Abstract
Variants in ABCB1 and CYP2C19 have been identified as predictors of cardiac events during clopidogrel therapy initiated after myocardial infarction (MI) or percutaneous coronary intervention (PCI). In addition, PON1 has recently been associated with stent thrombosis. The reported effects of these variants have not yet been replicated in a real-world setting. We used BioVU, the Vanderbilt DNA repository linked to de-identified electronic health records (EHRs), to find data on patients who were on clopidogrel treatment after an MI and/or a PCI; among these, we identified those who had experienced one or more recurrent cardiac events while on treatment (cases, n = 225) and those who had not experienced any cardiac event while on treatment (controls, n = 468). We found that CYP2C19*2 (hazard ratio (HR) 1.54, 95% confidence interval (CI) 1.16–2.06, P = 0.003) and ABCB1 (hr 1.28, 95% CI 1.04–1.57, P = 0.018), but not PON1 (HR 0.91, 95% CI 0.73–1.12, P = 0.370), were associated with recurrent events. In this population, genetic signals for clopidogrel resistance in ABCB1 and CYP2C19 were replicated, supporting the use of EHRs for pharmacogenomic studies. Our data do not show an association between PON1 and recurrent cardiovascular events.
Current guidelines recommend dual antiplatelet therapy to be initiated after a recent myocardial infarction (MI) or intracoronary stent placement to prevent future adverse cardiac events. Aspirin in conjunction with clopidogrel is the most frequently prescribed combination.1–6 Clopidogrel is a prodrug, bioactivated predominately by CYP2C19.7,8 A loss-of-function polymorphism in CYP2C19, CYP2C19*2 (rs4244285), results in the clopidogrel-poor-metabolizer trait and has been associated with reduced on-drug platelet inhibition and an increased risk of adverse cardiac events, particularly stent thrombosis.9–13 These results, from studies conducted in registries and large randomized clinical trials, have prompted the US Food and Drug Administration to include in the clopidogrel label a black-box warning that poor metabolizers merit consideration for alternative therapies. This risk appears to be present for patients who are either heterozygous or homozygous for the CYP2C19*2 loss-of-function allele.14,15
Other genetic polymorphisms have been implicated in decreased efficacy of clopidogrel therapy. CYP2C19*17 (rs12248560) encodes a protein with increased function as compared to CYP2C19*1 and has been associated with both improved efficacy of clopidogrel and increased risk of bleeding.16,17ABCB1 encodes the P-glycoprotein efflux transporter that is expressed at the apical membrane of intestinal mucosa (among other locations), and the C3435T (rs1045642) polymorphism has been associated with reduced bioavailability.18 Clinically, this variant has been associated with an increase in the rate of cardiac events during clopidogrel therapy.11,19,20 The common Paraoxinase-1 (PON1) polymorphism, Q192R (rs662), which converts 2-oxo-clopidogrel to the active thiol metabolite, has also recently been associated with stent thrombosis; however, this finding remains controversial.21,22 PON1 is believed to result in a lower rate of bioactivation of the compound and therefore to translate into an increased risk of stent thrombosis after percutaneous coronary intervention (PCI).21
Genetic associations such as those described above have been generally studied in clinical trial or registry settings. The extent to which findings in these research settings can be translated to real-world clinical care settings is uncertain. Biobanks linking DNA to electronic health records (EHRs) offer an approach to addressing this issue.23–26 To date, such EHR-based studies have focused on disease susceptibility or physiologic traits, but their utility for pharmacogenomics has not been evaluated. The Vanderbilt University Medical Center EHR includes comprehensive information on ∼1.7 million patients; as of June 2011, BioVU, the Vanderbilt DNA biobank27 included de-identified DNA samples from more than 120,000 individuals. From the BioVU database we identified individuals who were started on clopidogrel therapy after an MI and/or PCI with stent placement and then assigned “case” or “control” status depending on the occurrence of a recurrent cardiac event during a 1- to 2-year follow-up. We report here that the associations of adverse cardiac events with variants in CYP2C19 and ABCB1 were readily replicated, whereas the association with PON1 was not.
Results
Through the implementation of an electronic algorithm followed by manual review, we identified 807 individuals who had been started on clopidogrel therapy after an MI and/or intracoronary stent placement procedure (Figure 1). The primary outcome was a recurrent cardiovascular event, defined as a composite of all-cause mortality, MI, revascularization, and stroke. This primary outcome occurred in 260 individuals, and, after biobank opt-out and inadequate DNA samples were taken into account, 225 underwent genotyping. Among the genotyped cases, there were 14 deaths, 40 MIs (9 ST segment elevation MI, 28 non–ST segment elevation MI, and 3 MIs not categorized), 206 revascularization procedures, and 1 stroke. In addition, stent thrombosis occurred in 12 individuals. We identified 547 individuals with no second composite cardiac event, of whom 468 underwent genotyping. Of the controls, 76.7% had 2 years of follow-up, and the remaining 23.3% had at least 1 year of follow-up.
Figure 1.
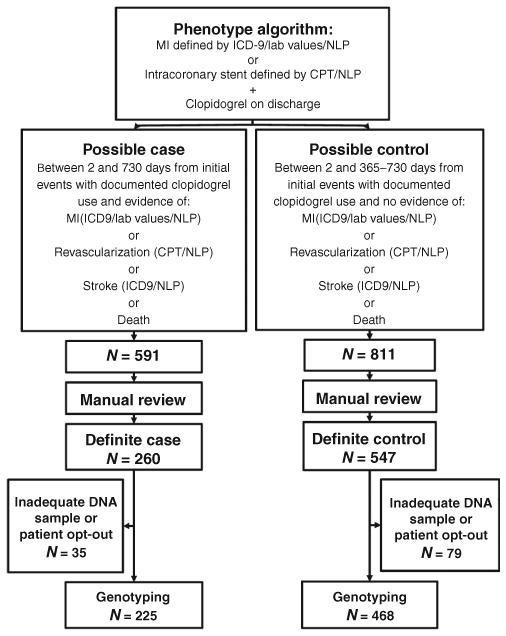
Phenotype algorithm for selection of cases and controls for genotyping. CPT, Current Procedural Terminology; ICD, International Classification of Disease; MI, myocardial infarction; NLP, natural language processing.
Cases and controls were similar in age, gender, body mass index, and cardiac risk factors, including hypertension, diabetes mellitus, hyperlipidemia, and smoking status (Table 1). The overall rates of hypertension, hyperlipidemia, and positive smoking status were higher than in previously reported studies. Cases and controls were also similar in their indication for study enrollment.
Table 1.
study population characteristics
| Controls | Cases | |
|---|---|---|
| N = 468 | N = 225 | |
| Demographics | ||
| Age (years) (± SD) | 69 ± 12 | 67 ± 11 |
| Gender (% male) | 62.8 | 64.9 |
| Race (%) | ||
| European American | 88 | 88.4 |
| African American | 9.4 | 10.7 |
| Other | 2.6 | 0.9 |
| Body mass index (kg/m2) (± SD) | 30.2 ± 6.6 | 30.5 ± 6.0 |
| Past medical history (%) | ||
| Hypertension | 84.6 | 72.9 |
| Diabetes | 34.8 | 34.8 |
| Hyperlipidemia | 95.5 | 87.1 |
| Smoking | ||
| Current | 16.4 | 15.6 |
| Ever | 47.1 | 38.7 |
| Never | 36.4 | 45.8 |
| Heart failure | 13.7 | 19.1 |
| Primary event (%) | ||
| MI | 4.1 | 6.7 |
| Stent | 67.9 | 70.2 |
| Both | 28 | 23.1 |
| Number of stents (%) | N = 449 | N = 210 |
| 1 stent | 65.2 | 56.6 |
| >1 stent | 32.7 | 43.3 |
| Unknown | 1.8 | 0.0 |
| Stent type | ||
| Bare metal | 33.6 | 21.4 |
| Drug-eluting | 60.8 | 67.1 |
| Mixed | 2.9 | 3.3 |
| Unknown | 2.7 | 8.1 |
| Stent locationsa | ||
| LMCA | 1.1 | 3.3 |
| LAD | 45.7 | 33.8 |
| LCX | 22.0 | 25.7 |
| RCA | 39.6 | 37.6 |
| Graft | 2.4 | 15.7 |
LAD, left anterior descending coronary artery; LCX, left circumflex coronary artery; LMCA, left main coronary artery; MI, myocardial infarction; RCA, right coronary artery.
Percentages total more than 100% because patients may receive more than one intervention.
We performed genotyping for six single-nucleotide polymorphisms (SNPs): rs1045642, ABCB1; rs4244285, CYP2C19*2; rs4986893, CYP2C19*3; rs28399504, CYP2C19*4; rs12248560, CYP2C19*17; and rs662, PON1. Two SNPs—rs28399504 (CYP2C19*4) and rs4986893 (CYP2C19*3)—were excluded from further analysis because of the very low frequency of the minor alleles. The genotype frequencies by case status for four SNPs—rs1045642 (ABCB1), rs4244285 (CYP2C19*2), rs12248560 (CYP2C19*17), and rs662 (PON1)—are available in Supplementary Table S1 online.
Kaplan–Meier curves (Figure 2) of the primary end point for CYP2C19 among cases and controls showed that carriers of at least one CYP2C19 loss-of-function allele (*1/*2 or *2/*2) had higher event rates than those without this allele (P = 0.005). For ABCB1, the recurrent cardiac event rate was greater among A/A genotypes as compared to G/G genotypes (P = 0.065). Neither the CYP2C19*17 nor the PON1 genotype affected the time to second cardiac event in dominant, recessive, or additive genetic models.
Figure 2.
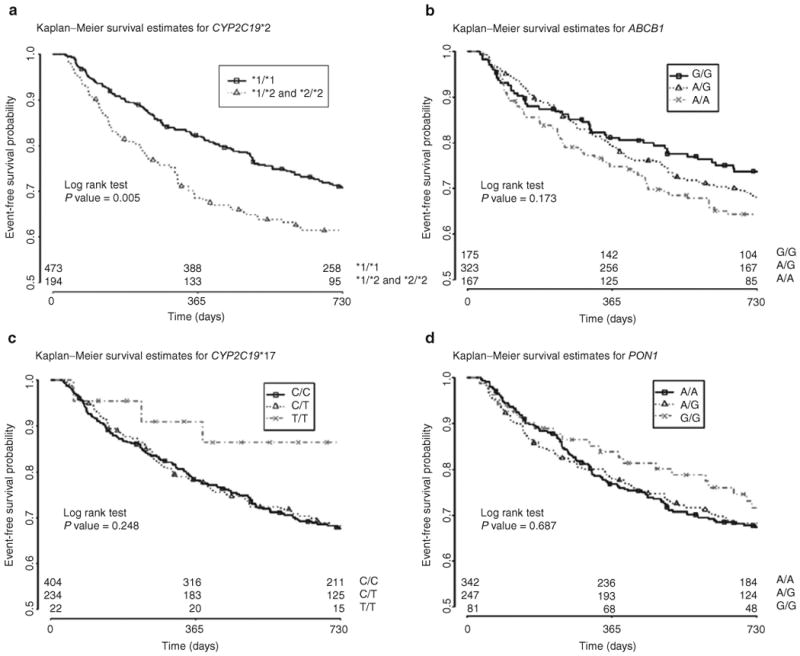
Kaplan–Meier curves for time to recurrent cardiovascular event. (a)CYP2C19*2, (b)ABCB1, (c)CYP2C19*17, (d)PON1.
Unadjusted and adjusted results for the primary analysis are shown in Table 2 for all individuals. According to the adjusted estimates, CYP2C19*2 and ABCB1 were significantly associated with the primary end point, with hazard ratios (HR) equal to 1.54 (95% CI 1.16–2.06, P = 0.003) and 1.28 (1.04–1.57, P = 0.018), respectively. Individuals who were CYP2C19*2 heterozygotes were also at increased risk as compared to homozygote normal metabolizers, the HR being 1.61 (1.20–2.17, P = 0.001). There were 15 individuals who were CYP2C19*2 homozygotes, and no increased risk compared to normal metabolizers was detected in this small group. There was no evidence to suggest an effect of CYP2C19*17 or PON1 on the primary outcome measure: HRs were 0.91 (0.71–1.18, P = 0.48) for CYP2C19*17 and 0.91 (0.73– 1.12, P = 0.37) for PON1. For both CYP2C19*2 and ABCB1, the primary end point was driven largely by repeat revascularization, the HR values being 1.54 (1.14–2.07, P = 0.004) and 1.21 (0.99–1.47, P = 0.067), respectively (Table 3). For the secondary end points (death, MI, and stent thrombosis considered individually), none of the SNPs was found to have a significant association. However, ABCB1 approached significance for stent thrombosis, the HR being 2.64 (0.84–8.29, P = 0.096), and CYP2C19*2 approached significance for MI, the HR being 2.02 (0.99–4.12, P = 0.054).
Table 2.
Unadjusted and adjusted tests of association for ABCB1, CYP2C19*2, CYP2C19*17, and PON1
| Unadjusted | Adjusted | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Gene | SNP | Chr | Loc | Coded allele | HR | 95% CI | P | HR | 95% CI | P |
| ABCB1 | rs1045642 | 7 | 86976581 | A | 1.20 | 0.99–1.46 | 0.061 | 1.28 | 1.04–1.57 | 0.018 |
|
| ||||||||||
| CYP2C19*2 | rs4244285 | 10 | 96531606 | A | 1.50 | 1.13–2.00 | 0.0049 | 1.54 | 1.16–2.06 | 0.003 |
|
| ||||||||||
| CYP2C19*17 | rs12248560 | 10 | 96511647 | T | 0.88 | 0.68–1.13 | 0.308 | 0.91 | 0.71–1.18 | 0.481 |
|
| ||||||||||
| PON1 | rs662 | 7 | 94775382 | G | 0.93 | 0.76–1.13 | 0.466 | 0.91 | 0.73–1.12 | 0.370 |
Analysis performed using Cox proportional hazards model. The adjusted model was stratified by gender and race, and adjusted for age, smoking status, and body mass index. Chr, chromosome; CI, confidence interval; HR, hazard ratio; Loc, location.
Table 3.
Test of association for secondary end points for ABCB1, CYP2C19*2, CYP2C19*17, and PON1
| HR | 95% CI | P | |
|---|---|---|---|
| Stent thrombosis (n = 12) | |||
| ABCB1 | 2.64 | 0.84–8.29 | 0.096 |
| CYP2C19*2 | 2.79 | 0.70–11.16 | 0.147 |
| CYP2C19*17 | 0.27 | 0.04–2.09 | 0.210 |
| PON1 | 0.77 | 0.26–2.22 | 0.622 |
| Revascularization (n = 206) | |||
| ABCB1 | 1.21 | 0.99–1.14 | 0.067 |
| CYP2C19*2 | 1.54 | 1.14–2.07 | 0.004 |
| CYP2C19*17 | 0.86 | 0.66–1.11 | 0.247 |
| PON1 | 0.93 | 0.75–1.14 | 0.470 |
| Myocardial infarction (n = 40) | |||
| ABCB1 | 1.33 | 0.80–2.22 | 0.270 |
| CYP2C19*2 | 2.02 | 0.99–4.12 | 0.054 |
| CYP2C19*17 | 0.63 | 0.31–1.29 | 0.207 |
| PON1 | 1.03 | 0.63–1.69 | 0.906 |
| Death (n = 14) | |||
| ABCB1 | 1.70 | 0.78–3.70 | 0.178 |
| CYP2C19*2 | 1.76 | 0.57–5.37 | 0.323 |
| CYP2C19*17 | 1.62 | 0.69–3.81 | 0.267 |
| PON1 | 0.57 | 0.23–1.44 | 0.231 |
All results report unadjusted Cox proportional hazard models. CI, confidence interval; HR, hazard ratio.
Supplementary Table S2 online shows the unadjusted and adjusted analyses individually by race. In European Americans, CYP2C19*2 and ABCB1 were significantly associated with the primary end point in both adjusted and unadjusted analyses, with adjusted HR equal to 1.44 (95% CI 1.05–1.97, P = 0.03) and 1.27 (1.03–1.57, P = 0.029), respectively. In African Americans, CYP2C19*2 was significantly associated with the primary outcome in the unadjusted analysis only, with an HR of 2.40 (1.04– 5.55, P = 0.04).
Discussion
This study validates the association of CYP2C19*2 (rs4244285) and ABCB1 3435 G>A (rs1045642) polymorphisms with recurrent cardiovascular events in patients on clopidogrel therapy. Because these associations were observed in a de-identified cohort compiled from an EHR populated with real-world phenotypic and treatment information, the clinical relevance of these associations, previously observed in controlled trials, is bolstered. This study, along with ongoing work with warfarin,28 is among the first to demonstrate the use of EHR data to replicate and discover pharmacogenomic associations.
Multiple studies have examined the associations of CYP2C19*2 and ABCB1 G>A with clopidogrel resistance. These studies have included registry trials of patients who have had an MI or undergone PCI, cohort studies with those receiving clopidogrel for stent thrombosis or PCI, post hoc analysis of randomized control trials, and a meta-analysis.9,11–15,20,29,30 For the primary end point (a composite of all-cause mortality, MI, revascularization, and stroke), our results are within the ranges previously reported for both CYP2C19*2 and ABCB1. Through the use of an electronic database, both study recruitment and follow-up can be acquired instantaneously, thereby decreasing the time, personnel, and cost required to perform a study. Both the cases and the controls were identified from the same population, reducing a selection bias between the groups. However, the use of the EHR for such studies does limit the outcomes assessed to those recorded within the EHR and to hard end points such as revascularization status from cardiac catheterization records. We were unable to replicate the association of CYP2C19*2 with stent thrombosis that has been reported in previous studies,14,29 possibly because of insufficient power due to the low numbers of cases. Because the indication for cardiac catheterization could not always be reliably assessed from EHR data, our data may underestimate the number of stent thromboses. In addition, given the nature of our study, the angiograms could not be reviewed in our de-identified cases to ascertain the validity of the diagnoses of stent thrombosis in line with the Academic Research Consortium criteria.31 However, the rate of stent thrombosis in our population was 1.7% over 2 years, which was within the range of 0.5–1% per year reported in previous studies.32 Future accrual of larger numbers of patients may yield adequately powered studies on stent thrombosis.
Bouman et al. recently reported a significantly increased risk of stent thrombosis associated with PON1 in a small cohort of 40 patients showing a gene–dose effect dependent on the number of major alleles.21 However, when a larger cohort of patients with stent thrombosis was examined, this relationship was not replicated.22 In addition, a previous genome-wide association study of clopidogrel's effect on adenosine diphosphate–induced platelet aggregation readily detected the CYP2C19*2 signal but failed to identify an association with PON1.30
Although our study was not powered to detect an association with stent thrombosis, we would have expected a trend toward association between PON1 and either the composite outcome or stent thrombosis if a strong association with stent thrombosis does indeed exist. Moreover, these PON1 variants have also been associated with adverse cardiac events independent of clopidogrel therapy, further weakening the association of PON1 and adverse cardiac outcomes in the context of clopidogrel efficacy.33,34
There are limitations to the results of the work we report here. Compliance with prescribed medication regimens is not readily assessable in the EHR. In our study, we used evidence of medication refills and visits with providers as evidence of ongoing adherence to prescribed medications. The strength of the signal in an EHR-based study may be reduced relative to randomized controlled trials that are characterized by strict medication adherence measurements such as pill counts. The fact that our findings are consistent with clinical trial data despite this limitation lends further support to the importance of clopidogrel pharmacogenomics as applied to clinical practice. Finally, because evidence of death status was obtained primarily through linkage to the Social Security Death Index, we were unable to assess cause of death and therefore could not ascertain the incidence of cardiovascular-related death.
In 2007, more than 25 million patients were prescribed clopidogrel.35 The demonstration of the relationship between recurrent cardiac events during clopidogrel therapy and common variants in CYP2C19*2 and ABCB1 as obtained from EHR data improves the generalizability of clinical trial results, highlighting the possibility that a large proportion of “real-world” clopidogrel users are at risk for treatment failure.36
The argument has been made that randomized clinical trials should be conducted to establish the optimal approach to therapy in patients with these variants; however, we believe that the totality of the available data to date make such a trial difficult to mount because of potential lack of clinical equipoise. In the absence of data from randomized clinical trials, we advocate personalized approaches to therapy. These may include platelet-function testing, early genotyping for patients likely to require clopidogrel, or point-of-care genotyping at the time of initiation of clopidogrel treatment.37
Methods
BioVU database and subject selection
Cases and controls were identified from the BioVU DNA biobank as described previously.23,24,27 In brief, BioVU links DNA extracted from blood samples (obtained during routine clinical care and about to be discarded) to a de-identified image of the Vanderbilt EHR, termed the Synthetic Derivative. Each DNA sample and associated de-identified health record is linked by a unique identifier generated by a one-way hash function.27 The resource has been and continues to be reviewed by the institutional review board; it has been considered as containing data for nonhuman subjects in accordance with the provisions of Title 45 of the Code of Federal Regulations part 46, as have the individual research studies utilizing the resource. Exclusion criteria include insufficient or poor-quality DNA, patient opt-out, and absence of signed consent to treat. In the Vanderbilt EHR, the race parameter is administratively assigned; we have previously shown that this is highly concordant with race defined by ancestry-informative markers.38
The selection of records for inclusion was a two-part process: an electronic algorithm was employed to identify possible cases and controls, followed by a manual review of all “possible” records by three physicians (Figure 2). Individuals who presented with an MI and/or intracoronary stent placement and who were subsequently placed on clopidogrel before discharge were identified through a combination of International Classification of Disease, 9th edition (ICD-9), and Current Procedural Terminology codes, laboratory values, medication orders, and natural-language processing in physician notes.
The primary outcome was defined as a composite of MI, revascularization, stroke, and/or all-cause mortality within a 2-year period starting 2 days after the initial event, while the patient was on clopidogrel treatment. Revascularization was defined as either intracoronary angioplasty or coronary bypass surgery. For the purpose of ascertaining all-cause mortality, all records in the Synthetic Derivative have been linked to the Social Security Death Index.26 In addition, in-hospital deaths are documented in the EHR and transferred into the Synthetic Derivative. Controls were defined as those with an absence of the primary outcome while on clopidogrel treatment for up to 2 years of observation after the primary event. In addition, we required all controls to have outpatient visits, with clinical documentation being available for at least 1 year after the primary event. The electronic algorithm was then used to assign the individual's status as possible case or possible control, and a manual review was employed to determine the validity of the status assignment. Covariates for age, gender, and body mass index were obtained through a combination of ICD-9 and Current Procedural Terminology codes, natural-language processing, and manual review.
Genotyping and data analysis
Prior to genotyping, the data for 114 subjects were removed from the study because of inadequate DNA concentration or opt-out from the DNA biobank. The Vanderbilt DNA Resources Core performed genotyping on 693 samples, using TaqMan (Applied Biosystems, Foster City, CA). Genotyping was performed for six SNPs: rs1045642, ABCB1; rs4244285, CYP2C19*2; rs4986893, CYP2C19*3; rs28399504, CYP2C19*4; rs12248560, CYP2C19*17; and rs662, PON1. Quality-control analysis was assessed with the PLINK, version 1.07, http://pngu.mgh.harvard.edu/∼purcell/plink/ and Plato software packages, version 1.1 https://chgr.mc.vanderbilt.edu/plato.39,40 SNP call rates were >95% for all six SNPs, with a concordance of >98% between duplicates and HapMap samples. One individual's sample was excluded because of poor genotyping efficiency. For all samples, the allele frequencies and tests of Hardy–Weinberg equilibrium were calculated. All SNPs were in Hardy–Weinberg equilibrium for European Americans, and all SNPs except rs662 were in Hardy–Weinberg equilibrium in African Americans. Four SNPs passed the quality-control criteria and were taken up for statistical analysis. The two SNPs excluded from further analysis were rs28399504 (CYP2C19*4), the minor allele of which was observed in only five Asian subjects, and rs4986893 (CYP2C19*3), which was monomorphic.
statistical analyses
Descriptive statistics were calculated for patient characteristics. Continuous variables were summarized as mean values and standard deviations, and categorical variables were expressed in percentages. The cumulative incidence of the primary event (stent thrombosis, revascularization, MI, or death) was estimated using the Kaplan–Meier product limit estimator, and unadjusted comparisons between target SNP–defined groups were compared using the log-rank test. Cox proportional hazards models, stratified by gender and race, were used to examine the effect of genotype on the primary event. Stratification was required because of violations to the proportional hazards assumption. We assumed an additive genetic model for rs1045642 (ABCB1), rs12248560 (CYP2C19*17), and rs662 (PON1). On the basis of information from the literature, we assumed a dominant genetic model for rs4244285 (CYP2C19*2) for the primary analysis.12,13 Prespecified covariates included age, gender, race (European American, African American, and other/unknown), BMI, and smoking status (current, ever, or never). BMI was imputed for 34 subjects in whom the values were missing, using race, height, and/or weight where available. Secondary end points were all-cause mortality, MI, revascularization, and stent thrombosis (definite and probable, as defined by ARC criteria).31 Statistical analyses were performed using the R programming language.
Supplementary Material
Acknowledgments
The Vanderbilt DNA Resources Core, which houses all the samples, performed the genotyping under the supervision of Cara Sutcliffe and Holli Dilks. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core, provided computational and analytical support for this work. This study was funded in part by RC2 GM092618 and U19 HL065962. The data sets used for the analyses described were obtained from Vanderbilt University Medical Center's BioVU, and the REDcap database, which is supported by institutional funding and by Vanderbilt CTSA grant 1UL1RR024975 from the National Center for Research Resources/National Institutes of Health (NCRR/NIH). The database used for this study was created with REDCap, a Web-based application, also supported by 1UL1RR024975 from NCRR/NIH.
Footnotes
Conflict of Interest: The authors declared no conflict of interest.
Supplementary Material is linked to the online version of the paper at http://www.nature.com/cpt
References
- 1.Chen ZM, et al. COMMIT (ClOpidogrel and Metoprolol in Myocardial Infarction Trial) collaborative group. Addition of clopidogrel to aspirin in 45,852 patients with acute myocardial infarction: randomised placebo-controlled trial. Lancet. 2005;366:1607–1621. doi: 10.1016/S0140-6736(05)67660-X. [DOI] [PubMed] [Google Scholar]
- 2.Sabatine MS, et al. CLARITY-TIMII 28 Investigators. Addition of clopidogrel to aspirin and fibrinolytic therapy for myocardial infarction with ST-segment elevation. N Engl J Med. 2005;352:1179–1189. doi: 10.1056/NEJMoa050522. [DOI] [PubMed] [Google Scholar]
- 3.Yusuf S, Zhao F, Mehta SR, Chrolavicius S, Tognoni G, Fox KK. Clopidogrel in Unstable Angina to Prevent Recurrent Events Trial Investigators. Efects of clopidogrel in addition to aspirin in patients with acute coronary syndromes without ST-segment elevation. N Engl J Med. 2001;345:494–502. doi: 10.1056/NEJMoa010746. [DOI] [PubMed] [Google Scholar]
- 4.A randomised, blinded, trial of clopidogrel versus aspirin in patients at risk of ischaemic events (CAPRIE).CAPRIE Steering Committee. Lancet. 1996;348:1329–1339. doi: 10.1016/s0140-6736(96)09457-3. [DOI] [PubMed] [Google Scholar]
- 5.Anderson JL, et al. ACC/AHA 2007 Guidelines for the Management of Patients With Unstable Angina/Non ST-Elevation Myocardial Infarction Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Writing Committee to Revise the 2002 Guidelines for the Management of Patients With Unstable Angina/Non ST-Elevation Myocardial Infarction) Developed in Collaboration with the American College of Emergency Physicians, the Society for Cardiovascular Angiography and Interventions, and the Society of Thoracic Surgeons Endorsed by the American Association of Cardiovascular and Pulmonary Rehabilitation and the Society for Academic Emergency Medicine. J Am Coll Cardiol. 2007;50:652–726. doi: 10.1016/j.jacc.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Kushner FG, et al. 2009 focused updates: ACC/AHA guidelines for the management of patients with ST-elevation myocardial infarction (updating the 2004 guideline and 2007 focused update) and ACC/AHA/SCAI guidelines on percutaneous coronary intervention (updating the 2005 guideline and 2007 focused update) a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2009;54:2205–2241. doi: 10.1016/j.jacc.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Kazui M, et al. Identifcation of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos. 2010;38:92–99. doi: 10.1124/dmd.109.029132. [DOI] [PubMed] [Google Scholar]
- 8.Hulot JS, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108:2244–2247. doi: 10.1182/blood-2006-04-013052. [DOI] [PubMed] [Google Scholar]
- 9.Trenk D, et al. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol. 2008;51:1925–1934. doi: 10.1016/j.jacc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 10.Harmsze A, et al. Besides CYP2C19*2, the variant allele CYP2C9*3 is associated with higher on-clopidogrel platelet reactivity in patients on dual antiplatelet therapy undergoing elective coronary stent implantation. Pharmacogenet. Genomics. 2010;20:18–25. doi: 10.1097/FPC.0b013e328333dafe. [DOI] [PubMed] [Google Scholar]
- 11.Simon T, et al. French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction (FAST-MI) Investigators. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 12.Collet JP, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373:309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 13.Sibbing D, et al. Cytochrome P450 2C19 loss-of-function polymorphism and stent thrombosis following percutaneous coronary intervention. Eur Heart J. 2009;30:916–922. doi: 10.1093/eurheartj/ehp041. [DOI] [PubMed] [Google Scholar]
- 14.Mega JL, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 15.Hulot JS, et al. Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration: a systematic meta-analysis. J Am Coll Cardiol. 2010;56:134–143. doi: 10.1016/j.jacc.2009.12.071. [DOI] [PubMed] [Google Scholar]
- 16.Tiroch KA, et al. Protective effect of the CYP2C19*17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am Heart J. 2010;160:506–512. doi: 10.1016/j.ahj.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 17.Sibbing D, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–518. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 18.Taubert D, et al. Impact of P-glycoprotein on clopidogrel absorption. Clin Pharmacol Ther. 2006;80:486–501. doi: 10.1016/j.clpt.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Mega JL, et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet. 2010;376:1312–1319. doi: 10.1016/S0140-6736(10)61273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiewak M, et al. Infuence of C3435T multidrug resistance gene-1 (MDR-1) polymorphism on platelet reactivity and prognosis in patients with acute coronary syndromes. Kardiol Pol. 2009;67:827–834. [PubMed] [Google Scholar]
- 21.Bouman HJ, et al. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat Med. 2011;17:110–116. doi: 10.1038/nm.2281. [DOI] [PubMed] [Google Scholar]
- 22.Sibbing D, et al. No association of paraoxonase-1 Q192R genotypes with platelet response to clopidogrel and risk of stent thrombosis after coronary stenting. Eur Heart J. 2011;32:1605–1613. doi: 10.1093/eurheartj/ehr155. [DOI] [PubMed] [Google Scholar]
- 23.Denny JC, et al. Identifcation of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie MD, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS ONE. 2010;5:e13011. doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denny JC, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roden DM, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu H, et al. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J Am Med Inform Assoc. 2011;18:387–391. doi: 10.1136/amiajnl-2011-000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harmsze AM, et al. CYP2C19*2 and CYP2C9*3 alleles are associated with stent thrombosis: a case-control study. Eur Heart J. 2010;31:3046–3053. doi: 10.1093/eurheartj/ehq321. [DOI] [PubMed] [Google Scholar]
- 30.Shuldiner AR, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efcacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cutlip DE, et al. Academic Research Consortium. Clinical end points in coronary stent trials: a case for standardized definitions. Circulation. 2007;115:2344–2351. doi: 10.1161/CIRCULATIONAHA.106.685313. [DOI] [PubMed] [Google Scholar]
- 32.Holmes DR, et al. Stent thrombosis. J Am Coll Cardiol. 2010;56:1357–1365. doi: 10.1016/j.jacc.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharyya T, et al. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA. 2008;299:1265–1276. doi: 10.1001/jama.299.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wheeler JG, Keavney BD, Watkins H, Collins R, Danesh J. Four paraoxonase gene polymorphisms in 11212 cases of coronary heart disease and 12786 controls: meta-analysis of 43 studies. Lancet. 2004;363:689–695. doi: 10.1016/S0140-6736(04)15642-0. [DOI] [PubMed] [Google Scholar]
- 35.Ellis KJ, Stoufer GA, McLeod HL, Lee CR. Clopidogrel pharmacogenomics and risk of inadequate platelet inhibition: US FDA recommendations. Pharmacogenomics. 2009;10:1799–1817. doi: 10.2217/pgs.09.143. [DOI] [PubMed] [Google Scholar]
- 36.Gurbel PA, Bliden KP, Hiatt BL, O'Connor CM. Clopidogrel for coronary stenting: response variability, drug resistance, and the effect of pretreatment platelet reactivity. Circulation. 2003;107:2908–2913. doi: 10.1161/01.CIR.0000072771.11429.83. [DOI] [PubMed] [Google Scholar]
- 37.Gurbel PA, Tantry US, Shuldiner AR, Kereiakes DJ. Genotyping: one piece of the puzzle to personalize antiplatelet therapy. J Am Coll Cardiol. 2010;56:112–116. doi: 10.1016/j.jacc.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Dumitrescu L, et al. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet Med. 2010;12:648–650. doi: 10.1097/GIM.0b013e3181efe2df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grady BJ, Torstenson E, Dudek SM, Giles J, Sexton D, Ritchie MD. Finding unique filter sets in plato: a precursor to efficient interaction analysis in gwas data. Pac Symp Biocomput. 2010:315–326. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


