Abstract
Piwi proteins, together with their bound Piwi-interacting RNAs, constitute an evolutionarily conserved, germline-specific innate immune system. The piRNA pathway is one of the key mechanisms for silencing transposable elements in the germline, thereby preserving genome integrity between generations. Recent work from several groups has significantly advanced our understanding of how piRNAs arise from discrete genomic loci, termed piRNA clusters, and how these Piwi-piRNA complexes enforce transposon silencing. Here, we discuss these recent findings, as well as highlight some aspects of piRNA biology that continue to escape our understanding.
The piRNA pathway
Germ cells are the only cell type of an organism that contribute genetic material to future progeny. It is therefore essential that the integrity of this genome is preserved to protect reproductive success. One threat placed on germ cells is the movement of mobile genetic elements, or transposons, which correspond to a large fraction of the eukaryotic genome. Although transposons provide some benefits in driving evolution, their uncontrolled expression can lead to loss of genome integrity [1]. One of the major ways in which transposable elements (TEs) are kept under control is via Piwi-interacting RNAs (piRNAs) [2,3]. piRNAs are a class of small RNAs bound by the Piwi clade of Argonaute (Ago) proteins. As with all members of the Ago family, Piwi clade proteins rely on sequence complementarity to identify their targets, which for piRNAs are most commonly transposable elements. The importance of this pathway is evident; Piwi proteins are highly conserved throughout evolution, and their loss of function leads to gross defects in gametogenesis and to sterility.
With many aspects of this pathway being studied in a range of organisms, it is impossible to summarize all recent insights. Therefore, we will focus specifically on the piRNA pathway in the ovary of Drosophila melanogaster, which has been one of the main model organisms in the study of this pathway and which has helped establish the framework for how it functions.
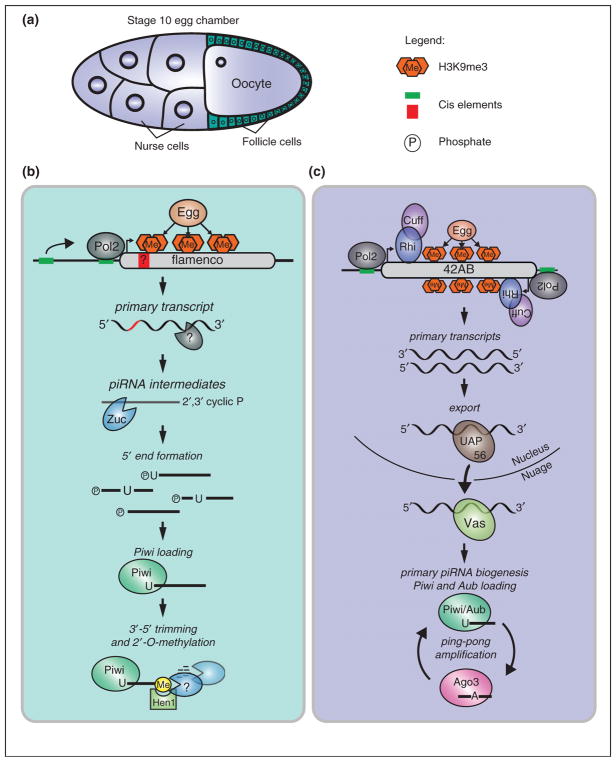
An intriguing aspect of piRNA biology in Drosophila ovaries is that there are two distinct iterations of the pathway active in this tissue: one in the germ cells and one in the follicle cells, cells of somatic origin that surround and support the developing germ cells [4,5] (Figure 1a). Controlling TEs in both of these cell types is important, since active transposons found within follicle cells, such as those from the gypsy family of retrovirus-like transposons, can form viral particles and infect the oocyte [6]. The somatic and germline piRNA pathways are distinct mainly because of the different expression patterns of the three fly Piwi proteins. While Aubergine (Aub) and Argonaute (Ago3) are exclusively found in the nuage of germ cells, Piwi is found in the nuclei of both germ cells and follicle cells [7–10]. Therefore, the somatic pathway acts only through piRNAs generated by primary biogenesis, while in germ cells, in addition to primary biogenesis, a more complex piRNA amplification loop exists that depends on the slicer activity of Aub and Ago3 [9,10]. Understanding the less complex primary piRNA pathway acting in somatic cells has provided a basic mechanistic framework of piRNA biogenesis that is likely shared between both somatic and germline piRNAs.
Figure 1.
A model for piRNA biogenesis in the Drosophila ovary. (a) Two distinct piRNA pathways are active in a stage 10 egg chamber of the Drosophila ovary. The nurse cells that provide nutrients to the oocyte and the oocyte itself make up the germ cells of the ovary, shown in blue. The monolayer of somatic follicle cells surrounding the oocyte is shown in green. Nuclei are indicated as circles within each cell. (b) In follicle cells, primary piRNAs arise from flamenco and are processed through a cascade of enzymatic cuts. Transcription by RNA polymerase II (Pol2) depends on deposition of Histone 3 Lysine 9 trimethyl marks (H3K9me3) by Eggless (Egg). Regulatory cis-acting elements, indicated as green boxes, upstream of the transcriptional start site could affect Pol2 recruitment and transcription. Additionally, clusters could carry cis elements within themselves, shown in red, that affect downstream processing. After processing of the primary cluster transcript by unknown activities, piRNA intermediates are cleaved by the nuclease, Zucchini (Zuc). After 5′ end formation, transcripts with a U at the first position are preferentially loaded into Piwi. Trimming activity, which could be carried out by redundant nucleases, shortens the transcript to its mature length. This process is coupled to 2′-O-methylation by Hen1. (c) The transcription of clusters in germ cells can occur bidirectionally. In addition to Egg, the HP1 homolog Rhino (Rhi) and Cutoff (Cuff) are essential for transcription. Subsequently, the helicase UAP56 binds the primary transcript and escorts it to the nuclear periphery. There, it is handed over to another RNA helicase Vasa (Vas) and arrives at its site of biogenesis, the nuage. After primary processing by similar machinery as in (a), primary piRNAs are loaded into Piwi and Aub, and potentially Ago3. These primary piRNAs can be used to kick-start the ping-pong amplification cycle, which silences transposons post-transcriptionally.
Taking advantage of the ease with which genetic manipulations can be done in Drosophila, studies of the small RNA populations in different piRNA mutants, together with other general molecular and cell biological analyses, such as localization studies and measurements of protein-protein interactions, have provided the main bulk of experimental data in the piRNA field [11]. The availability of cell lines derived from follicle cells (OSS/OSC) has also aided the study of the piRNA pathway [12–14]. To date, there are more than two-dozen proteins implicated in the piRNA pathway. However, many of the specific molecular steps that occur to generate a piRNA and that enable a piRNA to silence transposons remain unclear. In this review we will provide a brief summary of what is known about the piRNA pathway as well as discuss the open questions in the field.
How are piRNAs made?
The majority of piRNAs arise from specific genomic loci, known as piRNA clusters, which are found in pericentromeric heterochromatin [9]. Other sources of piRNAs do exist, such as the 3′ UTRs of protein coding genes and dispersed euchromatic copies of TEs [9,14,15]. piRNA clusters contain remnants of transposons and serve as a catalog of sequences previously defined as targets for silencing. Exposure to a new transposon can lead to the expansion of this catalog and control of the TE, while omission from the catalog can mean that the element escapes repression [16•]. Brennecke et al. defined over 140 such clusters in Drosophila and saw that these clusters could be uni-directionally or bidirectionally transcribed [9]. Most of these clusters are active specifically in germ cells, while only a single major cluster (flamenco) drives transposon silencing in the soma. In general, germline clusters have two promoters, one on either side of the cluster (e.g. cluster 42AB), and are transcribed bidirectionally, while flamenco is uni-directionally transcribed.
Little is understood about what defines a piRNA cluster, how clusters are transcribed, and how this process is regulated. To date, we have no knowledge of transcription factors that regulate cluster expression. Clusters seem to be expressed in a cell-type specific manner, so there must be cell-type specific transcription factors enforcing this pattern. The promoters of clusters and their regulatory elements have not been defined, but in the case of flamenco, existing evidence suggests a single, discrete promoter, since a P-element insertion at the beginning of the cluster abolishes piRNA production, even ~200 kb away from the insertion point [9,17].
Some studies suggest a role for chromatin context in regulating cluster transcription. Deposition of Histone 3 Lysine 9 trimethyl marks (H3K9me3) was proposed to be necessary for cluster transcription, since mutations in Eggless (Egg, dSETDB1), a histone methyltransferase, lead to decreases in H3K9me3 deposition, and in the levels of cluster transcripts within both germ cells and somatic cells [18] (Figure 1b and c). As expected, these decreases in cluster transcription led to a reduction of mature piRNAs and upregulation of TEs. Rhino, a Heterochromatin Protein 1 homolog, and Cutoff (Cuff), a yeast Rai1-like nuclease, physically interact, and together bind specifically to bidirectionally transcribed clusters in the germline to promote their transcription [19,20]. Both proteins are found in nuclear foci in germ cells and depend on each other for their nuclear localization. How these factors promote cluster transcription remains unclear. Although Rhino and Cutoff are predominantly nuclear, their depletion is sufficient to disrupt Aub and Ago3 localization in nuage [19,20].
In another study addressing the role of chromatin context in cluster identity, Muerdter and colleagues found that when a cluster was taken out of its normal heterochromatic genomic context and placed in a euchromatic locus, it is still able to produce piRNAs [21]. This implies that clusters themselves contain sufficient information, possibly through cis-elements or secondary structure, to trigger piRNA production. However, it is also possible that information in the modified cluster is capable of recreating the chromatin context necessary for its expression, since the authors did not verify the euchromatic status of the cluster after insertion. In summary, more research is needed to understand the determinants of cluster identity; whether it be the chromatin context of the cluster, sequences in or surrounding the cluster that are important for transcription, or if it is sequences recognized within the transcript after transcription that then mark it as a piRNA producing transcript.
Following cluster transcription, the current model states that the primary transcript is exported to the cytoplasm, where it is processed into primary piRNAs that are loaded into Piwi or Aub. A recent study by Zhang et al. shed some light on how cluster transcripts are escorted from the transcription site to the nuage where processing is thought to occur [22]. The study shows that UAP56, a putative helicase, co-localizes with Rhino in nuclear foci. Mutation of UAP56 leads to germline transposon upregulation, decrease of piRNAs mapping to germline clusters, and disruption of Aub, Ago3, and Vasa from nuage. Based on how the Rhino-UAP56 foci are positioned next to the nuclear pore, and the finding that UAP56 and Vasa bind germline cluster transcripts, the authors proposed a model in which UAP56 escorts the primary transcript through the nuclear pore to nuage, where the transcript is handed over to Vasa and funneled into the biogenesis machinery. Since UAP56 is believed to be germ cell specific, factors that mediate export in the follicle cells remain a mystery. Whether the cluster transcript is exported as one long RNA or if some processing occurs in the nucleus to generate smaller piRNA intermediates to be exported, remains unknown.
After the cluster transcript is exported, it must be processed into piRNAs. Since Piwi-bound piRNAs have a strong preference for a uridine at the 5′ end (1 U) [9], this suggests a model of primary piRNA biogenesis wherein the 5′ end of the piRNA is generated first, followed by preferential loading of piRNA intermediates with a 5′ U into Piwi, followed by 3′ trimming. The variable lengths of primary piRNAs (23–29nt) could result from a footprint specific to the Piwi protein into which the intermediate is loaded, since the size of the RNA binding pocket probably varies slightly between each protein, and Aub, Ago3 and Piwi associated piRNAs are of slightly different lengths.
The factors responsible for 5′ and 3′ end formation have yet to be uncovered. However, recent advancements were made in our understanding of one piRNA protein that may be involved with end formation. Nishimasu et al. and Ipsaro et al. both revealed the crystal structure of the piRNA pathway protein Zucchini (Zuc) [23••,24••]. Based on its structure, Zuc shows a preference for binding specifically single stranded RNA. In vitro studies demonstrated that both the mouse and Drosophila Zuc protein had endoribonuclease activity [23••,24••], contradictory to previous reports implicating Zuc as a phospholipase [25,26]. The cleaved RNA product bore a 5′-monophosphate group, a characteristic of mature piRNAs. These data make Zuc the principal candidate for 5′ end formation. Both studies failed to show association of Zuc with piRNA precursors, which would have made the argument for its role as the 5′ nuclease much stronger, given that it shows no sequence preferences. Unlike most other piRNA factors, Zuc localizes to the mitochondrial membrane, and loss of this nuclease in either the germline, or the soma, results in a dramatic reduction of piRNAs [4,25–28]. The role that mitochondria could play in the piRNA pathway remains enigmatic, though its ancient connections to antiviral responses, for example it serving as the location at which the RIG-I pathway operates, is provocative [29]. In flies and mice, Piwi proteins are localized to discrete cytoplasmic structures associated with mitochondria [3], but whether this is purely to allow compartmentalization of the pathway, or whether it implies a further role of mitochondrial activity in the piRNA pathway is unclear.
The precise biochemical mechanism of piRNA 3′ end formation remains a mystery. Recent work in a cell line derived from silkworm ovaries, BmN4, has brought the field closer to identifying the 3′ generating enzyme [30••]. Kawaoka and colleagues established an in vitro 3′ trimming assay using BmN4 cell extracts. The authors found that Siwi (silkworm Piwi) binds transcripts with a bias toward 1 U, and that extended precursor transcripts could be trimmed in extracts, in a Mg 2+ dependent manner, to mature piRNA length. It had been determined previously that piRNAs are 2′-O-methylated at their 3′ termini by Hen1, and the addition of this modification was observed to be coupled with the trimming activity [31,32]. The importance of the 3′ terminal modification remains uncertain, because mutants of Hen1 have no detectable phenotype [31,32]. These findings are in accordance with the model that piRNA precursors bind to Piwi in the cytoplasm, and then are trimmed and methylated at the 3′ terminus. Unfortunately, the molecular nature of the trimming activity remains enigmatic; ‘trimmer’ could not be purified due to its insoluble nature. Moreover, no exonuclease has yet emerged as a candidate trimmer from genetic screen, which could indicate that multiple redundant trimmers exist or that trimmer has essential functions that mask an ability to isolate it as a piRNA pathway mutant.
Our current model follows the idea that Piwi must be loaded with a mature piRNA in order to be imported into the nucleus. Successful loading of Piwi-family proteins with primary piRNAs requires several other players. Although there are some distinguishing factors between the loading process in somatic and germ cells, many proteins are shared between the two pathways. The common proteins involved in biogenesis are Armitage (Armi), an RNA helicase, Shutdown (Shu), a cochaperone, and Vreteno (Vret) a TUDOR domain containing protein [27,28,33–37]. Although we understand little of the precise role of any of these proteins, mutation of any one disrupts localization of Piwi, and levels of associated piRNAs decrease dramatically [4,28,34–36]. It is important to note that mutations in Shu and Vret lead to delocalization of all three Piwi proteins in the germline, while Zuc and Armi mutants delocalize Piwi, but not Aub and Ago3. This could mean that Shu and Vret play a more general role in primary biogenesis involving Piwi and Aub, while Armi only aids Piwi in the piRNA loading process.
In the soma, Yb, a TUDOR-domain protein that also contains an RNA helicase motif, is an important additional factor for primary biogenesis. This protein localizes to foci in the cytoplasm, together with all other known loading components [27,33,38]. Zuc, the putative 5′ nuclease, localizes to mitochondria, many of which are adjacent to Yb bodies, supporting the role of these structures in Piwi RISC assembly. In Zuc mutants, Vret, Armi, Shu, and Yb all accumulate in enlarged Yb bodies with Piwi, suggesting that when the 5′ end of the piRNA cannot be generated, the loading machinery accumulates in the foci in response to a stall in biogenesis [27,28,33,35]. In the germline, there are no Yb bodies, and Yb is not expressed. Current evidence suggests that two Yb-related proteins, Brother of Yb and Sister of Yb, might serve the role played by Yb in the cytoplasm [28].
In germ cells, the loading process seems to occur in the nuage, where Aub and Ago3 localize. The function of the nuage is unknown, but many piRNA factors are found there, suggesting an important role in the piRNA pathway. One important difference between germ cells and the soma is that in germ cells, Aub and Ago3 engage in an adaptive, slicer-dependent loop termed the ping-pong cycle, which specifically amplifies the piRNA response against active elements [9,10]. In this model, Aub, bound to cluster-derived piRNAs, recognizes an active transposon transcript and cleaves it, generating the 5′ end of a new sense piRNA, which associates with Ago3. Subsequently, sense strand piRNA-loaded Ago3 can recognize complementary sequences in cluster transcripts, and through its slicer activity can generate a new antisense Aub bound piRNA, completing the cycle. According to the ping-pong model of piRNA amplification, Aub and Ago3 must be catalytically active in order to cleave new piRNAs from expressed transposons or piRNA cluster transcripts. However, the phenotypes of catalytically inactive mutants have never been described. While Aub and Ago3 seem to be responsible for generating the 5′ end of each piRNA amplified through ping-pong, how the 3′ end is generated remains unknown, though it may proceed through the action of the same trimmer that is used for primary biogenesis.
In order to initiate the ping-pong cycle, piRNAs loaded into Aub are required. These come from two sources. One is primary biogenesis. The second is maternally deposited Aub, as the protein is loaded into developing oocytes along with associated piRNAs [8,39,40]. The importance of maternally deposited piRNAs is evident from analyses of hybrid dysgenesis models. In these cases, maternal deposition of piRNAs, produced by ping-pong and corresponding to the I-element or P-element, correlates with initiation of ping-pong in progeny and with effective element silencing [40]. For the I-element, as mothers age, their progeny have a reduced probability of being sterile even in the absence of the ability of the mother to use active I-elements as ping-pong substrates [16•,41]. For P-elements, even the dysgenic progeny can regain some fertility as the animals age. This suggests that perhaps primary piRNAs corresponding to those elements accumulate with age in the mother or offspring to a level sufficient to confer resistance.
How do piRNAs silence transposons?
It seems evident that in germ cells Aub and Ago3 silence transposons through post-transcriptional gene silencing (PTGS). These two proteins posses slicer activity and cleave active TE transcripts during the ping-pong amplification cycle. By using the cleavage products to make more piRNAs, this cycle is able to amplify its response to actively transcribed elements [9,10].
The mechanism by which Piwi silences transposons proved much more difficult to dissect. It had long been suspected that Piwi mediates transcriptional gene silencing (TGS) of TEs through impacts on chromatin, due mainly to several provocative clues. First, Piwi is a nuclear protein, and this localization is essential to its silencing capability. A mutant Piwi lacking its nuclear localization signal is found in the cytoplasm and is incapable of silencing TEs but binds piRNAs to wildtype levels [14,27,42]. In addition, Piwi’s slicer activity is not necessary for silencing, as a catalytically dead Piwi mutant rescues the null mutant phenotype [14,27].
Many studies have suggested that Piwi could silence transposons at a transcriptional level by inducing changes in histone marks, much like the mechanism by which small RNAs induce heterochromatin formation in yeast [43]. In fact, the murine piRNA pathway silences transposable elements by inducing chromatin changes, ultimately resulting in DNA methylation [44,45]. In Drosophila, several studies support a role for Piwi in acting through TGS in the ovary; multiple groups have reported changes in histone marks on a handful of transposons upon disruption of the piRNA pathway [42,46,47], and a study by Shpiz et al. detected an increase in several nascent TE transcripts upon Piwi knockdown (KD) [48]. However, it was the recent study by Sienski and colleagues that definitively demonstrated that Piwi silences transposons at the transcriptional level, triggering changes in chromatin state genome wide (Figure 2). The authors took advantage of the OSS/OSC cell line and did side-by-side comparisons of RNA Polymerase II (Pol2) occupancy, trimethylation of H3K9 (a common mark of heterochromatin), nascently transcribed RNA, and steady state RNAs at a global level in Piwi KD versus control cells [49••]. They observed that in the absence of Piwi, Pol2 occupancy on transposons increased, along with an increase of nascent TE transcripts and steady state RNA levels. Furthermore, levels of H3K9me3 marks on transposons dropped in the Piwi KD as compared to controls. Interestingly, the authors also observed that many TE sequences dispersed in euchromatin trigger the formation of an H3K9me3 island that is dependent on Piwi and on transcription of the locus. This strongly implicates an RNA-recognition mode for Piwi-dependent silencing. The study also identified Maelstrom (Mael), a protein previously implicated in the germline piRNA pathway, as playing a role in transcriptional silencing of transposons [50,51]. Upon Mael KD, there was no change in levels of mature piRNAs, but there were increases in Pol2 occupancy on TEs and nascent transcripts. Interestingly, levels of H3K9me3 did not decrease when Mael was depleted; rather, H3K9 methylation appeared to spread downstream of the TE insertion, in some cases for up to 30 kb. This places Mael downstream of Piwi in silencing of TEs. The precise mechanism by which Piwi influences chromatin state remains elusive. Other than Mael, no other effector protein has been identified. One likely candidate to play a role in this process is Heterochromatin Protein 1a (HP1a), which is believed to bind H3K9 methyl groups [52,53]. HP1a has been shown to interact with Piwi, and its depletion leads to TE derepression [47,54]. The current model of piRNA-mediated TGS proposes that Piwi RISC recognizes nascent transposon transcripts by sequence complementarity and then, with the help of Mael, recruits silencing machinery to trigger histone modifications at the site of transcription (Figure 2). The association of Piwi with chromatin seems to be unstable, as the authors were unable to map it to TE loci using chromatin immunoprecipitation. It is clear that other silencing effectors in addition to H3K9 are necessary because Mael mutants do not lose H3K9me3, but have upregulation of TE transcripts. Further experiments are needed to fully understand this process. Even though it seems likely that TGS is the main silencing mode for Piwi, there remains a possibility that it is also acting through PTGS at some level. This study did not address the role of Piwi in the germline nucleus but it seems likely that it will also silence TEs by TGS in that setting.
Figure 2.
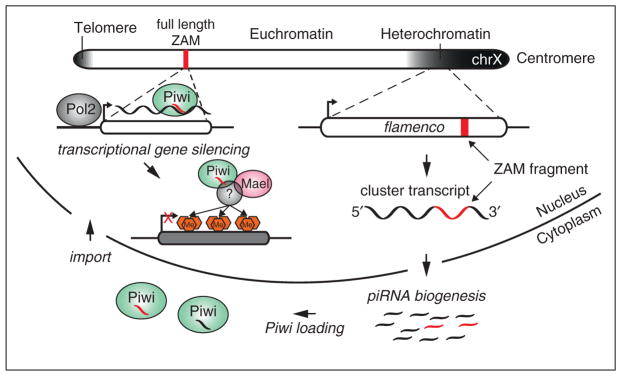
Transcriptional silencing of transposable elements by Piwi-piRNA complexes in the soma. The X chromosome of Drosophila melanogaster (chrX) is shown. A simplistic view of its chromatin state is indicated in shades of gray. The transcriptionally active euchromatin in white harbors a full-length copy of the retroelement, ZAM (indicated as a red box). An inactive remnant of the same element (in red) can be found within the flamenco piRNA cluster in pericentromeric heterochromatin. After transcription and processing of flamenco, this fragment gives rise to antisense piRNAs that are loaded into Piwi in the cytoplasm (indicated as red piRNA species). Upon reimport into the nucleus, these Piwi-piRNA complexes recognize active transcription of the full-length ZAM copy by RNA polymerase II (Pol2) based on sequence complementarity. This recognition leads to the recruitment of additional factors such as Maelstrom (Mael) and unknown chromatin remodelers. Ultimately, the deposition of H3K9me3 marks leads to loss of Pol2 occupancy and the transcriptional silencing of ZAM.
Germ cells might prove to be more complex because of the presence of Aub and Ago3. Although these two proteins are engaged in the ping-pong cycle in the nuage, spatially separated from Piwi in the nucleus, there seems to be a more intimate connection between these proteins than has been generally appreciated. A strong indication of this connection is that in Aub and Ago3 mutants, levels of Piwi protein decrease [5]. Furthermore, in an Ago3 mutant, the levels of Piwi-bound piRNAs decrease and there is a shift in their sense versus antisense bias [5]. Considered together, these data indicate that there is significant crosstalk between Piwi and the ping-pong cycle. One point to remember is that, although ping-pong is thought to occur mainly between Aub and Ago3, there are a significant number of Piwi:Ago3 ping-pong pairs detected in ovaries [9]. Further studies will be critical in understanding the relationship between Piwi and ping-pong, and which mechanisms are employed to silence TEs in the germline.
What is the function of maternally deposited Piwi RISC complexes?
Piwi and Aubergine, together with their bound piRNAs, are maternally deposited in the embryo and accumulate in the pole plasm, which gives rise to the future germline [8,39,40]. These maternally contributed complexes are thought to be essential in priming the piRNA pathway to be able to successfully silence elements. Previous studies have revealed that hybrid dysgenesis is caused by the failure to maternally deposit piRNAs corresponding to a paternally contributed transposon [40]. These maternally contributed Piwi and Aub RISCs may serve to jump-start the silencing pathway to target elements even before zygotic transcription has begun. Therefore, maternally deposited complexes could be one of the triggers to initiate the ping-pong cycle, which will continue throughout the life of the organism.
A recent study offers another important role for these inherited complexes. de Vanssay et al. found that maternally deposited piRNAs could be involved in the specification of a piRNA cluster [55•]. In a previous study, the group characterized a phenomenon known as trans-silencing effect (TSE) in which P-element derived transgenes inserted in a heterochromatic region can silence a distinct P-element derived transgene inserted at a euchromatic locus. Using this system the authors found that a transgene cluster that induces strong silencing can convert a separate, homologous locus that is normally incapable of trans-silencing, into a strong silencer, in a heritable manner. This effect is dependent on maternally deposited piRNA complexes. This implies that the inherited piRNA complexes are needed to reestablish piRNA cluster definitions in the progeny. Consequently, the piRNA pathway may completely reset and cluster identity be re-acquired between each generation. This concept is analogous to piRNA-driven transposon silencing in mammals; during primordial germ cell development, the germline is stripped of all DNA methylation, which is then reacquired on TEs through the action of piRNA-driven de novo methylation [45]. Since Drosophila lacks the ability to methylate DNA, maternally deposited piRNA complexes may serve a similar role in identifying TEs in the developing progeny. However, further work is necessary to evaluate this hypothesis. For instance, it would be interesting to specifically eliminate the maternally inherited pool of Piwi RISCs to observe if cluster definitions are lost.
In embryos, although maternally deposited Piwi and Aub are both localized to pole plasm in early embryogenesis, their localization patterns rapidly change during the cellularization of the embryo. While Aub continues to reside exclusively in pole cells, Piwi localizes to the nuclei of every cell of the embryo, and continues to do so until ~12 hours after egg laying [40,54]. What role might Piwi play in somatic nuclei during embryogenesis? One interesting possibility, especially considering the recent findings implicating Piwi in TGS, is that the protein is establishing silencing marks on transposons throughout the somatic compartment. In fact, many studies have implicated Piwi in positional effect variegation (PEV), a clearly somatic effect, and have observed Piwi binding on polytene chromosomes [54,56,57]. Perhaps the suppression of transgenes observed during PEV is mediated by Piwi-induced chromatin silencing in early embryogenesis, and is maintained throughout development. Extensive additional work will be necessary to fully understand the role of maternally deposited Piwi and Aub, but there is no doubt that there are many fascinating discoveries to be made in this area.
Conclusions
It has been almost a decade since the discovery of piRNAs, and many advances have been made toward understanding the general function of the pathway. However, surprisingly little is known about several key aspects of piRNA biology, such as the mechanistic details of piRNA biogenesis and how the downstream targets of the pathway are silenced. This is because many of the important players in the pathway still remain unknown. A genome-wide screen for piRNA pathway factors would aid in identifying all proteins involved, so that a full genetic framework could finally and conclusively be established. There is also an overwhelming need to develop biochemical assays that recapitulate several aspects of the piRNA pathway in vitro. These could bring much needed mechanistic insights into precisely how the pathway operates. Some progress has been made in this direction with the development of the silkworm trimming assay [30••]. Following the introduction of the Drosophila OSS/OSC cell lines by Niki et al., both genome wide screens and in vitro assays have become feasible [12]. Given these tools and recent advances described here, it is easy to imagine that we will see many more exciting discoveries and insights into how small RNAs provide an immune defense against mobile elements.
Acknowledgments
We would like to thank Clare Rebbeck and Leah Sabin for critical comments on the manuscript and helpful discussion. We would also like to thank Julius Brennecke for sharing of data before publication. P.M.G. is a NIH trainee on a CSHL WSBS NIH Kirschstein-NRSA pre-doctoral award (T32 GM065094), a William Randolph Hearst Scholar and a Leslie Quick Junior Fellow. Work in the Hannon laboratory is supported by grants from the NIH and by a kind gift from Kathryn W. Davis. G.J.H. is an investigator of the HHMI.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 4.Malone CD, Brennecke J, Dus M, Stark A, Mccombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lécher P, Bucheton A, Pelisson A. Expression of the Drosophila retrovirus gypsy as ultrastructurally detectable particles in the ovaries of flies carrying a permissive flamenco allele. J Gen Virol. 1997;78(Pt 9):2379–2388. doi: 10.1099/0022-1317-78-9-2379. [DOI] [PubMed] [Google Scholar]
- 7.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development (Cambridge, England) 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 8.Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development (Cambridge, England) 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- 9.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 10.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science (New York, NY) 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 11.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niki Y, Yamaguchi T, Mahowald AP. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci USA. 2006;103:16325–16330. doi: 10.1073/pnas.0607435103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau NC, Robine N, Martin R, Chung W-J, Niki Y, Berezikov E, Lai EC. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 15.Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. A broadly conserved pathway generates 3′ UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–1563. doi: 10.1016/j.cell.2011.11.042. This paper demonstrates how the piRNA system can evolve its silencing repertoire in response to challenge by a new transposon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robert V, Prud’homme N, Kim A, Bucheton A, Pelisson A. Characterization of the flamenco region of the Drosophila melanogaster genome. Genetics. 2001;158:701–713. doi: 10.1093/genetics/158.2.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. piRNA production requires heterochromatin formation in Drosophila. Curr Biol. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pane A, Jiang P, Zhao DY, Singh M, Schüpbach T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 2011;30:4601–4615. doi: 10.1038/emboj.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, Hannon GJ, Aravin AA. Production of artificial piRNAs in flies and mice. RNA (New York, NY) 2012;18:42–52. doi: 10.1261/rna.029769.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012 doi: 10.1038/nature11502. This paper reports the biochemistry and three dimensional structure of the mouse Zucchini protein and implicates it as the nuclease that forms piRNA 5′ ends. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012 doi: 10.1038/nature11509. Along with the Ipsaro paper, this report of the structure and biochemistry of Drosophila Zucchini may have solved the mystery of piRNA 5′ processing. [DOI] [PubMed] [Google Scholar]
- 25.Huang H, Gao Q, Peng X, Choi S-Y, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell. 2011;20:376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011;30:3977–3993. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnoult D, Soares F, Tattoli I, Girardin SE. Mitochondria in innate immunity. EMBO Rep. 2011;12:901–910. doi: 10.1038/embor.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. This paper reports one of the few successful attempts at addressing questions of piRNA biology using in vitro, biochemical approaches and provides a mechanism, if not the enzyme, for piRNA 3′ end formation. [DOI] [PubMed] [Google Scholar]
- 31.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 32.Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development (Cambridge, England) 2011;138:4039–4050. doi: 10.1242/dev.069187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olivieri D, Senti K-A, Subramanian S, Sachidanandam R, Brennecke J. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell. 2012;47:954–969. doi: 10.1016/j.molcel.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA (New York, NY) 2012;18:1446–1457. doi: 10.1261/rna.034405.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosakawa M, Reuter M, Yang Z, Berninger P, Palencia A, et al. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell. 2012;47:970–979. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 38.Szakmary A, Reedy M, Qi H, Lin H. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol. 2009;185:613–627. doi: 10.1083/jcb.200903034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 40.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science (New York, NY) 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grentzinger T, Armenise C, Brun C, Mugat B, Serrano V, Pelisson A, Chambeyron S. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 2012;22:1877–1888. doi: 10.1101/gr.136614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci USA. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huisinga K, Elgin S. Small RNA-directed heterochromatin formation in the context of development: what flies might learn from fission yeast. Biochim Biophys Acta. 2008;1789:3–16. doi: 10.1016/j.bbagrm.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aravin AA, Sachidanandam R, Girard A, Fejes Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science (New York, NY) 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 45.Aravin AA, Bourc’his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970–975. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang SH, Elgin SCR. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci USA. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011;39:8703–8711. doi: 10.1093/nar/gkr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. This paper provides a genome-wide view of changes in chromatin structure upon Piwi silencing and provides concrete evidence that Piwi operates in the soma by regulating transposons at the level of their transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development (Cambridge, England) 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- 51.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 53.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 54.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SCR, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.de Vanssay A, Bougé A-L, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. The authors implicate maternally deposited piRNAs as being important for specifying the identity of piRNA clusters. [DOI] [PubMed] [Google Scholar]
- 56.Haynes K, Caudy A, Collins L, Elgin S. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr Biol. 2006;16:2222–2227. doi: 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pal-Bhadra M. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science (New York, NY) 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]