secondary structures, etc.9,10) which precede the first cleavage of the transcript. Then the second period, corresponding to the exponential decay, is probably occupied by the cleavage alone. In conformity with our hypothesis, we can obtain new estimates for the half-life time of the primary transcript by adding the values of lag periods to the values of the half-life calculated from the exponents (Fig. 5). In this way, we find the “complete” half-life of pre-rRNA for HeLa cells: 2.29 + 0.84 = 3.1 ± 0.8 min; for LEP cells: 1.64 + 1.05 = 2.7 ± 0.3 min. In the case of the murine 3T3 cells, the lag period is much shorter, and we cannot exclude that it just equals the time required for the inhibitory action of ActD upon the pol I activity. Nevertheless, if we make similar calculation of the “complete” half-life time for these cells, i.e., 1.29 + 0.36 = 1.7 ± 0.5 min, the result will coincide with the data obtained earlier by other authors for the murine Ltk and hamster CHO cells.16 The human primary transcripts seem to be more stable (with the half-life about 3 min) than those of rodents. This supposition agrees with the data of earlier works25,29,30 indicating that the first processing steps are more rapid in murine cells than in human cells.
Following the expression of ETS-2 in the absence of transcription, we could shed some more light on the temporal course of the early steps of rRNA processing in human cells. The curves (Fig. 5A and B) show that the first cleavage of the primary transcript and the final degradation of the 5′ETS are spaced by three to five minutes. This period slightly exceeds the half-life of the primary transcript.
To summarize, our data suggest that in mammalian cells there is a preparatory period (about one minute in human cells) between the formation of primary transcript (47S rRNA) and its first cleavage. Considering this, we have the following rough estimates for the “complete” half-life times of the primary rRNA transcript: in human-derived HeLa and LEP cells, about three minutes; in the murine NIH 3T3 cells, about two minutes. Additionally, according to our results, the formation of 41S rRNA (the third or fourth step of the processing) from the primary human transcript takes five to eight minutes.
Materials and Methods
Cell culture and drug treatment
HeLa and primary LEP (human embryonic fibroblast, Sevapharm, Czech Republic) cells were cultivated at 37°C in Dulbecco modified Eagle's medium (DMEM, Sigma, #D5546) containing 10% fetal calf serum, 1% glutamine, 0.1% gentamycin and 0.85 g/l NaHCO3 in atmosphere supplemented with 5% CO2. Mouse NIH 3T3 cells were maintained in DMEM supplemented with 10% calf serum and 1 mg/ml penicillin/streptomycin. Sub-confluent cells (in 60 mm plastic dishes) were incubated with 0.05, 1.0 and 20 μg/ml actinomycin D (Sigma, #A1410) for various periods from one minute to two hours.
RNA isolation and reverse transcription
Total RNA was extracted from human HeLa, LEP and mouse 3T3 cells with TRIzol reagent (Invitrogen, #10296–028), dried and resuspended in diethyl pyrocarbonate-treated sterile water. Contaminating DNA was removed from the RNA samples by incubation for 45 min with RNase-free DNase I (Fermentas, # EN0525) according to manufacturer’s instructions.
Reverse transcription was performed using RevertAid Reverse Transcriptase (Fermentas, #EP0441). The cDNA was prepared from 2 μg DNA-free RNA in a 20 μl reaction mixture containing: reaction buffer [50 mM TRIS-HCl (pH 8.3 at 25°C), 50 mM KCl, 4 mM MgCl2 and 50mM DTT]; 1 mM each of dATP, dTTP, dCTP, dGTP; 20U rRNasin ribonuclease inhibitor; 100U moloney murine leukemia virus reverse transcriptase (M-MuLV RT); and 100 pmol of random hexamer primer. The reactions were incubated initially at 25°C (10 min) and then at 42°C for one hour. To stop the reaction, the mixture was heated at 65°C for 10 min.
Real-time PCR
Two different fragments of human 5′ETS (ETS-1 and ETS-2, Fig. 1) and one fragment of murine 5′ETS (mETS) were amplified after transcription inhibition with ActD. ETS-1 is positioned between +302 and +548 (246 bp). This fragment includes the first processing cleavage site at +414, so it was used for estimation of the pre-rRNA half-life time. ETS-2, positioned between +1317 and +1497 (180 bp), was selected for the evaluation of 5′ETS life span, because it is contained within that part of 5′ETS which is to be degraded after the cleavage at the 5′ end of the 18S segment. The murine mETS is situated between +444 and +683 (240 bp). This fragment included the first cleavage site at position +650. Fragments of human β-actin (253 bp) and mouse β-actin (238 bp) were used as the internal control for normalization of the relative quantification results, respectively in human and murine cells.
These fragments were amplified using BioRad CFX 1000 real-time PCR system and iQ SYBR Green Supermix (BioRad, #170–8880). The reaction mix volume (25 μl) included 10 pM of each primer and 2 μl of cDNA diluted 1:10. All samples had duplicates. Calibration curves based on five serial cDNA dilutions (1, 10−1, 10−2, 10−3 and 10−4) were used for calculation of the reactions efficiencies. Primers were designed with freely available Primer3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). The following primer sequences were used:
For ETS-1: 5′-gtgcgtgtcaggcgttct-3'(forward); 5′-gggagaggagcagacgag-3′(reverse).
For ETS-2: 5′-ccttccccaggcgtccctcg-3′ and 5′-ggcagcgctaccataacgga-3′.
For mETS: 5′-ttttggggaggtggagagtc-3′ and 5′-agagaactccggagcaccac-3′.
For human β-actin: 5′-tgcgtctggacctggctggc-3′ and 5′-gcctcagggcagcggaaccg-3′.
For murine β-actin: 5′-cgtgcaccgcaagtgctt-3′ and 5′-gctgtcgccttcaccgtt-3′.
Amplification of ETS-1 was performed at the following thermal conditions: initial DNA denaturation—94°C for 3 min followed by 40 cycles including denaturation at 94°C for 45 sec, annealing at 58°C for 30 sec, DNA synthesis at 72°C for 30 sec. The amplification program for ETS-2 was: 94°C for 3 min followed by 40 cycles including denaturation at 94°C for 45 sec, annealing at 62°C for 30 sec and 72°C for 30 sec. Fragment of human β actin was successfully amplified with both of these programs. For the murine primers, the amplification program was: 94°C for 3 min followed by 40 cycles including 45 sec at 94°C, 30 sec at 59°C and 30 sec at 72°C.
Incorporation of fluorouridine
Cell cultures were incubated two minutes with 10 μM of the 5-fluorouridine (FU) (Sigma, #F5130) and fixed 30 min in methanol at -20°C. Such fixation improves the accessibility of the halogenated epitope.31 Inc. FU was then visualized using mouse monoclonal anti-BrdU antibody (Sigma, #B8434) diluted 1/50 and secondary Cy3-conjugated goat anti-mouse antibody (Jackson Laboratories, A10521). After several rinses, the slides were mounted with mowiol.
For visualization of fibrillarin in the nucleoli, we used primary monoclonal antibody against mouse fibrillarin (clone 17C12), kindly donated by Kenneth M. Pollard (Scripps Research Institute). Nucleolin was stained with rabbit anti-nucleolin antibody (abcam, #ab22758).
Acknowledgments
The work was supported by the grants: P302/12/G157 and P302/12/1885 from the Czech Grant Foundation, Prvouk/1LF/1, Prvouk /3LF P34 and UNCE204022 from the Charles University in Prague and OPVK CZ.1.07/2.3.00/30.0030 from the Ministry of Education, Youth and Sport of the Czech Republic.
Glossary
Abbreviations:
- pol I
RNA polymerase I, ActD: actinomycin D, ETS: external transcribed spacer, ITS: internal transcribed spacer
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/23985
References
- 1.Kominami R, Urano Y, Mishima Y, Muramatsu M. Organization of ribosomal RNA gene repeats of the mouse. Nucleic Acids Res. 1981;9:3219–33. doi: 10.1093/nar/9.14.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalez IL, Sylvester JE. Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics. 1995;27:320–8. doi: 10.1006/geno.1995.1049. [DOI] [PubMed] [Google Scholar]
- 3.Eichler DC, Craig N. Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1994;49:197–239. doi: 10.1016/S0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- 4.Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–8. doi: 10.1016/S0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 5.Gerbi SA, Borovjagin AV. Pre-Ribosomal RNA Processing in Multicellular Organisms. In: Olson MOJ, ed. The Nucleolus: Springer, 2004:170-98. [Google Scholar]
- 6.Granneman S, Baserga SJ. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr Opin Cell Biol. 2005;17:281–6. doi: 10.1016/j.ceb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Raska I, Shaw PJ, Cmarko D. New insights into nucleolar architecture and activity. Int Rev Cytol. 2006;255:177–235. doi: 10.1016/S0074-7696(06)55004-1. [DOI] [PubMed] [Google Scholar]
- 8.Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–59. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nazar RN. Ribosomal RNA processing and ribosome biogenesis in eukaryotes. IUBMB Life. 2004;56:457–65. doi: 10.1080/15216540400010867. [DOI] [PubMed] [Google Scholar]
- 10.Mullineux ST, Lafontaine DLJ. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie. 2012;94:1521–32. doi: 10.1016/j.biochi.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Kass S, Craig N, Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol. 1987;7:2891–8. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strezoska Z, Pestov DG, Lau LF. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J Biol Chem. 2002;277:29617–25. doi: 10.1074/jbc.M204381200. [DOI] [PubMed] [Google Scholar]
- 13.Kent T, Lapik YR, Pestov DG. The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA. 2009;15:14–20. doi: 10.1261/rna.1384709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonnart C, Gérus M, Hoareau-Aveilla C, Kiss T, Caizergues-Ferrer M, Henry Y, et al. Mammalian HCA66 protein is required for both ribosome synthesis and centriole duplication. Nucleic Acids Res. 2012;40:6270–89. doi: 10.1093/nar/gks234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gérus M, Bonnart C, Caizergues-Ferrer M, Henry Y, Henras AK. Evolutionarily conserved function of RRP36 in early cleavages of the pre-rRNA and production of the 40S ribosomal subunit. Mol Cell Biol. 2010;30:1130–44. doi: 10.1128/MCB.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazdins IB, Delannoy M, Sollner-Webb B. Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma. 1997;105:481–95. doi: 10.1007/BF02510485. [DOI] [PubMed] [Google Scholar]
- 17.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155–66. [PMC free article] [PubMed] [Google Scholar]
- 18.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–26. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 19.Hadjiolova KV, Nicoloso M, Mazan S, Hadjiolov AA, Bachellerie JP. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur J Biochem. 1993;212:211–5. doi: 10.1111/j.1432-1033.1993.tb17652.x. [DOI] [PubMed] [Google Scholar]
- 20.Puvion-Dutilleul F, Mazan S, Nicoloso M, Pichard E, Bachellerie JP, Puvion E. Alterations of nucleolar ultrastructure and ribosome biogenesis by actinomycin D. Implications for U3 snRNP function. Eur J Cell Biol. 1992;58:149–62. [PubMed] [Google Scholar]
- 21.Puvion-Dutilleul F, Puvion E, Bachellerie JP. Early stages of pre-rRNA formation within the nucleolar ultrastructure of mouse cells studied by in situ hybridization with a 5’ETS leader probe. Chromosoma. 1997;105:496–505. doi: 10.1007/BF02510486. [DOI] [PubMed] [Google Scholar]
- 22.Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-F. [DOI] [PubMed] [Google Scholar]
- 23.Tollervey D, Hurt EC. The role of small nucleolar ribonucleoproteins in ribosome synthesis. Mol Biol Rep. 1990;14:103–6. doi: 10.1007/BF00360433. [DOI] [PubMed] [Google Scholar]
- 24.Reichow SL, Hamma T, Ferré-D’Amaré AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–64. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker KA, Bond U. Analysis of pre-rRNAs in heat-shocked HeLa cells allows identification of the upstream termination site of human polymerase I transcription. Mol Cell Biol. 1989;9:2500–12. doi: 10.1128/mcb.9.6.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry RP, Kelley DE. Persistent synthesis of 5S RNA when production of 28S and 18S ribosomal RNA is inhibited by low doses of actinomycin D. J Cell Physiol. 1968;72:235–46. doi: 10.1002/jcp.1040720311. [DOI] [PubMed] [Google Scholar]
- 27.Hadjiolova KV, Hadjiolov AA, Bachellerie JP. Actinomycin D stimulates the transcription of rRNA minigenes transfected into mouse cells. Implications for the in vivo hypersensitivity of rRNA gene transcription. Eur J Biochem. 1995;228:605–15. doi: 10.1111/j.1432-1033.1995.0605m.x. [DOI] [PubMed] [Google Scholar]
- 28.Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–11. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller KG, Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981;27:165–74. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- 30.Financsek I, Mizumoto K, Mishima Y, Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982;79:3092–6. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koberna K, Malínský J, Pliss A, Masata M, Vecerova J, Fialová M, et al. Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. J Cell Biol. 2002;157:743–8. doi: 10.1083/jcb.200202007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balda M. LMFnlsq - Solution of nonlinear least squares. MathWorks, MATLAB Central, File Exchange, Id=17534. 2008. [Online, http://www.mathworks.com/matlabcentral/fileexchange/17534, of 12.11.2012].