Abstract
The presence of adhesins is arguably an important determinant of pathogenicity for Uropathogenic Escherichia coli (UPEC). Antimicrobial susceptibilities were tested by agar dilution method, fifteen adhesin genes were detected by polymerase chain reaction, and multilocus sequence typing (MLST) was analyzed in 70 UPEC isolates and 41 commensal E. coli strains. Extended-spectrum β-lactamase (ESBL) was determined with confirmatory test. The prevalence of ESBL-producers in UPEC (53%, 37/70) was higher than the commensal intestinal isolates (7%, 3/41), and 97% (36/37) of the ESBL-producing UPEC harbored bla CTX-M genes. afa was present in 36% (10/28) UPEC isolates from recurrent lower urinary tract infection (UTI), and none in the acute pyelonephritis, acute uncomplicated cystitis or commensal strains (P<0.0001). papG was detected in 28% (20/70) of UPEC isolates, while 5% (2/41) of the commensal strains were papG positive (P = 0.0025), and the prevalence of papG was significantly higher in acute pyelonephritis group (71%) than the other two UTI groups (P<0.0001). The prevalence of flu, yqi, yadN and ygiL was significantly higher in UPEC isolates than in the commensal strains. ESBL-producing UPEC showed a lower prevalence of adhesin genes compared with non-ESBL-producing strains. The MLST profiles were different between UPEC and commensal strains, with ST131 (19%, 13/70) and ST10 (20%, 8/41) being the most common MLSTs, respectively. This study demonstrated that several adhesin genes were more prevalent in UPEC isolates than in commensal E. coli, and afa may be associated with recurrent lower UTI whereas papG is more frequently associated with acute pyelonephritis.
Introduction
Urinary tract infection (UTI) is one of the most common bacterial infections, and it is estimated that the overall lifetime prevalence of UTI in women is greater than 50% [1]. Further recurrent UTIs are reported in about 25% of women within 6 months of an acute UTI episode and pose a major health problem [2]. Uropathogenic Escherichia coli (UPEC) is the causative pathogen in 70–95% community-acquired UTI and over 50% nosocomial UTI [3]. Many virulence factors contribute to the pathogenesis of UPEC. The presence of adhesins is arguably the most important determinant of the pathogenicity for UPEC [4]. Adhesive organelles, including type 1, P, S/F1C, M, G and curli fimbriae, along with Afa/Dr adhesins, as well as temperature sensitive haemagglutinin (TSH), promote both bacterial attachment to and invasion of host tissues within the urinary tract [4]–[6].
E. coli expressing Dr/Afa adhesins may predispose the establishment of chronic or recurrent infections [7]. In previous studies, it was shown that UPEC entered the urinary epithelial cells through adhesins AfaD and AfaE, and therefore avoided the clearance of host immunosurveillance and antibiotic treatment. When the host cells shed, releasing a large amount of intracellular UPEC, a new round of infection could begin causing recurrence or relapse of UTI [8]. P fimbriae is encoded by the pap gene cluster, including papG encoding the tip adhesin PapG [9]. The three most studied PapG molecular variants, which are shown to bind distinct isoreceptors are PapG I, II and III [10]. Clinically the class II papG allele is primarily associated with human pyelonephritis, and class III papG allele with genitourinary infections in dogs and cats yet some cases of human cystitis [11]. In addition to P fimbriae, type 1 and S/F1C fimbriae, curli and M and G fimbriae which are encoded by the fim and sfa/foc, csg, bma and gaf gene clusters, respectively, may also be related to the adherence of UPEC [4], [6], [12]. More recently, it is discovered that adhesin Ag43, encoded by flu and abundant on the outer membrane of E. coli, enables bacterial cells to adhere to each other and form biofilm over the host urothelial cells, leading to poor clearance of the pathogens [13]. Other novel genes like yqi, yadN and ygiL were also found to be prevalent among UPEC strains and played an important role in colonization and pathogenesis [6], [14].
Previous studies have shown that the prevalence of adhesin genes is different between UPEC and fecal commensal strains of E. coli [15]. However, few epidemiologic data about adhesin genes of UPEC are available in China. The aim of the present study was to compare the prevalence of the adhesin genes and antimicrobial susceptibilities between UPEC and intestinal colonizing strains of E. coli, and adhesin genes between strains isolated from different types of UTIs and their association with a particular type of UTIs.
Materials and Methods
E. coli isolates
A total of 70 clinical isolates of UPEC were collected from clean midstream urine specimens of inpatients (63 female and 7 male) with urinary tract infections at Huashan Hospital, Shanghai from January 2008 to December 2010. The exclusion criteria were: 1) patients with indwelling catheter; 2) patients with urinary calculus or structural abnormalities. Isolation was performed according to standard laboratory protocols and UPEC were isolated from urine with >105 colony-forming units/ml.
Among the 70 UPEC isolates, 28 were from patients with recurrent lower UTIs. The recurrent lower UTIs were defined as at least two episodes of UTI within 6 month. Each time the patients had: a) typical clinical symptoms as urinary urgency, pain, and frequent voiding, b) pyuria, c) without fever, chill or other systematic symptoms. All of the 28 patients in recurrent lower UTI group had recurrences caused by UPEC. MLST typing of the UPEC strains from 11 randomly selected patients showed that the strains isolated from different episodes of each patient had the same MLST type.
Seventeen strains were isolated from acute pyelonephritis patients by the following criteria: a) the temperature >38.5°C, b) positive percussion pain on kidney area, c) an elevated leukocyte count (>11×109/L) and elevated percentage of neutrophils (>70%) in the blood routine test. The remaining 25 strains were from patients with acute uncomplicated cystitis.
The intestinal commensal E. coli isolates (n = 41) were collected from anal swab specimens of healthy volunteers. The inclusion criteria emphasized: 1) without diarrhea or abdominal pain, 2) without antibiotic administration within the three months before taking the specimen. After isolation, all isolates were kept frozen at −70°C in 20% glycerol.
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of imipenem, cefepime, ceftazidime, cefotaxime, cefuroxime, cefazolin, piperacillin/tazobactam, piperacillin, gentamicin, amikacin, ciprofloxacin, and nitrofurantoin for both UPEC and commensal isolates were determined by agar dilution according to CLSI standards [16]. The antimicrobial concentrations were from 0.06 to 128 µg/ml. E. coli ATCC 25922 was used as a quality control strain.
Detection of ESBL-producing bacterial strains and ESBL genes
Extended-spectrum β-lactamase (ESBL) was determined with the previously recommended CLSI disk diffusion ESBL screening and confirmatory test [17]. CTX-M- specific genes in ESBL-producing strains were identified by polymerase chain reaction (PCR) with the primers reported previously [18].
Adhesin gene detection
The presence of genes afa, draE, daaE, papG I, papG II, papG III, flu, fimH, sfaD, focG, bmaE, gafD, csgA, tsh, yqi, yadN and ygiL was investigated by PCR amplification using primers (Sangon, Shanghai, China) as described previously [12], [14], [19]–[22]. The sequence amplified by afa primers, located between afaB and afaC, was conservatively shared by many subtypes of Dr/Afa adhesins [19]. All the sfa and foc PCR fragments were confirmed by sequencing as sfa and foc are highly homologous. The positive isolates identified by sequencing of PCR products in the pre-test were used as positive control in the following screening.
Analysis of UPEC by multilocus sequence typing (MLST)
All of the 70 clinical UPEC and 41 intestinal commensal isolates were analyzed by MLST. The seven housekeeping genes (adk, fumC, icd, purA, gyrB, recA and mdh) for typing [23], as well as the primer sets and PCR conditions were according to the MLST website for E. coli (mlst.ucc.ie/mlst/dbs/Ecoli/documents/primersColi_html). The based upon related sequence types (BURST) clustering algorithm (eburst.mlst.net) was used to analyze the allelic profiles and define clonal complexes (CCs). CCs were identified according to the number of single-locus variants (SLVs) and double-locus variants (DLVs) shared between isolates, where only STs that shared six or more loci were assigned to a defined CCs.
Statistical analysis
The comparison of prevalence rates was performed using Pearson Chi-square test or Fisher's exact test (two-tailed) with the software SPSS 16.0, and the level of significance was set at P<0.05.
Results
Antimicrobial susceptibility and ESBL differences of the two groups of E. coli strains
UPEC isolates demonstrated remarkably lower susceptibility rates to piperacillin (14%), cefazolin (21%), cefuroxime (39%), cefotaxime (43%), ceftazidime (71%), cefepime (79%), gentamicin (43%), amikacin (90%) and ciprofloxacin (23%) than intestinal commensal isolates (P<0.05). However, 91%–100% of E. coli isolates in the two groups were susceptible to piperacillin/tazobactam, imipenem and nitrofurantoin and there was no significant difference between the two groups (P>0.05, Table 1).
Table 1. Comparison of the antimicrobials susceptibility between UPEC and commensal E. coli strains.
| Antimicrobial agents | UPEC isolates (n = 70) | Commensal E. coli strains (n = 41) | P value | ||||||
| MIC range (µg/ml) | MIC50 | MIC90 | S% | MIC range (µg/ml) | MIC50 | MIC90 | S% | ||
| Piperacillin | 1–256 | 128 | 256 | 14 | 0.5–256 | 2 | 64 | 73 | <0.0001 |
| Piperacillin/tazobactam | 0.25–256 | 2 | 4 | 91 | 0.5–16 | 1 | 2 | 100 | 0.0539 |
| Cefazolin | 0.06–256 | 128 | 256 | 21 | 0.5–256 | 1 | 64 | 83 | <0.0001 |
| Cefuroxime | 2–256 | 256 | 256 | 39 | 2–256 | 2 | 256 | 85 | <0.0001 |
| Cefotaxime | 0.06–256 | 8 | 256 | 43 | 0.06–128 | 0.06 | 8 | 85 | <0.0001 |
| Ceftazidime | 0.06–256 | 0.5 | 32 | 71 | 0.06–32 | 0.125 | 0.25 | 98 | 0.0007 |
| Cefepime | 0.06–256 | 1 | 16 | 79 | 0.06–8 | 0.06 | 1 | 100 | 0.0014 |
| Imipenem | 0.06–0.25 | 0.06 | 0.125 | 100 | 0.06–125 | 0.06 | 0.06 | 100 | - |
| Gentamicin | 0.25–256 | 64 | 128 | 43 | 0.5–256 | 1 | 64 | 85 | <0.0001 |
| Amikacin | 1–256 | 2 | 8 | 90 | 1–4 | 2 | 2 | 100 | 0.0364 |
| Ciprofloxacin | 0.06–256 | 32 | 128 | 31 | 0.06–16 | 0.06 | 1 | 90 | <0.0001 |
| Nitrofurantoin | 4–128 | 8 | 16 | 99 | 4–32 | 8 | 16 | 100 | 0.4420 |
Thirty-seven (53%) of 70 UPEC isolates, while 3 (7%) out of 41 intestinal commensal isolates were ESBL-producing (P<0.001). bla CTX-M genes were present in 97% (36/37) ESBL-producing isolates, and of these, 58% (21/36) carried bla CTX-M-1-group ESBL genes while 42% (15/36) belonged to bla CTX-M-9-group. ESBL genotype could not be determined in one strain. The CTX-M-2, CTX-M-8 and CTX-M-25/26 groups were not found.
Prevalence of adhesin genes
(1) Afa/Dr adhesin family member genes
afa was present only in 10 UPEC isolates that belonged to the recurrent lower UTIs group (10/28, 36%). None of the isolates from the acute pyelonephritis group or the acute uncomplicated cystitis group carried afa. No afa was found in 41 intestinal commensal E. coli isolates. Neither draE nor daaE was detected among all the strains we screened.
(2) Type P fimbriae related gene papG
papG was positive in 28% (20/70) of the UPEC isolates including 16 papG II and 4 papG III, and 5% (2/41) of the intestinal commensal isolates carried papG II (P = 0.0025). The prevalence of papG in acute pyelonephritis (71.4%, 5/7) was significantly higher than that of recurrent lower UTI group (14.3%, 3/21, P = 0.0004) and that of acute uncomplicated cystitis group (15.8%, 3/19, P = 0.0001). No papG I was detected in any of all the tested strains.
(3) Biofilm related adhesin gene flu and novel genes yqi, yadN and ygiL
A majority (77%, 54/70) of the UPEC isolates and 41% (17/41) of the intestinal commensal isolates were found to be flu positive (P = 0.0002). The prevalence of novel genes yqi, yadN and ygiL was higher in UPEC isolates than that of the commensal strains (27% vs 7%, 54% vs 32%, and 44% vs 17%, respectively, P<0.05). The prevalence differences of these four genes of flu, yqi, yadN and ygiL between the three UTI groups were not significant.
(4) Type 1 fimbrial gene fimH and curli fiber gene csgA
fimH was found positive in 86% (60/70) UPEC isolates and 73% (30/41) commensal isolates (P = 0.1034). The positive rates of fimH in the three UTI groups were 86%, 82% and 88%, respectively. 30% (21/70) UPEC isolates and 34% (14/41) commensal ones carried csgA (P = 0.6500).
(5) Other adhesin genes
sfaD was present in 5 isolates and focG in 2 isolates among the UPEC group. One of the sfaD carrying isolates was also detected focG positive. None of the isolates in the intestinal commensal group were sfaD/focG positive. No more than 5 isolates in UPEC or commensal groups produced amplicons with the bmaE, gafD or tsh primers. (Table 2)
Table 2. Comparison of the prevalence of adhesin genes among different groups of E. coli strains.
| Adhesin genesa | Recurrent lower UTI, n = 28 (%) | Acute pyelonephritis, n = 17 (%) | Acute uncomplicated cystitis, n = 25 (%) | Total UPEC, n = 70 (%) | Commensal isolates, n = 41(%) | P value (Total UPEC vs Commensal) |
| afa | 36b | 0 | 0 | 14 | 0 | <0.0001c |
| papG | 18 | 71d | 12 | 28 | 5 | 0.0025 |
| flu | 89 | 65 | 72 | 77 | 41 | 0.0002 |
| fimH | 86 | 82 | 88 | 86 | 73 | 0.1034 |
| sfaD | 4 | 12 | 8 | 7 | 0 | 0.0799 |
| focG | 0 | 12 | 0 | 3 | 0 | 0.2747 |
| bmaE | 4 | 6 | 4 | 4 | 5 | 0.8845 |
| gafD | 4 | 0 | 0 | 1 | 5 | 0.2794 |
| csgA | 21 | 24 | 44 | 30 | 34 | 0.6500 |
| tsh | 0 | 18 | 8 | 7 | 5 | 0.6357 |
| yqi | 21 | 35 | 28 | 27 | 7 | 0.0114 |
| yadN | 50 | 76 | 44 | 54 | 32 | 0.0212 |
| ygiL | 46 | 43 | 40 | 44 | 17 | 0.0035 |
, draE and daaE genes were not detected in any of the isolates.
, Compared to the other two UTI groups, P<0.0001;
. Recurrent lower UTI group compared to commensal isolates;
, Compared to the other two UTI groups, P<0.0001.
Comparison of the prevalence of fimbrial genes between ESBL-producing and non-ESBL-producing UPEC
The prevalence of fimH (97%, 32/33) in the non-ESBL-producing UPEC was significantly higher than that in the ESBL-producing UPEC (76%, 28/37, P = 0.0110). All the six strains carrying sfaD/focG were from the non-ESBL-producing UPEC group. The prevalence of the other fimbrial genes in the non-ESBL-producing UPEC group seemed a little higher than the ESBL-producing UPEC strains, however, there were no statistically significant differences (P>0.05, Table 3)
Table 3. Comparison of the prevalence of fimbrial genes between ESBL-producing and non-ESBL-producing UPEC strains.
| Fimbrial genes | ESBL-producing UPEC n = 37 (%) | Non-ESBL-producing UPEC n = 33 (%) | P value |
| afa | 14 | 15 | 0.8450 |
| papG | 22 | 36 | 0.1729 |
| flu | 73 | 82 | 0.3790 |
| fimH | 76 | 97 | 0.0110 |
| sfaD/focG | 0 | 18 | 0.008 |
| bmaE | 3 | 6 | 0.5990 |
| gefD | 0 | 3 | 0.4710 |
| csgA | 22 | 39 | 0.1053 |
| tsh | 5 | 9 | 0.6610 |
| yqi | 22 | 33 | 0.2714 |
| yadN | 49 | 61 | 0.3161 |
| yqiL | 43 | 45 | 0.8525 |
MLST analysis of UPEC and commensal isolates
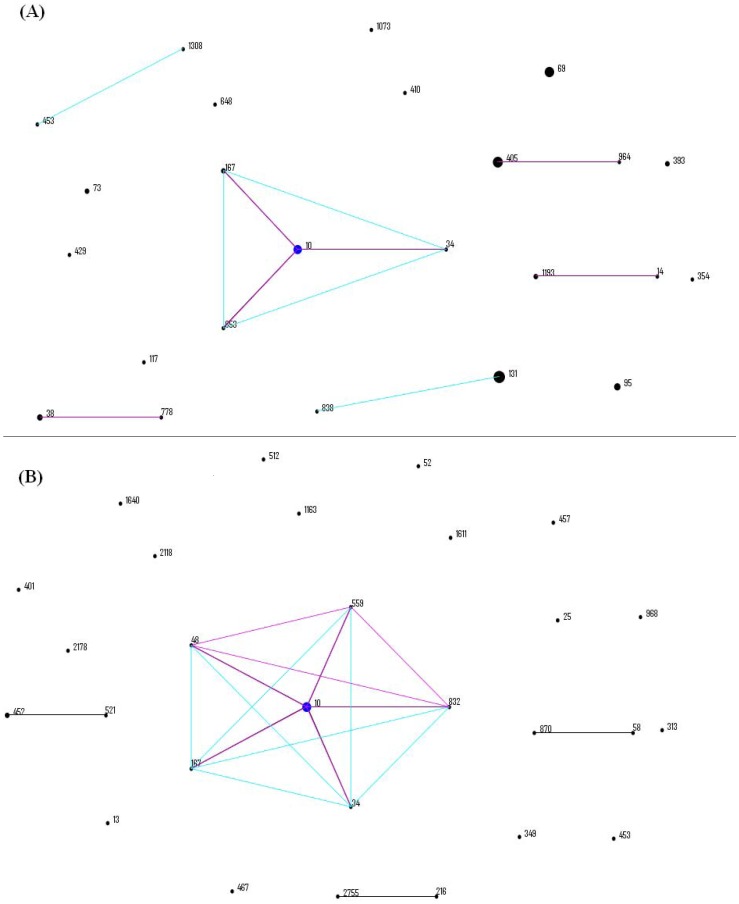
The 70 UPEC isolates analyzed were assigned to 26 distinct STs. The most common STs were ST131 (n = 13, 19%), followed by ST69 (9, 13%), ST405 (8, 11%), ST10 (7, 10%), ST393 (4, 6%), ST95 (4, 6%) and ST38 (3, 4%). The seven most common STs accounted for 69% (48/70) of the isolates, demonstrating the diversity of lineages. eBURST analysis revealed that 4 CCs (CC10, CC405, CC14 and CC38) encompassing 10 STs represented 25 (36%) UPEC isolates and the remaining 45 isolates included in the other 16 STs appearing as singletons (Figure 1A).
Figure 1. eBURST diagrams of UPECand intestinal commensal isolates showing related STs and individual STs.
(A) referred to UPEC isolates and (B) referred to intestinal commensal isolates. Each ST is represented by a circle, the size of which correlates to the frequency of the ST. Predicted founders are positioned centrally and shown in blue, and single-locus variants and double-locus variants are shown in pink and blue, respectively.
The 41 intestinal commensal isolates analyzed by MLST were grouped into 32 different STs. 28 were already included in the E. coli website database and 4 were new STs. ST10 (n = 8, 20%) was the most common group. A new ST had 2 strains, while the other 29 STs contained a single strain each. As analyzed by eBURST, ST10, ST34, ST167, ST48, ST559 and ST832 belonged to CC10 (Figure 1B).
Discussion
Most of the intestinal commensal isolates tested in this study were susceptible to all the tested antimicrobial agents, while the UPEC isolates were characterized by a varying degree of susceptibilities to different antimicrobials. Of 70 UPEC strains, 37 (53%) produced extended-spectrum β-lactamase (ESBL). In a previous study, 2.9% of E. coli isolates recovered by outpatient urine cultures in 2008 harbored ESBL in USA [24], which is much higher than previously reported but far lower than the rate in our study. The annual percentage of ESBL-producing UPEC with bla CTX-M genes changed from 35% in 2003 to 64% in 2008 with the average rate of 56% in that study [24]. However, in this study, bla CTX-M ESBL genes were detected in 36 (97%) of 37 ESBL positive strains, which is much higher than in the USA.
All 10 UPEC isolates harboring the afa belonged to recurrent lower UTI group, none was afa positive in isolates from other UTIs or intestinal carriage, indicating that UPEC carrying afa might correlate with the recurrence of UTI episodes. Blanco and colleagues screened UPEC for afa and found no afa positive strain in acute pyelonephritis isolates, however, afa was detected in 5 of the 116 isolates (4.3%) from cystitis patients and 1 of the 42 (2.4%) isolates from asymptomatic bacteriuria [19]. Previous studies showed that Afa/Dr fimbrial adhesins contributed to the ability of UPEC isolates to colonize and persist long term within the urinary tract and therefore more likely to cause the recurrence of UTI episodes [7]–[8].
The isoreceptors of adhesin PapG primarily are scattered within the kidney, including glomerulus, proximal tubulus, distal tubulus, and collecting duct. Therefore sitting on the top of type P fimbriae, adhesin PapG is thought to be associated with bacterial adhesion to the kidney [9]. The prevalence of papG in acute pyelonephritis isolates was significantly higher (71%) than that in recurrent lower UTIs (18%) and acute uncomplicated cystitis (12%) in the present study, illustrating high correlation between the papG and pyelonephritis. Our findings are consistent with some previous studies. Johnson et al found 69% (118/170) acute pyonephritis strains and 25% (21/83) of cystitis strains were papG positive [15].
Encoded by flu, Ag43 is a self-recognizing adhesin that is associated with cell aggregation and biofilm formation of UPEC [13]. We found a high prevalence (77%) of flu in UPEC isolates, while flu was detected in 41% of the intestinal commensal isolates. The finding is consistent with the results of Ulett and colleagues. They reported 83% (30/36) of the UPEC isolates and 56% (35/62) of the commensal strains carried flu [22]. However, there was no statistically significant difference in flu gene prevalence between recurrent lower UTI and non-recurrent lower UTI in the present study.
It was reported that the adhesin encoding gene yqi was prevalent among UPEC by more than 50% while absent in all the intestinal pathogenic E. coli [6]. In this study, yqi was detected in 7% of commensal isolates, but the prevalence was significantly lower than that in UPEC strains (27%, P = 0.0114).
yadN and ygiL were found to be more prevalent in UPEC (54% and 44%, respectively) than the intestinal commensal strains (32% and 17%) in the present study (P<0.05), which coincides with Mobley's work. It was reported that fimbriae related structure Yad and Ygi could enhance the virulence-related phenotypes, including biofilm formation and adherence to immortalized human epithelial cells [14]. In addition, we did not detect any significant prevalence difference of yadN or ygiL between UPEC strains causing recurrent lower UTI, acute pyelonephritis and uncomplicated cystitis.
The results showed that 86% of UPEC isolates and 73% of commensal isolates carried fimH, encoding the adhesin FimH as the tip fimbria for type 1 fimbriae. Wang et al detected 91% (72/79) fimH positive in UPEC isolates [25]. On the other hand, Schlager found that 81% (74/91) strains isolated from fecal specimen of healthy girls carried fimH [26]. A high prevalence of fimH was found in both UPEC and commensal isolates. However, the transcription and expression level of fimH might be different between UPEC and commensal strains, which made type 1 fimbriae an important virulence factor for UPEC [27].
In the present study, we found that ESBL-producing UPEC isolates showed a lower prevalence of fimbrial genes, fimH and sfaD/focG, compared with non-ESBL-producing ones. The results coincided with the previous findings that fimbrial genes were of reduced prevalence among UPEC resistant to extended-spectrum cephalosporins. A possible explanation is that: virulence traits like the fimbrial genes, as well as resistance genes, can be carried on conjugative plasmids; therefore, the incompatible resistance-encoding plasmids are outcompeting fimbrial factor encoding plasmids [28]. It is reported that the acquisition of antibiotic resistance may cause alterations in phenotypic and physiological characteristics, which are referred to as “biological fitness cost”. The biological fitness cost on antibiotics resistance generally results in reduced growth rates. More dramatic phenotypic changes, including poor fimbrial expression, were reported in an ampicillin-resistant mutant Acinetobacter sp. strain DR1A comparing with the wild type strain DR1 [29]. Therefore, the decreased fimbrial genes and adherence capability in ESBL-producing UPEC might also be a fitness trade-off for the ESBL to survive antibiotics exposure. The exact explanation for the lower incidence of fimbrial genes among resistant UPEC isolates needs additional study.
In this study, 69% of the UPEC isolates were included in the seven STs (ST131, ST69, ST405, ST10, ST393, ST95 and ST38) and ST131 was the most predominant (19%). Gibreel revealed a consistent profile of STs over a 3-year period with UPEC isolates (n = 300) in the Northwest region of England primarily from 8 lineages: ST73 (16.6%), ST131 (12.3%), ST69 (9%), ST95 (6.3%), ST10 (4.3%), ST127 (3.6%), ST14 (2.7%) and ST405 (1.7%) [30]. Most of these STs (ST131, ST69, ST95, ST10 and ST405) were also prevalent in our study. The 41 commensal isolates had a richer ST diversity (32 STs) than the 70 UPEC counterparts (26 STs). A ST10 E. coli strain was common in UPEC (10%) as well as in intestinal commensal isolates (20%). One isolate from each group was ST453. The remaining 24 STs of the UPEC isolates were different from the other 30 STs of the commensal strains.
This study indicated that afa of Afa/Dr adhesin family might be associated with lower UTI recurrence, while papG might be correlated with acute pyelonephritis. However, we only focused on detecting the prevalence of adhesin genes by PCR, instead of the mechanism of the related adhesins of UPEC. Therefore, we could not prove the correlation between the adhesins and the pathogenesis of the UPEC. Moreover, because of the “cross-talk” among the regulators of different adhesin systems of UPEC and their functional redundancy, when a certain type of adhesin is lost, it may be quite difficult to study the relationship between the UTI recurrence and a specific kind of adhesin. Studies on other factors and mechanisms involved in pathogenesis and persistence of UPEC are required.
Funding Statement
This work was supported by a grant from the National Natural Science foundation of China (number 81102509). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Griebling TL (2005) Urologic diseases in America project trends in resource use for urinary tract infections in women. J Urol 173: 1281–1287. [DOI] [PubMed] [Google Scholar]
- 2. Foxman B (1990) Recurring urinary tract infection-incidence and risk factors. Am J Public Health 80: 331–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Foxman B (2003) Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Dis Mon 49: 53–70. [DOI] [PubMed] [Google Scholar]
- 4. Mulvey MA (2002) Adhesion and entry of uropathogenic Escherichia coli . Cell Microbiol 4: 257–271. [DOI] [PubMed] [Google Scholar]
- 5. Rosen DA, Hooton TM, Stamm WE, Humphrey PA, Hultgren SJ (2007) Detection of intracellular bacterial communities in human urinary tract infection. Plos Medicine 4: 1949–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Antao EM, Wieler LH, Ewers C (2009) Adhesive threads of extraintestinal pathogenic Escherichia coli . Gut Pathog 1: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Korotkova N, Yarova-Yarovaya Y, Tchesnokova V, Yazvenko N, Carl MA, et al. (2008) Escherichia coli DraE adhesin associated bacterial internalization by epithelial cells is promoted independently by decay accelerating factor and carcinoembryonic antigen related cell adhesion molecule binding and does not require the DraD invasin. Infect Immun 76: 3869–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dhakal BK, Kulesus RR, Mulvey MA (2008) Mechanisms and consequences of bladder cell invasion by uropathogenic Escherichia coli . Eur J Clin Invest 38: 2–11. [DOI] [PubMed] [Google Scholar]
- 9. Lane MC, Mobley HLT (2007) Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int 72: 19–25. [DOI] [PubMed] [Google Scholar]
- 10. Stromberg N, Marklund BI, Lund B, Ilver D, Hamers A, et al. (1990) Host specificity of uropathogenic Escherichia coli depends on differences in binding specificity to gal-alpha1-4gal containing isoreceptors. Embo Journal 9: 2001–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson JR, O'Bryan TT, Low DA, Ling G, Delavari P, et al. (2000) Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papG allele III. Infect Immun 68: 3327–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson JR, Stell AL (2000) Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181: 261–272. [DOI] [PubMed] [Google Scholar]
- 13. van der Woude MW, Henderson IR (2008) Regulation and function of Ag43 (Flu). Annu Rev Microbiol 62: 153–169. [DOI] [PubMed] [Google Scholar]
- 14. Spurbeck RR, Stapleton AE, Johnson JR, Walk ST, Hooton TM, et al. (2011) Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of Ygi and Yad fimbriae. Infect Immun 79: 4753–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson JR, Owens K, Gajewski A, Kuskowski MA (2005) Bacterial characteristics in relation to clinical source of Escherichia coli isolates from women with acute cystitis or pyelonephritis and uninfected women. J Clin Microbiol 43: 6064–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CLSI (2010) Performance strandards, for antimicrobial susceptibility testing; Twentieth informational supplement M100-S20.Standard; Twentieth informational supplement M100-S20. Wayne, PA. [Google Scholar]
- 17.CLSI (2009) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically.Standard; approved standard, 8th ed M07-A8. Wayne, PA. [Google Scholar]
- 18. Wang P, Hu F, Xiong Z, Ye X, Zhu D, et al. (2011) Susceptibility of extended-spectrum-beta-lactamase-producing Enterobacteriaceae according to the new CLSI breakpoints. J Clin Microbiol 49: 3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blanco M, Blanco JE, Alonso MP, Mora A, Balsalobre C, et al. (1997) Detection of pap, sfa and afa adhesin-encoding operons in uropathogenic Escherichia coli strains: relationship with expression of adhesins and production of toxins. Res Microbiol 148: 745–755. [DOI] [PubMed] [Google Scholar]
- 20. Johnson JR, Russo TA, Brown JJ, Stapleton A (1998) papG alleles of Escherichia coli strains causing first episode or recurrent acute cystitis in adult women. J Infect Dis 177: 97–101. [DOI] [PubMed] [Google Scholar]
- 21. Sokurenko EV, Feldgarden M, Trintchina E, Weissman SJ, Avagyan S, et al. (2004) Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli . Mol Biol Evol 21: 1373–1383. [DOI] [PubMed] [Google Scholar]
- 22. Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, et al. (2007) Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun 75: 3233–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tartof SY, Solberg OD, Manges AR, Riley LW (2005) Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J Clin Microbiol 43: 5860–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qi C, Pilla V, Yu JH, Reed K (2010) Changing prevalence of Escherichia coli with CTX-M-type extended-spectrum beta-lactamases in outpatient urinary E. coli between 2003 and 2008. Diagn Microbiol Infect Dis 67: 87–91. [DOI] [PubMed] [Google Scholar]
- 25. Wang MC, Tseng CC, Wu AB, Huang JJ, Sheu BS, et al. (2009) Different roles of host and bacterial factors in Escherichia coli extra-intestinal infections. Clin Microbiol Infect 15: 372–379. [DOI] [PubMed] [Google Scholar]
- 26. Schlager TA, Whittam TS, Hendley JO, Bhang JL, Wobbe CL, et al. (2003) Variation in frequency of the virulence factor gene in Escherichia coli clones colonizing the stools and urinary tracts of healthy prepubertal girls. J Infect Dis 188: 1059–1064. [DOI] [PubMed] [Google Scholar]
- 27. Sokurenko EV, Chesnokova V, Doyle RJ, Hasty DL (1997) Diversity of the Escherichia coli type 1 fimbrial lectin - Differential binding to mannosides and uroepithelial cells. J Biol Chem 272: 17880–17886. [DOI] [PubMed] [Google Scholar]
- 28. Zhao L, Chen X, Zhu X, Yang W, Dong L, et al. (2009) Prevalence of virulence factors and antimicrobial resistance of uropathogenic Escherichia coli in Jiangsu province (China). Urology 74: 702–707. [DOI] [PubMed] [Google Scholar]
- 29. Kang YS, Park W (2010) Trade-off between antibiotic resistance and biological fitness in Acinetobacter sp. strain DR1. Environ Microbiol 12: 1304–1318. [DOI] [PubMed] [Google Scholar]
- 30. Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, et al. (2012) Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 67: 346–356. [DOI] [PubMed] [Google Scholar]