Abstract
Background
Febrile illnesses are pre-eminent contributors to morbidity and mortality among children in South-East Asia but the causes are poorly understood. We determined the causes of fever in children hospitalised in Siem Reap province, Cambodia.
Methods and Findings
A one-year prospective study of febrile children admitted to Angkor Hospital for Children, Siem Reap. Demographic, clinical, laboratory and outcome data were comprehensively analysed. Between October 12th 2009 and October 12th 2010 there were 1225 episodes of febrile illness in 1180 children. Median (IQR) age was 2.0 (0.8–6.4) years, with 850 (69%) episodes in children <5 years. Common microbiological diagnoses were dengue virus (16.2%), scrub typhus (7.8%), and Japanese encephalitis virus (5.8%). 76 (6.3%) episodes had culture-proven bloodstream infection, including Salmonella enterica serovar Typhi (22 isolates, 1.8%), Streptococcus pneumoniae (13, 1.1%), Escherichia coli (8, 0.7%), Haemophilus influenzae (7, 0.6%), Staphylococcus aureus (6, 0.5%) and Burkholderia pseudomallei (6, 0.5%). There were 69 deaths (5.6%), including those due to clinically diagnosed pneumonia (19), dengue virus (5), and melioidosis (4). 10 of 69 (14.5%) deaths were associated with culture-proven bloodstream infection in logistic regression analyses (odds ratio for mortality 3.4, 95% CI 1.6–6.9). Antimicrobial resistance was prevalent, particularly in S. enterica Typhi, (where 90% of isolates were resistant to ciprofloxacin, and 86% were multi-drug resistant). Comorbid undernutrition was present in 44% of episodes and a major risk factor for acute mortality (OR 2.1, 95% CI 1.1–4.2), as were HIV infection and cardiac disease.
Conclusion
We identified a microbiological cause of fever in almost 50% of episodes in this large study of community-acquired febrile illness in hospitalized children in Cambodia. The range of pathogens, antimicrobial susceptibility, and co-morbidities associated with mortality described will be of use in the development of rational guidelines for infectious disease treatment and control in Cambodia and South-East Asia.
Introduction
Febrile illness in children is a common cause of admission to hospital globally, with significant associated morbidity and mortality [1]. In developing countries this is frequently compounded by low rates of immunisation, untreated co-morbidities, and late presentations [2]. Febrile illnesses are caused by diverse pathogens, presenting with non-specific symptoms to healthcare facilities with limited diagnostic capacity [3], [4]. Clinical management guidelines for acute febrile illness are available [5], [6], but rarely supported by knowledge of the locally prevalent causative agents.
The Kingdom of Cambodia lies in South-East Asia and has a mortality rate in children aged <5 years of 54/1000 live births [7]. This has halved over the last decade but remains one of the highest in the region. The prevalence of undernutrition in children <5 years of age (less than 2 SD of weight for age) is 28% [7]. There is little published information on the causes of fever in Cambodian children.
We characterised the causes of febrile illness in children in Cambodia. We hypothesised that in addition to globally common childhood pathogens such as Streptococcus pneumoniae and influenza virus, other infections that require specific management would be identified, such as typhoid, dengue, leptospirosis, melioidosis and rickettsial disease [4], [8]–[19].
Materials and Methods
Ethics Statement
Parents of all children recruited to the study gave witnessed, written, informed consent before study enrolment. The Oxford Tropical Research Ethics Committee and Angkor Hospital for Children Institutional Review Board approved the study protocol on 24th September 2009 and 2nd October 2009 respectively.
Study Site and Population
This prospective, year-long study of the causes of fever in children was based at Angkor Hospital for Children (AHC), Siem Reap province, Cambodia ( Figure 1 ). AHC is a 50-bed paediatric hospital providing free universal inpatient and outpatient care to children <16 years of age from urban and rural settings. It has critical care capacity, including mechanical ventilation and inotropic support, and is one of two paediatric hospitals serving Siem Reap city and province.
Figure 1. Location and map of Cambodia.
AHC is in Siem Reap (underlined). Map: United Nations, 2004 (http://www.un.org/Depts/Cartographic/map/profile/cambodia.pdf). Accessed 2nd January 2013.
The national immunisation schedule included Bacillus Calmette-Guérin (BCG) and hepatitis B virus (HBV) at birth, and diphtheria-pertussis-tetanus, oral poliovirus and measles virus vaccines. 79% of children aged 12–23 months have received these vaccines [7]. Haemophilus influenzae type b immunisation was introduced during the study period.
Patients and Clinical Methods
Patients admitted to AHC between 12th October 2009 and 12th October 2010 were considered for enrolment. Eligibility criteria were age <16 years, documented axillary temperature ≥38.0°C within 48 h of admission, and caregiver consent. Febrile post-surgical patients were excluded. All children, except emergencies, were assessed using locally-modified Integrated Management of Childhood Illness (IMCI) guidelines [5] prior to a decision to admit. Admission information was recorded on a study-specific clinical record form. Admissions were reviewed twice daily for eligibility.
Sampling and Laboratory Methods
Blood was taken aseptically from all enrolled patients for culture, complete blood count, blood film (including malaria smear), biochemistry, and nucleic acid amplification tests (NAATs). In addition, study patients aged ≥60 days had admission blood samples taken for serology and Leptospira spp. culture, and a convalescent serology sample taken on discharge, or after 7 days of admission. Whole blood and serum samples were stored at –80°C until analysed.
Nasal and throat swabs were taken for respiratory virus detection in patients with a recent history of cough or sore throat, and increased respiratory effort. Cerebrospinal fluid (CSF) analysis was performed on patients with suspected central nervous system (CNS) infection. Urinalysis was performed on all children; other sampling (e.g. HIV serology, gastric aspirates) and imaging were performed as clinically indicated.
Bacterial Culture
Blood was taken for culture, when possible before in-hospital antimicrobial therapy and within 48 h of admission. Blood was inoculated into a pre-weighed blood culture bottle and the vented bottles were incubated aerobically at 37°C for 7 days [20]. Sub-culture onto sheep blood and chocolate agar was undertaken at 24 h, 48 h, 7 days, or if the culture was turbid. CSF samples were separated immediately upon receipt into aliquots for staining and microscopy, cell count and biochemistry, culture, and storage at –80°C. Cultured organisms were identified using routine methods [20], including API test kits (bioMérieux, France), disc diffusion antimicrobial susceptibility testing [21] and Etests™ (AB Biodisk, Sweden) performed as appropriate. Samples from other sterile sites (e.g. abscesses, pleural fluid) were cultured using routine methods [20] and were reviewed daily for clinical relevance by the infectious diseases team. Whole blood samples were cultured for Leptospira spp. [22].
Serology
The Panbio Japanese encephalitis virus (JEV) and dengue virus (DENV) IgM Combo enzyme-linked immunosorbent assay (ELISA) (Panbio, Australia) was used to detect anti-JEV and anti-DENV specific IgM antibodies in sera. An ELISA (Standard Diagnostics, Korea) was used to detect DENV NS1 antigen [23]. A capture IgM ELISA assay (Venture Technologies, Malaysia) was used to detect anti-JEV and anti-DENV specific IgM antibodies in CSF specimens [24]. Table S1 describes the result interpretation.
Separate ELISAs incorporating Orientia tsutsugamushi (Karp and Gilliam strain) and Rickettsia typhi (Wilmington strain) antigens were used to detect O. tsutsugamushi and R. typhi IgM antibodies in serum samples respectively [25]. A positive ELISA result was equivalent to a (conservative) 1∶200–1∶400 indirect immunofluorescence assay (IFA) titre [26] (SD Blacksell: unpublished data). “Acute positive serology” was diagnosed in paired samples with a ≥4-fold increase in IFA IgM antibody titres. Serum pairs with <4-fold increase in IFA IgM antibody titres, or a single sample with a titre ≥1∶400, were recorded as “acute/recent positive serology” [25].
Nucleic Acid Amplification Tests
DNA was extracted from whole blood samples with the QIAamp® DNA mini-kit (QIAGEN, Germany), with extended incubation for 30 minutes at 56°C. Probe-based real-time polymerase chain reaction (rPCR) assays were used to detect Leptospira spp. [27], O. tsutsugamushi [28] and R. typhi [29]. Low-positive plasmid or sample controls determined adequate detection limits of each assay.
Total nucleic acid was isolated from 100 µL of CSF specimens using the automated easyMAG® system (bioMérieux), and diagnostic NAATs were performed [30]. Four rPCR protocols were used for detection of S. pneumoniae, H. influenzae type B, Neisseria meningitidis, and Streptococcus suis [30]. rRT-PCRs were used to detect herpes simplex virus (HSV) 1 and 2 [31], varicella zoster virus (VZV) [32], enteroviruses (generic and 71-specific) [33], and human parechoviruses (generic) [34].
Nasal and throat swabs were stored in viral transport medium (Becton Dickinson, USA) at 4°C for ≤72 h, then divided and stored at –80°C. RNA was extracted from one aliquot of the pooled swab medium using the QIAamp® Viral RNA mini-kit (QIAGEN). Specimens were tested for influenza virus (types A, B; and subtypes H1N1-1977, H1N1-pdm09, H3N2 and H5N1) by real-time reverse-transcription PCR (rRT-PCR), and for RSV, parainfluenza virus (PIV)1, PIV2 and PIV3 using multiplex RT-PCR (QIAGEN) [35]–[38].
Full details are in the supplementary material. All nucleic acid extractions and assays were done according to the manufacturer’s instructions.
Clinical Data
For each episode, the first recorded parameter after the time of admission was used. The clinical syndrome was categorised by a senior paediatrician (VK) at hospital discharge according to the localising focus of infection, e.g. lower respiratory tract infection (LRTI), or non-infectious cause for fever. On final analysis of all data, a microbiological diagnosis was given when results were consistent with the presenting clinical syndrome. When microbiological results were inconsistent with the clinical syndrome, both were given as a final diagnosis. Two clinicians (KE and MC) made these judgements independently with disagreements resolved by discussion.
Data Management and Analysis
Data was managed on a study-specific database and analysed using Stata 12 (Stata Corp., USA), with weight-for-age z-scores calculated using the WHO Anthro 3.2.2 Stata macro (World Health Organisation). Multivariate logistic regression was used to analyse the effects of comorbid undernutrition, heart disease, HIV infection, and anaemia; adjusted for each other and age group.
Results
Baseline Characteristics
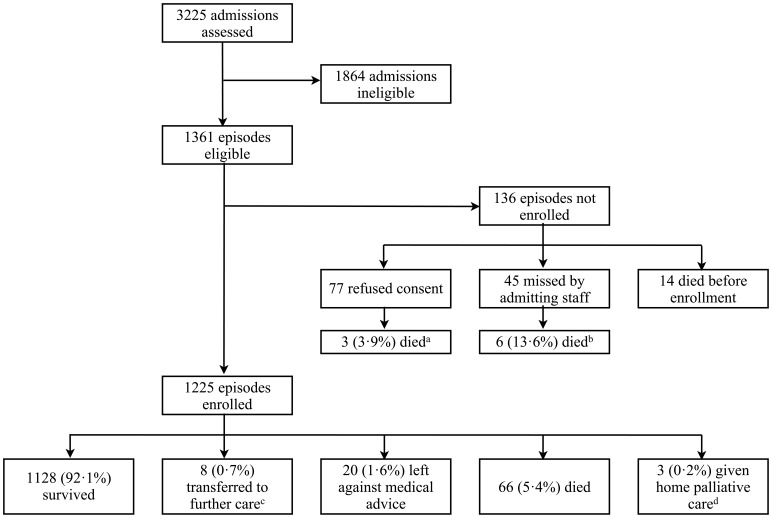
There were 3225 patient admissions during the study year, of which 1361 (42.2%) met the inclusion criteria. Of these, 136 (10.0%) were not enrolled, leaving 1225 febrile episodes in 1180 children ( Figure 2 ). 1144 children had a single episode, 31 children had two episodes, one child had three episodes and four children had four episodes. The median (inter quartile range [IQR]) age was 2.0 (0.8–6.4) years, with 850 (69.4%) episodes in children <5 years of age. The median (IQR) duration of illness prior to admission was 3 (2–5) days. Medication was given by the caregiver prior to admission in 53% of episodes. In 43% of these the medication was a known antibacterial, anti-malarial or steroid. Other baseline characteristics are in Table 1 .
Figure 2. Flow chart showing enrolment to the study.
Notes: aincluding one home palliative care; bincluding one home palliative care; cexcluded from analyses of outcome (e.g. in odds ratios); dincluded as “died” in analyses.
Table 1. Baseline, comorbidity and outcome characteristics in 1225 disease episodes among 1180 children admitted to hospital with fever over one year.
| Characteristic | Number or median (IQRa, range) |
| Demographics | |
| Number of episodes | 1225 |
| Male | 668 (54.5%) |
| Median (IQR, range) age (years) | 2.0 (0.8–6.4, 0.0–16.0) |
| Numbers in each age range: | |
| <28 days | 32 (2.6%) |
| 28 days to <1 year | 330 (26.9%) |
| ≥1 year to <5 years | 488 (39.8%) |
| ≥5 years to <16 years | 375 (30.6%) |
| Origin | |
| Acute transfer from other healthcare facility | 161 (13.2%) |
| Residence in Siem Reap district | 269 (22.0%) |
| Residence in Siem Reap province (not district) | 542 (44.2%) |
| Residence in neighbouring provinces | 371 (30.3%) |
| Residence in distant provinces | 43 (3.5%) |
| Known comorbidity b | |
| HIV infection | 58 (4.7%) |
| Hepatitis B infection | 5 (0.4%) |
| Hepatitis C infection | 1 (<0.1%) |
| Mean weight-for-age z-score (95% CI) | –1.97 (–4.5–+0.3) |
| Undernutrition (weight-for-age z-score <2 standard deviations below mean in those <5 yrs of age) | 371 (43.4%) |
| Congenital or rheumatic heart disease (on echocardiograpy) | 81 (6.6%) |
| Severe anaemia (<7.0 g/dL) on admission | 99 (8.1%) |
| Inpatient episode | |
| Median (IQR) duration of fever before hospitalisation (days) | 3 (2–5)c |
| Median (IQR, range) duration of hospitalisation (days) | 4 (3–8, 0–123) |
| Admitted to critical care unit at least once during admission | 300 (24.5%) |
| Requiring respiratory support | 192 (15.7%) |
| Needing CPAP alone | 81 (7.3%) |
| Needing mechanical ventilation | 111 (9.1%) |
| Mortality | |
| Died or discharged to die at home | 69 (5.6%) |
| Median (IQR, range) time to death (n = 69) (days) | 4 (2–10, 0–123) |
IQR interquartile range.
n = 1180 children except the z-score which was generated for those children <5 years of age using WHO (2006) data where n = 855.
3 children had caregiver reported fever for >1 month.
Clinical Syndrome Diagnoses
A total of 1333 presenting clinical syndromes were noted in 1225 febrile episodes, with the most common being LRTI (38.3% of episodes), undifferentiated fever (25.5%) and diarrhoeal disease (19.5%) ( Table 2 ). 575 (46.9%) episodes were positive for a microbiological cause of fever results by any method (culture, NAAT, direct stain, serology), with 731 microbiological causes of fever diagnosed within these 575 episodes ( Table 3 ). A consistent microbiological diagnosis was made in 125 of 472 (26.5%) episodes of LRTI, 77 of 267 (28.8%) episodes of diarrhoeal disease, and 66 of 148 (44.6%) episodes of undifferentiated fever.
Table 2. The presenting clinical syndromes diagnosed for 1225 episodes of febrile illness over the one-year study period.
| Age groups | |||||
| Clinical syndrome | <28 days (n = 32) | 28–365 days (n = 330) | 1–5 years (n = 488) | ≥5 years (n = 375) | Total (n = 1225) |
| Lower respiratory tract infection | 8 (25.0%) | 183 (55.5%) | 218 (44.7%) | 60 (16.0%) | 469 (38.3%) |
| Undifferentiated fever | 13 (40.1%) | 36 (10.9%) | 98 (20.1%) | 165 (44.0%) | 312 (25.5%) |
| Diarrhoeal disease | 4 (12.5%) | 104 (31.5%) | 110 (22.5%) | 21 (5.6%) | 239 (19.5%) |
| Skin/soft tissue/bone/joint infection | 4 (12.5%) | 10 (3.0%) | 28 (5.7%) | 46 (12.3%) | 88 (7.2%) |
| Upper respiratory tract infection | 0 | 14 (4.2%) | 36 (7.4%) | 14 (3.7%) | 64 (5.2%) |
| CNS infection | 1 (3.1%) | 12 (3.6%) | 10 (2.0%) | 33 (8.8%) | 56 (4.6%) |
| Genitourinary | 0 | 14 (4.2%) | 12 (2.4%) | 15 (4.0%) | 41 (3.4%) |
| Abdominal disease/surgical abdomen | 0 | 0 | 3 (0.6%) | 18 (4.8%) | 21 (1.7%) |
| Non-infectious cause of fever | 2 (6.3%) | 6 (1.8%) | 16 (3.3%) | 19 (5.1%) | 43 (3.5%) |
| Total clinical diagnoses | 1333 | ||||
| Acute mortality | 6 (18.8%) | 23 (7.0%) | 18 (3.7%) | 22 (5.9%) | 69 (5.6%) |
Within these 1225 episodes, a total of 1333 clinical syndromes were diagnosed: 1120 episodes had a single clinical syndrome diagnosed, 102 episodes had two separate syndromes diagnosed, and three episodes had three separate syndromes diagnosed (i.e. 1333 syndromes in total).
Table 3. Microbiological diagnoses.
| Age groups | ||||||
| <28 days | 28 days–<1 year | ≥1–<5 years | ≥5–<16 years | Total number | ||
| Number of children | 32 | 330 | 488 | 375 | 1225c | |
| Gram-positive bacteria | ||||||
| Staphylococcus aureus | 3 | 3 | 11 | 20 | 37 (3.0%) | |
| Streptococcus pneumoniae | 0 | 2 | 8 | 8 | 18 (1.5%) | |
| Streptococcus pyogenes | 1 | 1 | 1 | 1 | 4 (0.3%) | |
| Gram-negative bacteria | ||||||
| Salmonella enterica serovar Typhi | 0 | 1 | 6 | 15 | 22 (1.8%) | |
| Burkholderia pseudomallei | 0 | 1 | 6 | 7 | 14 (1.1%) | |
| Escherichia coli | 1 | 4 | 5 | 3 | 13 (1.1%) | |
| Haemophilus influenzae | 0 | 5 | 4 | 0 | 9 (0.7%) | |
| Klebsiella pneumoniae | 1 | 3 | 1 | 1 | 6 (0.5%) | |
| Neisseria meningitides | 0 | 2 | 0 | 2 | 4 (0.3%) | |
| Acinetobacter baumannii | 1 | 1 | 1 | 0 | 3 (0.2%) | |
| Pseudomonas aeruginosa | 1 | 1 | 0 | 1 | 3 (0.2%) | |
| Non-typhoid Salmonella spp. | 0 | 0 | 0 | 1 | 1 (<0.1%) | |
| Pantoea spp. | 1 | 0 | 0 | 0 | 1 (<0.1%) | |
| Blood isolate of uncertain significance | 3 | 9 | 9 | 5 | 26 (2.1%) | |
| Cultured bacterial pathogens (blood, pus and CSF) excluding blood isolates of uncertain significance | 135 (11.0%) | |||||
| Rickettsioses and other bacteria | ||||||
| Orientia tsutsugamushi | 0 | 12 | 21 | 63 | 96 (7.8%) | |
| Rickettsia typhi | 0 | 4 | 8 | 15 | 27 (2.2%) | |
| Rickettsia of indeterminate species | 0 | 0 | 5 | 6 | 11 (0.9%) | |
| Viruses | ||||||
| Dengue virus | 0 | 54 | 63 | 81 | 198 (16.2%) | |
| Japanese encephalitis virus | 0 | 29 | 27 | 15 | 71 (5.8%) | |
| Indeterminate flavivirus | 0 | 22 | 26 | 17 | 65 (5.3%) | |
| Parainfluenza virus 3 | 0 | 9 | 8 | 1 | 18 (1.5%) | |
| Respiratory syncytial virus | 0 | 6 | 7 | 1 | 14 (1.1%) | |
| Influenza virus A/H1N1-pdm09 | 0 | 3 | 5 | 3 | 12 (1.0%) | |
| Parainfluenza virus 1 | 1 | 3 | 5 | 1 | 10 (0.7%) | |
| Enteroviruses (non-71) | 0 | 1 | 1 | 6 | 8 (0.7%) | |
| Influenza virus A/H3N2 | 0 | 0 | 5 | 2 | 7 (0.6%) | |
| Influenza virus B | 0 | 3 | 2 | 0 | 5 (0.4%) | |
| Parainfluenza virus 2 | 0 | 1 | 1 | 0 | 2 (0.2%) | |
| Influenza virus A/H1N1-1977 | 0 | 0 | 0 | 1 | 1 (<0.1%) | |
| Herpes simplex virus | 0 | 0 | 1 | 0 | 1 (<0.1%) | |
| Malaria | ||||||
| Plasmodium falciparum only | 0 | 2 | 6 | 6 | 14 (1.1%) | |
| P vivax only | 0 | 2 | 3 | 2 | 7 (0.6%) | |
| P vivax and P falciparum | 0 | 0 | 0 | 3 | 3 (0.2%) | |
| Other | ||||||
| Leptospira spp | 0 | 3 | 8 | 6 | 17 (1.4%) | |
| Mycobacterium tuberculosis a | 0 | 0 | 1 | 5 | 6 (0.5%) | |
| Yeast (non-cryptococcal) | 0 | 1 | 0 | 1 | 2 (0.2%) | |
| Cryptococcus neoformans | 0 | 0 | 0 | 1 | 1 (<0.1%) | |
| Non-infectious cause of fever b | 17 (1.4%) | |||||
Ziehl-Neelsen stain only, no culture facilities available.
Non-infectious causes of fever included 11 episodes attributed to haematological malignancy, five episodes attributed to burns and one episode attributed to juvenile idiopathic arthritis.
Blood and Sterile Site Sampling
Blood was cultured within 48 h of admission in 1212 (99.0%) episodes. The mean volume of blood drawn was 1.7 mL (95% confidence interval [CI] 1.0–3.3) for children aged <28 days, and 2.0 mL (95% CI 1.0–4.0) for those ≥60 days of age. The blood culture was positive in 162 (13.4%) samples, of which 76 (6.3%) were considered true positives, 26 (2.1%) were isolates of uncertain significance ( Table 4 ), and 60 (5.0%) considered contaminants. 10 of 69 (14.5%) deaths were associated with a true positive blood culture (Odds Ratio [OR] for mortality 3.4, 95% CI 1.6–6.9). 5 of 69 (7.2%) deaths were associated with isolates from blood culture of uncertain significance (OR adjusted for age 3.8, 95% CI 1.4–10.4). Contaminants from blood culture were not associated with mortality (OR adjusted for age 0.8, 95% CI 0.2–2.6). Pus was sampled in 127 (10.8%) of episodes, with the most numerous isolates being S. aureus (31) and B. pseudomallei (8).
Table 4. Isolates from blood cultures (n = 1212).
| Isolate from blood culture | Number of positive cultures |
| S. enterica Typhi | 22a (1.8%) |
| S. pneumoniae | 13b (1.1%) |
| E. coli | 8c (0.7%) |
| H. influenzae | 7 (0.6%) |
| B. pseudomallei | 6e (0.5%) |
| S. aureus | 6d (0.5%) |
| K. pneumoniae | 4f (0.3%) |
| Acinetobacter baumannii | 3 (0.2%) |
| N. meningitidis | 2 (0.2%) |
| S. pyogenes | 2 (0.2%) |
| Yeast | 1 (<0.1%) |
| Pantoea spp. | 1 (<0.1%) |
| P. aeruginosa | 1 (<0.1%) |
| Total true isolates | 76 (6.3%) |
| Isolates of uncertain significance | |
| Unidentified Gram-negative bacilli | 5 (0.4%) |
| Pseudomonas spp. | 5 (0.4%) |
| Acinetobacter calcoaceticus | 4 (0.3%) |
| Ochrobactum anthropi | 3 (0.2%) |
| Burkholderia cepacia | 2 (0.2%) |
| Moraxella spp. | 2 (0.2%) |
| Alcaligenes faecalis | 1 (0.1%) |
| Gemella morbillorum | 1 (0.1%) |
| Sphingobacterium spiritivorum | 1 (0.1%) |
| Stenotrophomonas maltophilia | 1 (0.1%) |
| Streptococcus sanguinis | 1 (0.1%) |
| Total isolates of uncertain significance | 26 (2.1%) |
| Contaminants | 60g (5.0%) |
19/21 (90.5%) had intermediate susceptibility to ciprofloxacin; 18/21 (85.7%) were multi-drug resistant (resistant to chloramphenicol, ampicillin and co-trimoxozole).
8 strains were available for testing, all were susceptible to ceftriaxone.
4 strains were available for testing, one was ESBL producing.
6 strains available for testing, one was MRSA.
Constitutively resistant to ceftriaxone.
2 strains available for testing, one was ESBL producing.
33 coagulase-negative staphylococci, 27 Gram-positive bacilli.
CSF Sampling
A CSF sample was examined by microscopy and culture from children in 174 (14.2%) of disease episodes. 52 (30.0%) CSF samples showed white cell pleocytosis, and 11 (6.3%) were culture or CSF stain positive. There was sufficient CSF for NAATs and serology from 107 (62.6%) samples, with 15 (14.0%) positive by NAAT and 7 (6.5%) positive by serology ( Table 5 ). All CSF samples positive by culture for were also positive by NAATs. NAATs also identified an additional three S. pneumoniae and one H. influenzae positive episodes.
Table 5. CSF microscopy and culture (n = 174), NAAT (n = 107) results, and total episodes positive (n = 174) by all methods for children with suspected CNS infection.
| CSF isolate | Culture positive samples | NAAT positive samples | Serology positive samples | Episodes positive |
| H. influenzae | 4 (2.3%) | 1 (1.9%) | – | 5 (2.9%) |
| S. pneumoniae | 2 (1.1%) | 4 (3.7%) | – | 5 (2.9%) |
| N. meningitides | 3 (1.7%) | 1 (0.9%) | – | 3 (1.7%) |
| P. aeruginosa | 1 (0.6%) | – | – | 1 (0.5%) |
| C. neoformans | 1 (0.6%) | – | – | 1 (0.5%) |
| Enterovirus | – | 8 (7.5%) | – | 8 (4.6%) |
| Herpes simplex virus | – | 1 (0.9%) | – | 1 (0.5%) |
| Japanese encephalitis virus | – | – | 6 (5.6%) | 6 (3.4%) |
| Dengue virus | – | – | 1 (0.9%) | 1 (0.5%) |
| Total CSF isolates | 11 (6.3%) | 15 (14.0%) | 7 (6.5%) | 31 (19.5%) |
Leptospira spp. and Rickettsial Disease
Leptospira spp. culture was performed for 1068 of 1149 (93.0%) disease episodes in children ≥60 days of age, and two (0.2%) were positive. NAAT for Leptospira spp. was performed in 1179 episodes (96.2%) of all ages and 17 (1.4%) were positive. Both episodes positive by culture were positive by NAAT ( Table 6 ).
Table 6. Positive results by culture (n = 1068) and NAAT (n = 1179) for Leptospira spp infections; by serology (n = 1125) and NAAT (n = 1179) for rickettsial infections; and total episodes positive for Leptospira spp and rickettsial infections.
| Organism | Culture positive | NAAT positive | Episodes positive |
| Leptospira spp. | 2 (0.2%) | 17 (1.4%) | 17 |
| Serology positive | NAAT positive | ||
| Acute O. tsutsugamushi | 5 (0.4%) | 17a (1.4%) | 96 |
| Acute/recent O. tsutsugamushi | 90 (8.0%) | ||
| Acute R. typhi | 3 (0.3%) | 2b (0.2%) | 27 |
| Acute/recent R. typhi | 22 (2.0%) | ||
| Acute/recent Rickettsia of indeterminate species | 11 (1.0%) | – | 11 |
| Total rickettsial infections | 131c |
“Acute rickettsial serology” denotes a >4-fold dynamic rise in specific IgM between acute and convalescent (7-days) samples; “acute/recent serology” denotes either static IgM titres or raised IgM in a single acute sample.
One additional to serological testing.
Two additional to serological testing.
Three additional to serological testing.
Serology for O. tsutsugamushi and R. typhi was performed in 1125 (98.0%) disease episodes in children ≥60 days of age, and NAATs for O. tsutsugamushi and R. typhi were done in 1179 (96.3%) of episodes of all ages ( Table 6 ). 95 (8.4%) samples were seropositive for anti-O. tsutsugamushi IgM. 17 (1.4%) samples were positive by NAAT for O. tsutsugamushi, of which 16 were also IgM seropositive. 25 (2.2%) samples were seropositive for anti-R. typhi IgM. 2 (0.2%) samples were positive by NAAT for R. typhi, neither of which were IgM seropositive. Seropositivity for anti-O. tsutsugamushi IgM was negatively associated with seropositivity for anti-R. typhi IgM (4.7% for O. tsutsugamushi alone versus 4.0% for both; McNemar’s test, p = 0.0005).
Virology
Serological evidence of a flavivirus infection was determined in 1125 (98.0%) disease episodes in children ≥60 days of age ( Table 7 ); DENV NS1 antigen was assayed in 1105 (96.4%) of episodes in children ≥60 days of age and 60 (5.4%) samples were positive. There was pronounced seasonality in “acute serology” to DENV (NS1 antigen positive, dynamic rise in anti-DENV IgM titres, or CSF anti-DENV IgM positive) in contrast to lack of seasonality in “acute/recent serology” to DENV or other flaviviruses (Figure S2 and Figure S3).
Table 7. Serological results for flaviviruses for episodes in children of 60 days or older (n = 1125).
| Serological interpretation | NS1 positive | NS1 negative | Number of positive samples |
| Acute dengue virus infection | 59 | 55 0 | 114 (10.1%) |
| Acute/recent dengue virus infection | 0 | 84 4 | 84 (7.5%) |
| Total dengue virus infections | 198 (17.6%) | ||
| Acute JEV | 1a | 37 1 | 38 (3.4%) |
| Acute/recent JEV | 0 | 34 1 | 34 (3.0%) |
| Total JEV infections | 72 (6.4%) | ||
| Acute indeterminate flavivirus infection | 0 | 0 | 0 |
| Acute/recent indeterminate flavivirus infection | 0 | 65 2 | 65 (5.8%) |
| Total indeterminate flavivirus infections | 65 (5.8%) | ||
| Total positive to flaviviruses | 60 | 275 8 | 324 (28.8%) |
| Negative to flaviviruses | 0 | 801 23 | - |
By definition, all NS1 positive samples were denoted “acute dengue virus serology”, and all CSF IgM positive samples were “acute serology” (although we acknowledge that NS1 antigen assay is not a serological test). Numbers in superscript denote samples with insufficient serum for NS1 antigen assay.
CSF positive for anti-JEV IgM, serum positive for NS1 and anti-DENV IgM in same episode (diagnosed with both).
There were 389 febrile episodes that met our criteria to have oral and nasopharyngeal swabs sent for the analysis of respiratory viruses ( Table 8 ).
Table 8. Positive results by NAAT for respiratory viruses in children presenting with sore throat or cough and increased respiratory rate or effort (n = 389).
| Respiratory virus | NAAT positive |
| Parainfluenza virus 3 | 18 (4.6%) |
| Respiratory syncytial virus | 14 (3.6%) |
| Influenza virus A/H1N1-pdm09 | 11 (2.8%) |
| Parainfluenza virus 1 | 10 (2.6%) |
| Influenza virus A/H3N2 | 7 (1.8%) |
| Influenza virus B | 5 (1.3%) |
| Parainfluenza virus 2 | 2 (0.5%) |
| Influenza virus A/H1N1-1977 | 1 (0.3%) |
| Total | 68 (17.5%) |
Diagnostic Methods and Multiple Positivity
144 (20.0%) of 731 microbiological causes of fever were diagnosed from direct culture (76 from blood), 440 (60.3%) from serological testing, 115 (15.7%) from NAATs, and 32 (4.4%) from direct stain of CSF, pus or gastric aspirates (for M tuberculosis). Of 440 IgM seropositive tests for flaviviruses and rickettsias, 157 (35.7%) showed clear “acute serology”, with the remainder showing “acute/recent serology” (as defined above). Median time between acute and discharge/convalescent samples was 4 days (IQR 2–7).
444 (77.2%) of 575 microbiologically positive episodes had one microbiological cause of fever, 108 (18.8%) had two microbiological causes, 21 (3.7%) had three microbiological causes, and 2 (0.3%) had four microbiological causes of fever (excluding bacteria of uncertain significance from blood culture).
10 (5.1%) episodes with anti-DENV IgM seropositivity were also positive for invasive bacterial disease from blood culture (7 with S. enterica Typhi, 1 each with H. influenzae, K. pneumoniae and S. aureus). DENV infection was positively associated with malaria (McNemar’s test, p<0.0001) with 11 (5.6%) episodes seropositive for anti-DENV IgM also malaria positive (6 of which P. falciparum positive).
Mortality
During the one-year study, 69 (5.6%) children died during their acute illness ( Table 9 ). Common associations ( Table 1 for definitions and prevalence) with mortality in our cohort (adjusted for comorbid undernutrition, HIV infection, and heart disease), were comorbid undernutrition in children <5 years of age (OR 2.1 [95% CI 1.1–4.2]; population attributable fraction [PAF] = 0.31 [95% CI 0.10–0.52]), comorbid heart disease (OR = 3.3 [95% CI 1.5–6.9]; PAF = 0.14 [95% CI 0.02–0.25]), comorbid HIV infection (OR = 3.9 [95% CI 1.2–12.8]; PAF = 0.06 [–0.02–0.13]). 68 of 69 (98.9%) children who died were admitted to the critical care unit.
Table 9. Primary diagnosis, and contributing diagnoses, for the 69 children who died during the study.
| Primary diagnosis | Contributing diagnoses | Number of deaths(n = 69) |
| Invasive organisms | ||
| B. pseudomallei bacteraemia | DENV infection (1), O. tsutsugamushi (1) | 4 (5.8%) |
| E. coli bacteraemia | – | 3 (4.3%) |
| S. pneumoniae meningitis | O. tsutsugamushi (1) | 2 (2.9%) |
| S. aureus pyomyositis/arthritis | – | 2 (2.9%) |
| Acinetobacter spp bacteraemia | HIV (1) | 2 (2.9%) |
| H. influenzae bacteraemia | – | 1 (1.4%) |
| K. pneumoniae bacteraemia | – | 1 (1.4%) |
| Pantoea spp (surgical case) | – | 1 (1.4%) |
| M. tuberculosis pneumonia | HIV | 1 (1.4%) |
| C. neoformans meningitis | HIV | 1 (1.4%) |
| O. tsutsugamushi rickettsaemia | DENV infection | 1 (1.4%) |
| Total invasive organisms | 19 (27.5%) | |
| Viral infections | ||
| DENV infection | O. tsutsugamushi (1) | 5 (7.2%) |
| Influenza virus A/H1N1-pdm09 | – | 2 (2.9%) |
| Parainfluenza 1 bronchiolitis | S. aureus cellulitis (1), O. tsutsugamushi and uncertain flavivirus (1)a | 2 (2.9%) |
| JEV infection | Tetralogy of Fallot | 1 (1.4%) |
| RSV bronchiolitis | – | 1 (1.4%) |
| Total viral infections | 11 (15.9%) | |
| Primary clinical diagnoses | ||
| Clinical pneumonia | –b | 12 (17.4%) |
| DENV infection | 4 (5.8%) | |
| HIV | 2 (2.9%) | |
| Clinical diarrhoea | 1 (1.4%) | |
| Unknown source of fever | – | 11 (15.9%) |
| Leukaemia | 2 (2.9%) | |
| Clinical diarrhoea | –c | 1 (1.4%) |
| Clinical TB, HIV | 1 (1.4%) | |
| HIVd | 1 (1.4%) | |
| Clinical CNS infection | –d | 1 (1.4%) |
| Clinical tetanus (neonatal) | – | 1 (1.4%) |
| Clinical UTI | – | 1 (1.4%) |
| Clinical parotitis | – | 1 (1.4%) |
| Total primary clinical diagnoses | – | 39 (56.5%) |
These diagnoses are based on all the available results: hence are not identical to “clinical syndrome diagnosed” in Table 2 . Numbers in parentheses indicate the number of patients with the contributing diagnosis in addition to the primary diagnosis. Superscripts give further information about individual morbid patients:
Also with organism of uncertain significance from blood (Burkholderia cepacia) and pulmonary hypertension secondary to congenital heart disease;
also with organism of uncertain significance from blood (Acinetobacter calcoaceticus);
also with organism of uncertain significance from blood (A. calcoaceticus);
also with organism of uncertain significance from blood (Ochrobactrum anthropi);
also with organism of uncertain significance from blood (unidentified Gram-negative bacilli).
Importantly, 23 children of 136 eligible but non-enrolled episodes died (16.9%), and their microbiological data are therefore unavailable ( Figure 2 ). 10 of 19 enrolled children (52.6%) who died with a primary diagnosis of “clinical pneumonia” ( Table 9 ) had samples for respiratory viruses sent for analysis; and 1 of 11 enrolled children (9.1%) who died with a primary diagnosis of “unknown source of fever” ( Table 9 ) had CSF samples analysed.
Discussion
With comprehensive laboratory investigation we identified a microbiological cause in almost 50% of febrile illness in this one-year study of febrile Cambodian children requiring admission to hospital. The acute mortality rate was 5.6% of children enrolled (6.7% of all eligible). Nurse-led triage using IMCI [5] guidelines and paediatric review prior to admission excluded less-severe infections. Children died despite availability of parenteral broad-spectrum antimicrobials, critical care, and laboratory facilities, all unavailable to the majority of the Cambodian population. Prevalence of comorbid undernutrition was 43.7%, and doubled the odds of mortality. Comorbid heart disease, and known HIV infection were also associated with large increases in odds of mortality.
LRTIs, diarrhoeal disease or undifferentiated fever were the main presentations, but a microbiological diagnosis was achieved in only 27%, 29%, and 45% of episodes with these syndromes, respectively. The absence of microbiological examination of faeces, limited use of urine culture, and lack of M. tuberculosis culture (or NAAT) facilities were limitations. More inclusive criteria for the analysis of respiratory viruses, HIV testing, and greater emphasis on CSF sampling may have increased our diagnostic yield. In contrast to a recent report [33], we found no enterovirus-71 in 8 episodes of CSF enterovirus-positive meningoencephalitis.
The serological data from our study is consistent with similar studies in Cambodia [8], [14], and neighbouring urban Laos [10]. There was evidence of DENV infection in 16% of episodes, but clinical differentiation from bacterial septic shock was difficult, with children frequently treated for both. This strategy is supported by the co-incidence of both invasive bacterial disease (particularly S. enterica Typhi) and malaria, with DENV infection in our cohort. Serological evidence of infection by O. tsutsugamushi and R. typhi was also common. Although the interpretation of serological tests on cohorts of unselected febrile children can be difficult [25], [26] even with conservative cut-offs for IgM titres against O. tsutsugamushi and R. typhi, we estimated that 10% of children were infected with these microorganisms. NAATs may increase the specificity of diagnosis, but may lack sensitivity due to short periods of rickettsaemia.
Invasive community-acquired bacterial disease was common, frequently resistant to commonly used antimicrobials, and associated with significant mortality. The most common isolate from blood was S. enterica Typhi, with 90% of isolates with intermediate resistance to ciprofloxacin, and 85% multi-drug resistant (MDR) [18], contrasting with the decline in drug-resistant phenotypes seen in adjacent countries [12]. B. pseudomallei (inherently resistant to ceftriaxone and penicillins) was confirmed as a pathogen of major local public health significance [16], and E. coli (of which 1 of 7 isolates demonstrated extended-spectrum β-lactamase activity) was also prevalent. S. aureus was the commonest isolate from all sterile sites, with one meticillin-resistant isolate [19]. Invasive disease caused by S. pneumoniae, H. influenzae, and N. meningitidis is present and potentially vaccine-preventable. The small numbers of neonates in this cohort (not due to refusal of parental consent) and their high mortality emphasizes the need for accessible perinatal care in Cambodia [7].
We are investigating whether pre-treatment with incomplete or sub-therapeutic antimicrobial regimens, especially in an unregulated private sector, contributes to low blood culture yield and high-levels of resistance in Cambodia. Resolution of conflicting demands for rational antibiotic prescribing to help prevent further emergence of antimicrobial resistance, and the urgent need for successful treatment of patients, would be aided by a network of sentinel microbiology laboratories throughout South-East Asia [3], [4]. Without such laboratories, efforts to treat severe infections in low and middle-income countries will flounder in the dark [39].
Supporting Information
Flowchart summarising methods for the analysis of cerebrospinal fluid (CSF) from children with suspected meningoencephalitis enrolled in the study.
(PDF)
Number of admissions with “acute serology” (see main text for definition) to DENV against period of admission (30-day intervals starting from study start date 12th October 2012).
(PDF)
Number of admissions with “acute/recent serology” (see main text for definition) to DENV against period of admission (30-day intervals starting from study start date 12th October 2012).
(PDF)
Classification of DENV and JEV serology in samples. a Dynamic rise of ≥2 Panbio units between acute and discharge samples. b Dynamic fall of ≤2 Panbio units between acute and discharge samples. c Dynamic rise or fall of ≤2 panbio units between acute and discharge samples. d Considered negative by manufacturer’s criteria.
(PDF)
(DOC)
Acknowledgments
We thank the children and parents, without whom this study would have been impossible and the nursing, medical, laboratory and administrative staff of AHC, especially the resident doctors and the laboratory staff. Dr. Ly Sovann and team at the Communicable Disease Control Division of the Cambodian Ministry of Health allowed us to collaborate with their severe acute respiratory infection surveillance project. Chin Savuth, Sok Siyeatra, Unn Thy and Nuth Dara at the Cambodian National Institute of Public Health performed the respiratory virus assays with the authors. Prof. Steve Allen and an anonymous reviewer gave helpful comments on the manuscript. Drs. Shunmay Yeung and Rathi Guahadasan helped in the original conception of the study. Dr. Allan Richards gifted O. tsutsugamushi and R. typhi antigens for ELISA testing. Sabine Dietrich and Tippawan Anantatat performed the rickettsia and Leptospira spp. NAATs together with the authors. Ampai Tanganuchitcharnchai performed the serology testing and Sayan Langla performed the Leptospira spp. culture together with the authors. Standard Diagnostics, Korea donated NS1 ELISAs, Panbio Pty Ltd part-donated on JEV/Dengue IgM ELISAs. Other specific support was from Dr Luy Lyda, Line Denker, Sarah Fryxelius, Louisa Bethell, Margaret Chang, Clare Baker, Reet Nijjar, Nuttapol Panachuenwongsakul, Sue Lee, Jeff William, Sun Sopheary, Jon Morgan, and the registration team at AHC. Friends Without A Border helped in the development of the AHC-MORU Collaborative Laboratory. Finally, Dr. Bill Housworth gave his unstinting support.
Funding Statement
This study was supported by the Wellcome Trust of Great Britain, the Li Ka Shing–University of Oxford Global Health Programme, EMPERIE (European Management Platform for Emerging and Re-emerging Infectious Disease Entities). Respiratory virus assays were supported by the Influenza Division, US Centers for Communicable Disease Control as part of ongoing public health surveillance for severe acute respiratory infection (PK and BS). UK National Institute of Health Research provided grants for academic clinical fellowships to KE and MJC through the University of Oxford, and University College London, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Liu L, Johnson HL, Cousens S, Perin J, Scott S, et al. (2012) Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet 379: 2151–61. [DOI] [PubMed] [Google Scholar]
- 2. Bhutta ZA, Ali S, Cousens S, Ali TM, Azra Haider B, et al. (2008) Alma-Ata: Rebirth and revision 6 interventions to address maternal, newborn, and child survival: What difference can integrated primary health care strategies make? Lancet 372: 972–89. [DOI] [PubMed] [Google Scholar]
- 3. Peacock SJ, Newton PN (2008) Public health impact of establishing the cause of bacterial infections in rural Asia. Trans R Soc Trop Med Hyg 102(1): 5–6. [DOI] [PubMed] [Google Scholar]
- 4. Deen J, von Seidlein L, Andersen F, Elle N, White NJ, et al. (2012) Community-acquired bloodstream infections in developing countries in south and southeast Asia: a systematic review. Lancet Inf Dis 12: 480–87. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization (2005) Handbook: Integrated management of childhood illness. Geneva, Switzerland: World Health Organization.
- 6.World Health Organization (2005) Pocket book of hospital care for children: guidelines for the management of common illnesses with limited resources. Geneva, Switzerland: World Health Organization.
- 7.National Institute of Statistics, Directorate General for Health, ICF Macro (2011) Cambodia: Demographic and health survey 2010. Phnom Penh, Cambodia and Calverton, Maryland, USA: National Institute of Statistics, Directorate General for Health and ICF Macro.
- 8. Kasper MR, Blair PJ, Touch S, Sokhal B, Yasuda CY, et al. (2012) Infectious etiologies of acute febrile illness among patients seeking health care in south-central Cambodia. Am J Trop Med Hyg 86: 246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leelarasammee A, Chupaprawan C, Chenchittikul M, Udompanthurat S (2004) Etiologies of acute undifferentiated febrile illness in Thailand. J Med Assoc Thai 87(5): 464–72. [PubMed] [Google Scholar]
- 10. Phetsouvanh R, Phongmany S, Soukaloun D, Rasachak B, Soukhaseum V, et al. (2006) Causes of community-acquired bacteremia and patterns of antimicrobial resistance in Vientiane, Laos. Am J Trop Med Hyg 75(5): 978–85. [PMC free article] [PubMed] [Google Scholar]
- 11. Punjabi NH, Taylor WRJ, Murphy GS, Purwaningsih S, Picarima H, et al. (2012) Etiology of acute, non-malaria, febrile illnesses in Jayapura, northeastern Papua, Indonesia. Am J Trop Med Hyg 86(1): 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nga TV, Parry CM, Le T, Lan NP, Diep JS, et al. (2012) The decline of typhoid and the rise of non-typhoid salmonellae and fungal infections in a changing HIV landscape: Bloodstream infection trends over 15 years in southern Vietnam. Trans R Soc Trop Med Hyg 106(1): 26–34. [DOI] [PubMed] [Google Scholar]
- 13. Chhour YM, Ruble G, Hong R, Minn K, Kdan Y, et al. (2002) Hospital-Based diagnosis of hemorrhagic fever, encephalitis, and hepatitis in Cambodian children. Emerg Infect Dis 8: 485–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vong S, Khieu V, Glass O, Ly S, Duong V, et al. (2010) Dengue incidence in urban and rural Cambodia: results from population-based active fever surveillance, 2006–2008. PLoS Negl Trop Dis 4(11): e903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kasper MR, Wierzba TF, Sovann L, Blair PJ, Putnam SD (2010) Evaluation of an influenza-like illness case definition in the diagnosis of influenza among patients with acute febrile illness in Cambodia. BMC Inf Dis 10: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pagnarith Y, Kumar V, Thaipadungpanit J, Wuthikanun V, Amornchai P, et al. (2010) Emergence of pediatric melioidosis in Siem Reap, Cambodia. Am J Trop Med Hyg 82(6): 1106–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wijedoru LPM, Kumar V, Chanpheaktra N, Chheng K, Smits HL, et al. (2012) Typhoid fever among hospitalized febrile children in Siem Reap, Cambodia. J Trop Paediatr 58: 68–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emary K, Moore CE, Chanpheaktra N, An KP, Chheng K, et al. (2012) Enteric fever in Cambodian children is dominated by the multidrug resistant H58 Salmonella enterica serovar Typhi with intermediate susceptibility to ciprofloxacin. Trans R Soc Trop Med Hyg 106(12): 718–24. [DOI] [PubMed] [Google Scholar]
- 19. Chheng K, Tarquinio S, Wuthiekanun V, Sin L, Thaipadungpanit J, et al. (2009) Emergence of community-associated methicillin-resistant Staphylococcus aureus associated with pediatric infection in Cambodia. PloS One 4(8): e6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Tenover FC (2007) Manual of Clinical Microbiology (9th Edition Revised). Washington DC, USA: American Society for Microbiology.
- 21.Clinical and Laboratory Standards Institute (2012) Performance standards for antimicrobial susceptibility testing. Supplement M100-S22. Wayne, PA, USA: Clinical and Laboratory Standards Institute.
- 22. Wuthiekanun V, Chierakul W, Limmathurotsakul D, Smythe LD, Symonds ML, et al. (2007) Optimization of culture of Leptospira from humans with leptospirosis. J Clin Microbiol 45(4): 1363–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blacksell SD, Mammen MP, Thongpaseuth S, Gibbons RV, Jarman RG, et al. (2008) Evaluation of the Panbio dengue virus nonstructural 1 antigen detection and immunoglobulin M antibody enzyme-linked immunosorbent assays for the diagnosis of acute dengue infections in Laos. Diagn Microbiol Infect Dis 60(1): 43–9. [DOI] [PubMed] [Google Scholar]
- 24. Cardosa MJ, Wang SM, Sum MS, Tio PH (2002) Antibodies against prm protein distinguish between previous infection with dengue and Japanese encephalitis viruses. BMC Microbiol 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blacksell SD, Jenjaroen K, Phetsouvanh R, Tanganuchitcharnchai A, Phouminh P, et al. (2010) Accuracy of rapid IgM-based immunochromatographic and immunoblot assays for diagnosis of acute scrub typhus and murine typhus infections in Laos. Am J Trop Med Hyg 83(2): 365–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blacksell SD, Bryant NJ, Paris DH, Doust JA, Sakoda Y, et al. (2007) Scrub typhus serologic testing with the indirect immunofluorescence method as a diagnostic gold standard: A lack of consensus leads to a lot of confusion. Clin Infect Dis 44(3): 391–401. [DOI] [PubMed] [Google Scholar]
- 27. Thaipadunpanit J, Chierakul W, Wuthiekanun V, Limmathurotsakul D, Amornchai P, et al. (2011) Diagnostic accuracy of real-time PCR assays targeting 16S rRNA and lipl32 genes for human leptospirosis in Thailand: A case-control study. PloS One 6(1): e16236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang J, Chan TC, Temenak JJ, Dasch GA, Ching WM, et al. (2004) Development of a quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi . Am J Trop Med Hyg 70(4): 351–6. [PubMed] [Google Scholar]
- 29. Henry KM, Jiang J, Rozmajzl PJ, Azad AF, Macaluso KR, et al. (2007) Development of quantitative real-time PCR assays to detect Rickettsia typhi and Rickettsia felis, the causative agents of murine typhus and flea-borne spotted fever. Mol Cell Probes 21(1): 17–23. [DOI] [PubMed] [Google Scholar]
- 30. Le VT, Phan TQ, Do QH, Nguyen BH, Lam QB, et al. (2010) Viral etiology of encephalitis in children in southern Vietnam: Results of a one-year prospective descriptive study. Plos Negl Trop Dis 4(10): e854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Doornum GJ, Guldemeester J, Osterhaus AD, Niesters HG (2003) Diagnosing herpesvirus infections by real-time amplification and rapid culture. J Clin Microbiol 41(2): 576–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Jong MD, Weel JF, Schuurman T, Wertheim-van Dillen PM, Boom R (2000) Quantitation of varicella-zoster virus DNA in whole blood, plasma, and serum by PCR and electrochemiluminescence. J Clin Microbiol 38(7): 2568–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khanh TH, Sabanathan S, Thanh TT, Thoa LPK, Thuong TC, et al. (2012) A large outbreak of Enterovirus 71 associated hand, foot and mouth disease in southern Vietnam, September-November 2011. Emerg Infect Dis 18(12): 2002–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benschop K, Molenkamp R, van der Ham A, Wolthers K, Beld M (2008) Rapid detection of human parechoviruses in clinical samples by real-time PCR. J Clin Virol 41(2): 69–74. [DOI] [PubMed] [Google Scholar]
- 35.WHO Collaborating Centre for Influenza (2009) CDC rRT-PCR Protocol for Detection and Characterization of Influenza; version 2009 CDC Ref. # I-007–05. Geneva, Switzerland: World Health Organization.
- 36. Gueudin M, Vabret A, Petitjean J, Gouarin S, Brouard J, et al. (2003) Quantification of respiratory syncytial virus RNA in nasal aspirates of children by real-time RT-PCR Assay. J Virol Methods 109: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karron RA, Froelich JL, Bobo L, Belshe RB, Yolken RH (1994) Rapid detection of parainfluenza virus type 3 RNA in respiratory specimens: use of reverse transcription-PCR-enzyme immunoassay. J Clin Microbiol 32: 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Echevarría JE, Erdman DD, Swierkosz EM, Holloway BP, Anderson LJ (1998) Simultaneous detection and identification of human parainfluenza virus 1, 2 and 3 from clinical samples by multiplex PCR. J Clin Microbiol 36: 1388–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Becker JU, Theodosis C, Jacob ST, Wira CR, Groce NE (2009) Surviving sepsis in low-income and middle-income countries: new directions for care and research. Lancet Infect Dis 9: 577–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flowchart summarising methods for the analysis of cerebrospinal fluid (CSF) from children with suspected meningoencephalitis enrolled in the study.
(PDF)
Number of admissions with “acute serology” (see main text for definition) to DENV against period of admission (30-day intervals starting from study start date 12th October 2012).
(PDF)
Number of admissions with “acute/recent serology” (see main text for definition) to DENV against period of admission (30-day intervals starting from study start date 12th October 2012).
(PDF)
Classification of DENV and JEV serology in samples. a Dynamic rise of ≥2 Panbio units between acute and discharge samples. b Dynamic fall of ≤2 Panbio units between acute and discharge samples. c Dynamic rise or fall of ≤2 panbio units between acute and discharge samples. d Considered negative by manufacturer’s criteria.
(PDF)
(DOC)