Abstract
Gene loops have been described in different organisms from yeast to human and form through interaction between components of the transcription pre-initiation complex and Ssu72, a member of the 3′ end cleavage and polyadenylation complex. A recent study by Tan-Wong et al. reports a new role for gene loops in promoting ORF transcription directionality from otherwise bidirectional promoters.
Keywords: non-coding RNA, gene loop, histone deacetylase, bidirectional promoters, antisense RNA
Many yeast promoters are bidirectional and produce, in addition to the protein coding mRNA, non-coding RNAs (ncRNAs) from the opposite strand in a divergent orientation.1,2 This feature, shared by about 50% of S. cerevisiae promoters, contributes to the genome pervasive transcription. While a small subset of these ncRNAs has been attributed a regulatory role, genetic evidence for functionality is lacking for the vast majority of these transcripts. It is therefore reasonable to think that the cell has evolved mechanisms to keep this potential transcriptional noise under control.
Different strategies to maintain transcriptome steady-state levels have been described, which mainly involve RNA decay pathways. Indeed many non-coding RNAs are unstable and detected only in strains defective in the machineries responsible for their rapid elimination. Thus, a large fraction of ncRNAs called cryptic unstable transcripts (CUTs) are degraded by the 3′ to 5′ exonuclease activity of the nuclear exosome component Rrp6,1-3 while others are degraded by the cytoplasmic exonuclease Xrn1 (XUTs).4 Another proposed option to reduce the amount of divergent transcripts is to force the transcription orientation of a bidirectional promoter toward the coding sequence. Evidence supports that regulation of chromatin modifications and nucleosome remodelling may influence transcription directionality.5,6
The recent study by Tan-Wong et al. reports a new mechanism able to restrain divergent ncRNA synthesis from bidirectional promoters.7 The study provides evidence that gene loops resulting from the transcription-induced interaction of the promoter with the 3′ end of protein coding units8 enhance transcriptional directionality. Based on chromatin conformation capture (3C) experiments and transcript quantifications, the authors show that disruption of the gene loop in the ssu72–2 mutant leads to increased divergent transcription from the FMP27 promoter region. Ssu72 is a phosphatase part of the 3′ end cleavage and polyadenylation factor (CPF) that was implicated in the maintenance of the gene loop structure through its ability to also interact with promoter elements.9
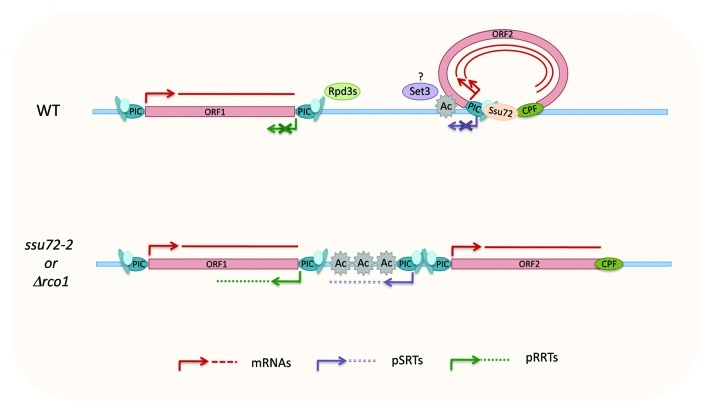
Tan-Wong et al. extend the observation on FMP27 to a more global analysis using tiling arrays in ssu72–2 and Δrrp6 single and double mutants. They identify a series of new transcripts defined as SRTs (Ssu72-restricted transcripts) in addition to the CUTs revealed by loss of the nuclear exosome component Rrp6. The authors then restrict the analysis to pairs of spaced tandem genes to demonstrate that the promoter associated SRTs (pSRTs) originate from the bidirectional promoter of the downstream ORF and are distinct from antisense transcripts potentially initiating within the transcription termination region of the upstream ORF (Fig. 1). Furthermore RNA PolII occupancy experiments indicate that the pSRTs appearing in ssu72–2 result from de novo transcription initiation.
Figure 1. Ssu72-dependent gene loops form upon ORF transcription, which results in reduced (Set3 dependent?) histone H4 acetylation at the promoter, restricting divergent pSRT transcription. pSRTs arise in ssu72–2 and are distinct from RRTs generated in the Rpd3s mutant Δrco1 at the 3′ end of genes in antisense orientation.
Interestingly, inspection of published genome-wide histone acetylation levels10 reveals that pSRT-associated promoters show significantly reduced histone H4 acetylation. Moreover, the authors detect an increase in promoter acetylation when abrogating gene loop formation in a ssu72–2 mutant background. These observations suggest that gene loops may favor the recruitment of a histone deacetylase (HDAC) in order to maintain the promoter in a deacetylated state limiting firing of divergent pSRTs.
The identification of the HDAC responsible for this deacetylation is not addressed in this paper, but the authors exclude the involvement of the Rpd3 small (Rpd3s) H4 deacetylation complex. Rpd3s is recruited via its Eaf3 or Rco1 subunits on histone H3 methylated on lysine 36 (H3K36me) by Set2 in the body of transcribed genes.5,11 Using a nascent transcript sequencing (NET-Seq) approach in Δrco1, Rpd3s was recently proposed by Churchman et al.12 to contribute to promoter directionality by repressing divergent transcription following its recruitment at the 3′ region of the upstream transcribed gene methylated on H3K36. However, comparing the entire set of pSRTs with the Rco1-restricted transcripts (RRTs) identified by Churchman et al. or by performing additional tiling arrays in Δrco1 and Δrco1Δrrp6 mutants, Tan-Wong et al. conclude that these transcripts have different features. While the pSRTs are associated with the transcription start site (TSS) of the downstream ORF, the RRTs are linked and antisense to the transcription termination site (TTS) of the upstream ORF in tandem pairs (Fig. 1).
Although the distinction between the two classes can only be established when the tandem genes are more than 400 bp apart, the results indicate that pSRTs, but not RRTs, derive from bidirectional promoters identifying Ssu72 rather than Rco1 as a major contributor to promoter directionality. In support of this view, the ssu72–2 mutation also induces a weak downregulation of the downstream ORF in a pair when a pSRT is present in between. Although the reported decrease in mRNA levels is modest, this observation suggests that promoters have an intrinsic ability to fire in both directions but the loop structure imposes transcription directionality limiting the assembly of two adjacent divergent PICs as recently proposed.13 Thus a single PIC may form in nucleosome free regions associated with promoters engaged in loops. In sufficiently spaced tandem ORFs, a second independent PIC may form that promotes synthesis in the opposite direction to generate antisense RRTs associated with the 3′ end of the upstream ORF. This scenario is also supported by recent data from Murray et al. showing that, despite the compact nature of the yeast genome, 3′ regions of genes can drive antisense transcription independently of a downstream bidirectional promoter. These observations imply the existence of a possible autonomous control of antisense transcription that may act as a widespread component of gene expression regulation.14 However, depending on their length and the distance between tandem genes, a fraction of divergent ncRNAs originating at bidirectional promoters may also be antisense to the upstream ORF.15
Excluding an effect of Rpd3s on the gene-loop enhanced transcriptional directionality, the authors propose Set3, an HDAC recruited to the 5′ region of transcribed genes by di-methylated H3K4 deposited by the methyltransferase Set1,16,17 as a potential HDAC to promote histone deacetylation at bidirectional promoters. Although no experiments were performed to confirm this hypothesis, Set3 appears as a plausible candidate due to its localization and already demonstrated ability to repress cryptic promoters in 5′-transcribed regions.17 This possibility raises the question of the signal responsible for recruiting this HDAC in the context of gene loops. Overall, the findings from both Churchman et al. and Tan-Wong et al. highlight the importance and the generality of deacetylation activities to repress either intragenic antisense transcription or divergent transcription from bidirectional promoters in favor of a more controlled mRNA production.
Of note, the reduced mRNA levels observed in ssu72–2 may not only be due to gene loop abrogation paralleled by increased acetylation and divergent transcription. Indeed, Ssu72 is a phosphatase involved in the dephosphorylation of Ser5P in the RNAPII C-terminal domain (CTD). This Ssu72 activity enhances productive transcription elongation by negatively regulating the association of the Nrd1-Nab3-Sen1 (NNS) early termination complex with RNAPII CTD.18,19 Furthermore, recent PAR CLiP data reveal the presence of NNS at the 5′ end of many ORFs.20 Thus, the observed repressive effect of the ssu72–2 mutant on ORFs could be due, at least in part, to the effect of this mutation on transcription elongation and NNS dependent termination.
ssu72–2 is not the only mutant resulting in gene loop disruption and increased pSRT transcription. Indeed, Tan-Wong et al. tested other polyadenylation complex components with similar results, underlining the importance of a functionally active 3′ end processing machinery for the maintenance of the gene loop structure and hence absence of divergent transcription. Moreover, the authors show that exchanging the polyA signals with an endonucleolytic Rnt1 cleavage site similarly disrupts the loop configuration and increases pSRT production. The effect of this cis-mutation was observed both on plasmid and in a yeast genomic context as well as on an integrated β-globin gene construct in human HEK 293 cells, indicating a similar and conserved gene loop-dependent transcriptional directionality in higher eukaryotes. The observation that abrogation of normal 3′ end formation results in gene loop disruption and concomitant 3-fold upregulation of divergent pSRTs, also points to the existence of a coordinated recycling of factors from the terminator back to the promoter that favors ORF transcription, in agreement with previously published data.21
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/24236
References
- 1.Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–42. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 2.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–7. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–37. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 4.van Dijk EL, Chen CL, d’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–7. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 5.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–5. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 7.Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, et al. Gene loops enhance transcriptional directionality. Science. 2012;338:671–5. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, et al. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–8. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 9.Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–78. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 11.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 12.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–73. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray SC, Serra Barros A, Brown DA, Dudek P, Ayling J, Mellor J. A pre-initiation complex at the 3′-end of genes drives antisense transcription independent of divergent sense transcription. Nucleic Acids Res. 2012;40:2432–44. doi: 10.1093/nar/gkr1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Z, Wei W, Gagneur J, Clauder-Münster S, Smolik M, Huber W, et al. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol. 2011;7:468. doi: 10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–72. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–69. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol Cell. 2004;14:387–94. doi: 10.1016/S1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 19.Singh N, Ma Z, Gemmill T, Wu X, Defiglio H, Rossettini A, et al. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol Cell. 2009;36:255–66. doi: 10.1016/j.molcel.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Creamer TJ, Darby MM, Jamonnak N, Schaughency P, Hao H, Wheelan SJ, et al. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet. 2011;7:e1002329. doi: 10.1371/journal.pgen.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell. 2010;40:410–22. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]