Abstract
The most significant recent advance in biomedical research has been the discovery of the~22-nt long class of non-coding RNAs designated as microRNAs (miRNAs). These regulatory RNAs provide a unique level of post-transcriptional gene regulation that modulates a range of fundamental cellular processes. Several viruses, including especially herpesviruses, also encode miRNAs and over 200 viral miRNAs have now been identified. Current evidence indicates that viruses use these miRNAs to manipulate both cellular and viral gene expression. Furthermore, viral infection can exert a profound impact on the cellular miRNA expression profile, and several RNA viruses have been reported to interact directly with cellular miRNAs and/or to use these miRNAs to augment their replication potential. Here we discuss our current knowledge of viral miRNAs and virally-influenced cellular miRNAs, and their relationship to viral infection.
Keywords: miRNAs, herpesvirus, latency, viral miRNA targets, host-pathogen interactions
INTRODUCTION
MicroRNAs (miRNAs) are ~19–24 nt non-coding RNAs that post-transcriptionally regulate gene expression by binding to target messenger RNAs (mRNAs). First identified in Caenorhabditis elegans, miRNAs are expressed by all metazoans and plants, as well as by several DNA viruses, and function as regulators of cellular processes such as development, differentiation, growth, homeostasis, stress responses, apoptosis and immune activation (6, 103). To date, >10,000 miRNAs have been annotated in 96 species, including over 700 human miRNAs (miRBase v14.0) (43). Remarkably, >45,000 miRNA target sites are computationally predicted in the 3′ untranslated regions (3′UTRs) of human mRNAs, indicating that miRNAs regulate >60% of all human protein-coding genes (35). Single miRNAs can potentially target >300 different transcripts (6, 35), thus illustrating the impact miRNAs can have on patterns of gene expression.
microRNA biogenesis
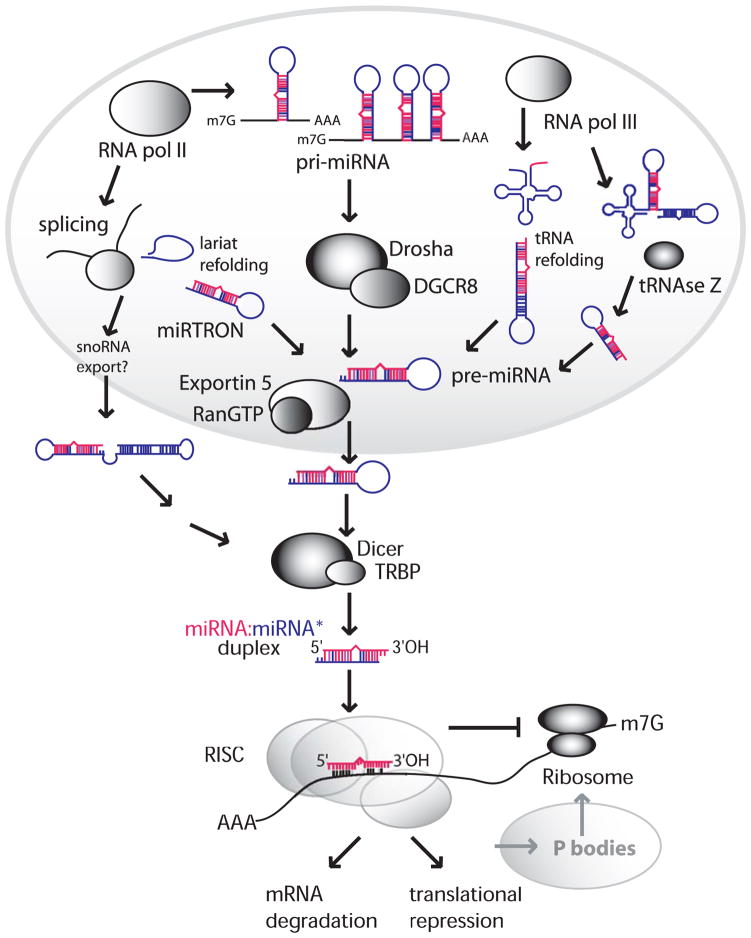
Canonical miRNA biogenesis (Fig. 1) initiates with the nuclear transcription of long primary miRNAs (pri-miRNAs) by RNA polymerase II (Pol II) (reviewed in 27). Pri-miRNAs contain a 5′ cap, are polyadenylated, and fold to produce one or more ~80-nt hairpin structures, each consisting of a ~32 bp imperfect stem and large terminal loop. These stem-loops are recognized by the RNase III enzyme Drosha, together with its co-factor DGCR8, which cleave ~22 bp down the stem to yield ~60 nt precursor miRNAs (pre-miRNA) containing 2 nt 3′ overhangs (27, 110). Exportin 5 transports these pre-miRNAs to the cytoplasm, where the terminal loops are removed by Dicer, a second RNase III enzyme, acting in association with TRBP. This produces ~22 bp miRNA duplex intermediates bearing 2 nt 3′ overhangs at each end (24, 50).
Figure 1. miRNA biogenesis pathways.
Pre-miRNAs can be generated (i) by Drosha/DGCR8 cleavage of long pri-miRNAs, or independent of Drosha/DGCR8, (ii) following debranching of lariat-structures known as miRtrons, (iii) through alternative folding of tRNAs or snoRNAs, or (iv) by tRNAse Z cleavage of pri-miRNAs containing tRNA-like structures linked to pre-miRNA stem-loops. Once exported to the cytoplasm, pre-miRNAs are cleaved by Dicer to generate a miRNA duplex, one strand of which is incorporated into RISC to target mRNAs.
One strand of the miRNA duplex is incorporated into the RNA-induced silencing complex (RISC) to function as a mature miRNA and guide RISC to target mRNAs, while the passenger strand, termed miRNA*, is degraded. Strand selection depends on the degree of base pairing at the duplex 5′ ends—the strand less stably base paired at its 5′ end is preferentially incorporated into RISC (91). In humans, RISC minimally consists of the mature miRNA and one of four different Argonaute proteins (Ago1-4). Only Ago2 exhibits endonuclease activity and has the ability to cleave bound target mRNAs (57, 64).
microRNA-directed mRNA silencing
Mature miRNAs typically bind to complementary sequences found in the 3′ UTRs of target mRNAs, and can repress translation and/or induce mRNA degradation. Important for this targeting are 5′ nucleotides 2–7 of the mature miRNA, termed the “seed” (6). Compensatory binding to the miRNA 3′ end has been demonstrated for some active target sites that show imperfect seed binding (6). The fate of a targeted mRNA is dependent on the degree of complementarity. Perfect complementarity, observed often in plants, generally results in endonucleolytic cleavage of the mRNA. Imperfect complementarity, observed for most mammalian and viral miRNA targets, results in translational repression, which can then lead to mRNA destabilization (5, 80). RISC-bound mRNAs often localize to cytoplasmic processing bodies (P bodies), which exclude the translational machinery and contain proteins involved in mRNA remodeling, decapping, and deadenylation, as well as exonucleases (8, 34). Additional P body components, such as GW182 and the RNA helicase RCK/p54, have been reported to play critical roles in miRNA-mediated repression. P bodies themselves, however, are not necessary for translational silencing by miRNAs (34).
VIRALLY-ENCODED MICRORNAS
miRNAs are potentially ideal tools for viruses to modulate gene expression. In contrast to viral proteins, miRNAs are non-immunogenic, require less coding capacity, and can evolve rapidly to target new transcripts. Point mutations in the miRNA seed region can alter target specificity while mutations within the pre-miRNA might affect strand-loading into RISC. Additionally, miRNAs not only have the capability of targeting mRNAs with high specificity but can also regulate multiple transcripts to varying degrees. By taking advantage of a conserved gene regulatory mechanism within the host cell, viral miRNAs can help establish a cellular environment conducive to viral replication.
Given these unique attributes, it is not surprising that a number of DNA viruses encode miRNAs. To date, >200 viral miRNAs have been identified, predominantly in herpesviruses, but also in polyomaviruses, ascoviruses, and adenoviruses (Table 1). miRNAs have several features that may have led these particular nuclear DNA viruses to evolve viral miRNAs. First, these viruses have access to the nuclear Drosha and DGCR8 pri-miRNA processing factors. In fact, viral miRNA biogenesis appears to be mediated solely by cellular factors as no viral proteins involved in miRNA processing have been described so far. Second, these dsDNA viruses exhibit bi-directional transcription, and the sequence specific regulation of viral transcripts is therefore easily achieved by expressing antisense miRNAs. Finally, the unique ability of DNA viruses, particularly herpesviruses, to establish long-term, latent infections means that these viruses need to block protective host innate or adaptive immune responses over the long term while minimizing the expression of potentially antigenic viral proteins.
Table 1.
Viral pre-miRNAs
| Virus family | Virus | Host | # pre-miRNAs | Reference |
|---|---|---|---|---|
| α-herpesviruses | HSV-1 | Human | 8 | (26, 95, 96) |
| HSV-2 | Human | 6 | (88, 89, 97) | |
| MDV-1 | Avian | 14 | (17, 108) | |
| MDV-2 | Avian | 18 | (98, 107) | |
| HVT | Avian | 17 | (98) | |
| ILTV | Avian | 7 | (98) | |
| β-herpesviruses | hCMV | Human | 11 | (32, 40, 72) |
| mCMV | Mouse | 18 | (16, 31) | |
| γ-herpesviruses | EBV | Human | 25 | (20, 73, 111) |
| rLCV | Primate | 32 | (20, 99) | |
| KSHV | Human | 12 | (19, 44, 72, 76) | |
| RRV | Primate | 15 | (79) | |
| MHV-68 | Mouse | 9 | (72) | |
| Polyomaviruses | multiple | Human, Primate, Mouse | 1 | (22, 81, 82, 86, 87) |
| Adenoviruses | hAV | Human | 2 | (2, 3, 78, 105) |
| Ascoviruses | HvAV | Insect | 1 | (49) |
Herpesviruses
Herpesviruses are large, enveloped dsDNA viruses that are classified into three subfamilies (α, β, and γ) based on genome sequence and virus biology. α-herpesviruses, such as human herpes simplex virus 1 (HSV-1), establish latent infections in neurons of the sensory ganglia and, upon reactivation, cause lesions on adjacent mucosal surfaces. Human cytomegalovirus (hCMV), a β-herpesvirus, is the leading cause of congenital birth defects and establishes latency primarily in hematopoietic progenitor cells. Human γ-herpesviruses establish latent infections primarily in B cells and are linked to several human malignancies. Epstein-Barr virus (EBV), for example, is associated with Hodgkin’s and Burkitt’s lymphomas, and nasopharyngeal carcinomas (NPC) while Kaposi’s sarcoma-associated herpesvirus (KSHV) is the etiologic agent of primary effusion lymphoma (PEL) and Kaposi’s sarcoma (KS).
Expression of EBV and KSHV microRNAs
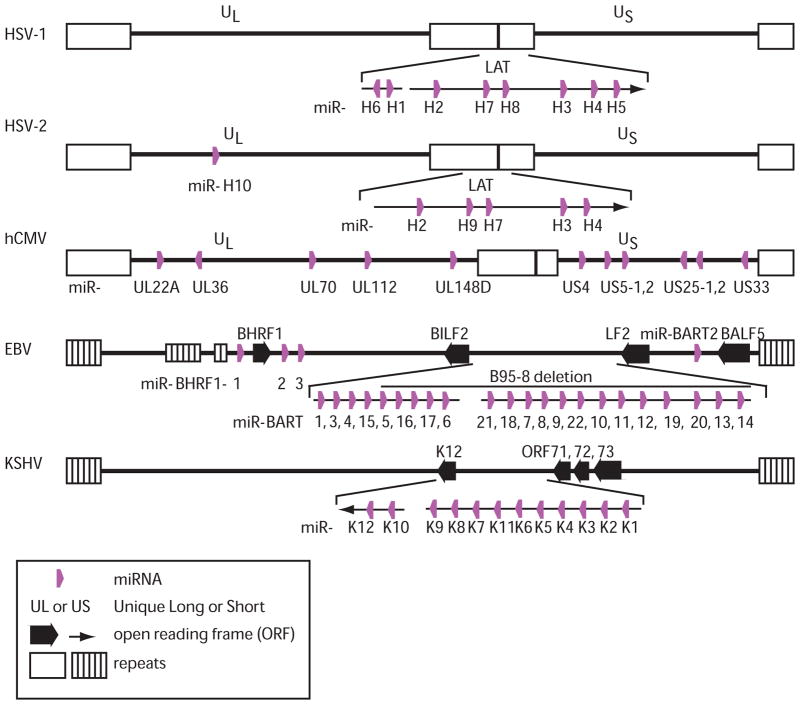
In 2004, Pfeffer and colleagues identified the first virus-encoded miRNAs expressed in B cells latently infected with EBV (73). A total of 25 EBV pre-miRNAs (Table 1) have since been reported in B cells and NPCs (20, 44, 111). Three EBV BHRF1 pre-miRNAs reside adjacent to the bhrf1 gene, and are derived from an intron generated through splicing of viral EBNA transcripts that are exclusively expressed in latency III EBV-infected B cells (104) (Fig. 2). In contrast, the 22 BART pre-miRNAs reside within two clusters in introns found in the viral BART transcripts. BART miRNAs are expressed primarily in latency II EBV-infected epithelial cells, although they are detectable at low levels in infected B cells (20, 44, 111). Strikingly, the majority of EBV miRNAs are deleted in the common B95-8 EBV laboratory isolate, yet B95-8 still immortalizes primary B cells in culture (20, 44). This indicates that most of the BART miRNAs are not required for EBV latency in B cells, although they may still play a key role in infected epithelial cells.
Figure 2. Locations of the viral pre-miRNAs encoded by human herpesviruses.
Both α and γ herpesvirus pre-miRNAs primarily reside in latency-associated clusters (HSV-1, HSV-2, EBV, and KSHV) while hCMV miRNAs are scattered throughout the genome.
The second human γ-herpesvirus, KSHV, encodes 12 pre-miRNAs which are clustered in the latency-associated region and highly expressed in latently infected PEL cells. Ten pre-miRNAs (miR-K1 through K9 and miR-K11) are clustered within a viral intron located between ORF71 and kaposin; the remaining two miRNAs reside within the coding region (miR-K10) or the 3′UTR (miR-K12) of the KSHV K12 gene (44, 72, 76) (Fig. 2). Pri-miRNA expression is regulated by three RNA Pol II promoters, one latent and the other two lytic (20, 106). Deletion of the ten intronic KSHV miRNAs does not inhibit lytic viral replication (106). Conversely, induction of lytic KSHV replication does not enhance expression of these intronic KSHV miRNAs, implying a primary function during latency (20, 38, 72, 76). In contrast, miR-K10 and miR-K12 are both upregulated in response to lytic induction; however, their biological role during any productive KSHV replication remains undetermined (19, 44). Of note, pre- miR-K10 is partly modified by a cellular adenosine deaminase acting on RNA (ADAR), which edits adenosine to inosine at nt 2 of the miRNA seed. While this is predicted to change the mRNA target specificity of miR-K10 (37, 72, 94), the functional consequences of miR-K10 editing remain unclear.
MHV-68 and non-canonical microRNA processing
Murine herpesvirus-68 (MHV-68) shares a similar genome structure with KSHV, and provides a small animal model to study γ-herpesvirus biology. MHV-68 encodes seven pri-miRNAs clustered within a 6 kb region (72). Uniquely, all MHV-68 pri-miRNAs are transcribed by RNA Pol III from internal tRNA promoters (12, 29, 72). The resultant ~130–200 nt long viral pri-miRNAs each consist of a ~60 nt 5′ tRNA moiety connected to one or two ~70 nt 3′ stem-loops (Fig. 1). These pri-miRNAs are cleaved by tRNAseZ, not by Drosha, to release the viral pre-miRNAs which are then processed by Dicer into at least nine mature miRNAs (12, 29, 72).
Interestingly, several cellular miRNAs also deviate from the canonical biogenesis pathway and are able to bypass the need for Drosha cleavage prior to nuclear export (Fig. 1) (4, 68, 75). For example, miRNAs arising from miRtrons, ~70-nt lariat-derived RNAs that mimic pre-miRNA hairpins after debranching (10), have recently been described. Some snoRNA and tRNA precursors can adopt alternative structures that also mimic pre-miRNAs and give rise to functional miRNAs after Dicer processing (4, 33). Thus, while there are several minor pathways available to generate pre-miRNAs independent of Drosha cleavage, there are currently no known miRNAs that do not require Dicer processing.
Most Herpes Simplex Virus 1 and 2 microRNAs are latency-associated
HSV-1 and HSV-2 primarily establish latency in sensory neurons of the trigeminal or sacral ganglia, respectively. During latency, viral gene expression is largely restricted to the non-coding latency associated transcripts (LATs). Deep sequencing of miRNAs expressed in latently HSV-1 infected mouse and human trigeminal ganglia revealed seven HSV-1 miRNAs, six of which are encoded in LAT (95, 96) (Fig. 2). One additional HSV-1 miRNA, miR-H1, is encoded 5′ of LAT and expressed in cells undergoing productive replication (26). Five LAT-associated HSV-2 miRNAs were identified in cells expressing a HSV-2 LAT construct and/or by deep sequencing of HSV-2 latently infected human sacral ganglia (88, 89, 97). Marek’s disease virus (MDV) types 1 and 2, which are avian α-herpesviruses, also encode miRNAs that are clustered in the viral LAT regions and expressed in latently infected cells (17, 98, 107). Thus α-herpesvirus miRNAs are thought to play a major role in facilitating stable latent infections.
Cytomegalovirus miRNAs are expressed during productive infection
In contrast to α and γ herpesvirus miRNAs, hCMV miRNAs were initially isolated from primary fibroblasts undergoing lytic replication (32, 40, 72). Furthermore, hCMV miRNAs are scattered across the genome in small clusters containing three or fewer miRNAs (Fig. 2). At least five are located within intergenic regions, while one is located within an intron (32, 40, 72). All 11 hCMV pre-miRNAs display expression kinetics of immediate early or early genes, indicating a role during the lytic replication cycle (40). Due to the paucity of tractable in vitro latency models for hCMV, it currently remains unclear whether any hCMV miRNAs are expressed during latency.
Conservation of herpesvirus miRNAs
Unlike their cellular counterparts, most herpesvirus miRNAs show little sequence similarity to other viral or cellular miRNAs. For example, Rhesus monkey rhadinovirus (RRV), a relative of KSHV, encodes 15 pre-miRNAs expressed from the latency-associated region (Umbach et al., unpublished observations, 79). While the genomic location of the 15 RRV and 12 KSHV miRNAs adjacent to ORF71 is conserved, there are no sequence similarities (79). Known γ-herpesvirus miRNA sequence conservation is currently limited to the more closely related γ-herpesviruses EBV and rhesus lymphocryptovirus (rLCV). 22 of the 25 EBV pre-miRNAs exhibit some degree of conservation with rLCV; however, several of the miRNA seed regions differ, suggesting evolutionary relatedness but divergent functions (20, 99). Similarly, HSV-1 and HSV-2 miRNAs are highly conserved in their genomic location but only partially conserved at the sequence level. For example, HSV-1 and HSV-2 miR-H3 exhibit 85% sequence identity yet lack seed conservation. In contrast, HSV-1 and HSV-2 miR-H2 do share an identical seed (96, 97), suggesting these miRNAs might share common targets. However, MDV-1 and MDV-2 miRNAs, which diverged ~26 million years ago, show no apparent sequence conservation (17, 107). Despite sequence differences, the fact that the genomic locations of most α and γ herpesvirus miRNAs are conserved (19, 20, 44, 76, 88, 89, 95–97) indicates that viral pre-miRNAs are subject to ongoing evolutionary selection. Furthermore, viral miRNAs may share conserved functions even when they lack sequence homology, i.e. they might target different sites on the same mRNA or even different components of the same cellular pathway.
Intriguingly, a handful of herpesvirus miRNAs exhibit homology to cellular miRNAs. KSHV miR-K11 and miR-155, for example, share identical seeds and, consequently, regulate an overlapping set of cellular transcripts (39, 83). Dysregulated miR-155 expression in vivo is linked to hematopoetic malignancies as well as alterations in lymphocyte development and innate and adaptive immune responses (74, 90, 103). Interestingly, the oncogenic MDV-1 also encodes a miR-155 ortholog (17). Furthermore, EBV induces the expression of cellular miR-155 in latency III B cells (109). Reticuloendotheliosis virus strain T (REV-T), an oncogenic chicken retrovirus, also induces miR-155 expression (13). Accordingly, the enhanced expression of cellular miR-155 or viral miR-155 orthologs may confer a selective advantage on these viruses. Given the role of miR-155 in many malignancies (103), the exploitation of existing miR-155 regulated pathways by viruses may contribute to viral oncogenesis.
Seed sequence homology to cellular miRNAs is observed for several other herpesvirus miRNAs. EBV miR-BART5, rLCV miR-rL1-8, and MHV-68 miR-M1-7-5p share seed homology with cellular miR-18a/b, a member of the miR-17-92 cluster which is often amplified in B cell lymphomas (20, 72, 83). MHV-68 miR-M1-4 shares seed homology with miR-151-5p, a discriminator of T cell lineages (72, 103). EBV-BART1-3p, rLCV miR-rL1-6-5p, and MDV-2 miR-M21 share seed homology with miR-29abc, a conserved miRNA with roles in apoptosis (20, 72, 98). Finally, HVT miR-H14-3p and chicken miR-221 share 21 out of 23 nucleotides and similarities in the pre-miRNA flanking region, indicating that this viral miRNA may have been captured from its host genome (98). The miR-221/miR-222 seed is also shared by MDV-1 miR-M32 (17). Future deep sequencing efforts to identify new viral and cellular miRNAs are likely to identify additional viral orthologs of cellular miRNAs.
microRNAs encoded by other viruses
In addition to herpesviruses, some other dsDNA viruses also encode miRNAs. Adenoviruses express two structured ~160 nt RNA Pol III transcripts (VAI and VAII) that are produced at high levels (>10^8 copies/cell) during infection. While the VA RNAs function primarily as inhibitors of the host innate immunity factor PKR, they also reduce miRNA biogenesis by inhibiting Dicer. Nevertheless, both are processed at low efficiency by Dicer into RISC-associated miRNAs (2, 3, 59, 105). To date, no target genes for these VA-derived miRNAs have been identified, and it remains unclear whether they have a physiologically relevant function.
Heliothis virescens ascovirus (HvAV), an insect dsDNA virus, encodes a miRNA which targets the viral DNA polymerase mRNA at late stages of infection (49). As a consequence, viral replication is down-regulated. Interestingly, Hz-miR-24, an insect cellular miRNA, is induced during HvAv infection and has been reported to interact directly with viral mRNAs, downregulating both the ascovirus RNA polymerase and β-subunit (48). Thus, insect DNA viruses also appear to usurp the cellular miRNA machinery to regulate their life cycles.
Finally, simian virus 40 (SV40), a primate polyomavirus, encodes a single pre-miRNA, miR-S1, which is expressed late in infection (86). Polyomaviruses infect a range of vertebrate hosts and can induce tumorigenesis. Human polyomavirus infections are generally asymptomatic but persist life-long and can be life-threatening in immunosuppressed patients. Both the genomic location of miR-S1 and its temporal expression are conserved amongst polyomaviruses, including human BKV, JCV, and Merkel cell virus (MCV), the primate virus SA12, and mouse polyomavirus (mPyV) (22, 81, 82, 87).
No Papillomavirus, Poxvirus, or RNA virus miRNAs are currently known
Several other viruses have been interrogated for viral miRNAs. Some of these viruses are predicted to form RNA structures that are conceivable Drosha or Dicer substrates. The 59-nt HIV-1 TAR RNA, for instance, forms a stem-loop structure similar to a pre-miRNA. Two groups have reported in vitro Dicer processing of TAR (53, 70), although no HIV-1 miRNAs have been identified in infected cells using direct cloning methods (56, 72). To date, no viral miRNAs have been identified in human papilloma virus (HPV), hepatitis C virus (HCV), yellow fever virus or human T-cell leukemia virus I infected cells using standard sequencing (18, 56, 72) or in HPV, coxpox virus, dengue virus or influenza virus infected cells using deep sequencing techniques (Skalsky, Umbach, and Cullen, unpublished results, 60), However, several of these viruses replicate exclusively in the cytoplasm and may lack access to nuclear Drosha. Furthermore, excision of a miRNA from a nuclear RNA virus, such as HIV-1, would mean the cleavage and, ultimately, destruction of the viral genomic RNA. Presumably, this confers a strong selection against the evolution of miRNAs in nuclear RNA viruses, although it remains possible that examples will be discovered in the future.
FUNCTIONS OF VIRAL MICRORNAS
While defined roles for viral miRNAs are just beginning to emerge, it is clear that viral miRNAs can target both viral and cellular transcripts. Viral miRNAs, like other viral factors, are involved in cellular reprogramming in order to (i) regulate the latent-lytic switch, (ii) support viral replication by promoting cell survival, proliferation, and/or differentiation, and (iii) modulate immune responses. Modulation of the host cell environment is achieved by multiple and partly redundant mechanisms as viral miRNAs and proteins work synergistically to promote a cellular environment favorable to completion of the viral life cycle.
A number of challenges exist in discerning viral miRNA targets. First, the potential requirement of only an ~7-nt seed match in a 3′UTR immediately creates a large pool of possible targets. Moreover, while many miRNA targets do feature full seed complementarity, 3′ compensatory binding can also occur (6). Some functional target sites also lack both perfect seed pairing and 3′ compensatory binding (6, 35). In addition to 3′UTRs, miRNAs can also target mRNA coding sequences (6, 95). Furthermore, different viral miRNAs can bind to multiple target sites on a single transcript, as has been suggested for the KSHV miRNAs (46, 77). Thus, the inhibitory effect of a single viral miRNA on a single mRNA target site may not be easily discerned unless additional miRNAs and their target sites in the same mRNA are also considered.
Viral targets of viral miRNAs
Identifying viral targets of viral miRNAs is more straightforward than identifying cellular targets simply because viral genomes encode fewer candidate mRNAs. Additionally, several viral miRNAs lie antisense to viral transcripts, which are obvious potential targets. The SV40 miRNAs, for example, lie antisense to the viral T-antigen mRNAs and mediate their cleavage late in infection (86). Disruption of miR-S1 activity not only increases T-antigen levels but also enhances the susceptibility of infected cells to killing by T-antigen-specific cytotoxic T cells in culture (86). While the sequences of polyomavirus miRNAs differ, the pre-miRNA location and hence, antisense viral mRNA target is conserved in different family members (22, 81, 82), indicating that these miRNAs confer a selective advantage to polyomaviruses through T-antigen downregulation. Furthermore, given their lack of sequence homology, it is unlikely that polyomavirus miRNAs target the same cellular mRNAs (82).
To examine the effect of polyomavirus miRNAs in vivo, Sullivan et al. (87) deleted the pre-miRNA from mouse polyomavirus (mPyV). Surprisingly, this knockout virus replicated indistinguishably from wild type in immunocompetent mice. Additionally, no differences in immune responses were observed, leading the authors to hypothesize that the effects of viral miRNA targeting might occur outside the limits of the experimental system, such as during natural virus transmission.
EBV miR-BART2 lies antisense to the viral DNA polymerase (BALF5) (72). During productive EBV replication, the BALF5 mRNA is cleaved (36) due to miR-BART2 activity (7). Interestingly, miR-BART2 is expressed during latent infection; however, BALF5 is a lytic gene. Induction of lytic replication reduces miR-BART2 levels and miR-BART2-mediated cleavage (7). Hence, miR-BART2 may regulate the latent-lytic switch by preventing premature BALF5 expression.
hCMV miR-UL112-1 lies antisense to UL114, the viral DNA glycosylase. Studies in the Nelson lab suggest that UL114 RNA is resistant to miR-UL112-1 directed cleavage (42); however, UL114 protein levels may be reduced with little apparent consequence to viral replication (85). At least two other hCMV miRNAs, miR-UL148D and miR-US33, lie anti-sense to viral transcripts, and might also target their complementary genes (US29 and UL150) (40, 72).
HSV-1 and HSV-2 use viral miRNAs to downregulate two immediate early gene products, ICP0 and ICP34.5 (88, 89, 95). HSV-1 and HSV-2 miR-H2 are both transcribed antisense to ICP0 and downregulate ICP0 protein expression (89, 95, 97). ICP34.5 is situated antisense to two HSV-1 and HSV-2 miRNAs, miR-H3 and miR-H4, and targeting of ICP34.5 has been confirmed for HSV-2 miR-H3 (88). In addition to ICP0 and ICP34.5, ICP4 is targeted by HSV-1 miR-H6. While not antisense to ICP4, HSV-1 miR-H6 exhibits extensive sequence complementarity to a site within the ICP4 mRNA, and inhibits ICP4 protein expression (95). Interestingly, a chicken α-herpesvirus, infectious laryngotracheitis virus (ILTV) encodes two miRNAs that are situated anti-sense to ILTV ICP4, and may also downregulate ICP4 expression (98). ICP0 and ICP4 are both immediate early viral transactivators with key roles in the induction of lytic replication. The targeting of these transcripts by viral miRNAs is thought to mediate entry into latency and render the latent state more robust (93, 95). ICP34.5, on the other hand, is a viral pathogenicity factor which inhibits PKR activity and contributes to neurovirulence (reviewed in 28). Consequently, ICP34.5 repression via viral miRNAs may protect infected neurons from cytotoxicity (28, 88).
The targeting of viral transactivators is an emerging theme for viral miRNAs. hCMV miR-UL112-1 targets the IE72/IE1 transactivator, which contains two miR-UL112-1 seed-match sites in its 3′UTR (41, 65). Introducing a miR-UL112-1 mimic prior to hCMV infection attenuates productive hCMV replication, indicating that delayed expression of miR-UL112-1 is necessary for optimal viral replication (41, 42). Additionally, KSHV miR-K9* targets RTA (ORF50), the key KSHV protein involved in the activation of lytic viral gene expression, and targeting of RTA by miR-K9* has been proposed to prevent premature entry into the lytic cycle (9).
Finally, EBV LMP1 is targeted by three EBV miRNAs, miR-BART1-5p, miR-BART16, and miR-BART17-5p (58). The 3′ UTR of LMP1 is highly conserved in NPC virus isolates. LMP1 is a constitutively active viral mimic of the tumor necrosis factor receptor (TNFR) family and induces cell proliferation during latency (58). However, when over-expressed, LMP1 can inhibit growth and stimulate apoptosis. Thus, fine-tuning LMP1 expression levels by EBV miRNAs may promote the proliferation of latently infected cells.
Cellular targets of viral microRNAs
Viral miRNAs are now known to target several cellular genes involved in cell proliferation and survival, stress responses, and antiviral defense pathways. In the strictest sense, a virus needs to keep a host cell alive long enough to complete its life cycle. This time period is greatly extended for viruses which establish latent infections. Thus, prolonging cell survival and evading immune recognition are at least two ways in which viral miRNAs can promote virus replication.
To date, the most fully characterized cellular targets of viral miRNAs are those of the KSHV miRNAs. Thrombospondin 1 (THBS1), identified through gene-expression profiling of cells engineered to stably express 10 KSHV pre-miRNAs (77), is targeted by multiple KSHV miRNAs. One function of THBS1 is to inhibit angiogenesis and cell growth by activating TGFβ. THBS1 expression was reduced at both the mRNA and protein level in KSHV miRNA-expressing cells, which may promote cell survival and proliferation. In line with these findings, KS tumors exhibit reduced THBS1 activity (77).
Other regulators of cell survival and growth are also targeted by viral miRNAs. BCLAF1, a protein involved in apoptosis, was identified as a target of KSHV miR-K5 in both B cells and endothelial cells (112). Intriguingly, siRNA inhibition of BCLAF1 resulted in an increase in lytic KSHV replication, suggesting that modulation of BCLAF1 by KSHV miRNAs might promote the reversibility of latent infection (112). PUMA, a p53-regulated pro-apoptotic Bcl2-family member triggered by stress, is targeted by EBV miR-BART5. In EBV-infected NPCs, inhibition of miR-BART5 leads to an increase in PUMA-mediated apoptosis (25). One function of miR-BART5, then, may be to rescue EBV infected epithelial cells from apoptotic removal initiated by innate cellular defense mechanisms. Finally, MAF, a bZIP transcription factor involved in terminal differentiation of many cell types, is specifically targeted by KSHV miR-K11 and miR-K6 in endothelial cells. Viral miRNA-mediated inhibition of MAF results in lymphatic endothelial cell transcriptional reprogramming. As such, KSHV miRNAs can regulate the differentiation of infected endothelial cells, which may contribute to oncogenesis (46).
KSHV miR-K11 is an ortholog of miR-155 (39, 83). One shared cellular target of these miRNAs is BACH1, a transcriptional repressor involved in regulating oxidative stress (39, 83). BACH1 binds NF-E2 sites on DNA and coordinates with MAF proteins to repress transcription of heme oxygenase 1 (HO-1); thus, miR-K11/miR-155 targeting of BACH1 is predicted to enhance HO-1 activity. While the consequences of BACH1 targeting during KSHV infection remain unclear, HO-1 upregulation can enhance the survival of endothelial cells infected de novo with KSHV (63).
A few viral miRNAs directly target antiviral signaling molecules. EBV miR-BHRF1-3 downregulates CXC-chemokine ligand 11 (CXCL11), an IFN-inducible T- cell chemoattractant (102). T cell immunity plays a major role in host defenses against EBV, as EBV-infected B cells are efficiently targeted by cytotoxic T cells. Hence, suppressing CXCL11 may aid infected cells in avoiding T-cell recognition.
Finally, the MICB (major histocompatibility complex class I-related chain B) 3′UTR is selectively targeted at three non-overlapping sites by hCMV miR-UL112-1, KSHV miR-K7, and EBV miR-BART2 (66, 84). MICB, which is upregulated in response to oncogenic stress and viral infection, is a ligand for NK cells and CD8+ T cells. Viral miRNA-mediated MICB downregulation leads to a reduction in NK cell mediated killing of infected cells (66, 84). Interestingly, hCMV miR-UL112-1 also targets the viral IE72 transcript (41, 65), making it the first viral miRNA known to target both viral and cellular mRNAs. Additionally, MICB is the first gene to be targeted by multiple herpesvirus miRNAs with no apparent sequence homology, and therefore exemplifies the possibility of convergent evolution by herpesvirus miRNAs.
MICRORNAS INFLUENCED BY VIRAL INFECTION
Cellular miRNA expression is profoundly influenced by viral infection, which can be attributed to both host antiviral defenses and viral factors altering the cellular environment. For example, in addition to miR-155, EBV induces miR-146a expression in B cells (21). Both miR-155 and miR-146a are also induced following bacterial lipopolysaccharide (LPS) stimulation of monocytes. Interesting, miR-146a can target LPS-activated components, TRAF6 and IRAK1, of the TLR signaling pathway, suggesting a negative feedback loop to limit innate immune responses (reviewed in 103). During latency III, EBV LMP1 activates the miR-146a promoter, leading to a miR-146a-induced reduction in the expression of several interferon-responsive genes (21). LMP1 also induces miR-29b, which results in miR-29b-mediated downregulation of T-cell leukemia gene 1 (TCL1), a protein with roles in cell survival and proliferation (1). Notably, LMP1 is targeted by EBV-encoded miRNAs (58). Thus, EBV may coordinate viral and cellular miRNAs in order to alter anti-viral interferon signaling as well as extend the persistence of infected cells.
Oncogenic human papillomaviruses encode two viral proteins, E6 and E7, which inhibit the p53 and Rb pathways, respectively. Consequently, cellular miRNAs controlled by these pathways are directly influenced by these viral proteins. E6 downregulates the expression of miR-34a, a p53 regulated miRNA, leading to an increase in cell growth (101). Additionally, E6 reduces miR-218 and thereby increases laminin 5 β3 (LAMB3), a miR-218 target, in HPV-16 infected cells, which may enhance cell migration and tumorigenicity (62).
Finally, hCMV infection downregulates miR-100 and miR-101, which are proposed to regulate mTOR signaling components (100). The mTOR signaling pathway controls a range of key cellular functions involved in metabolism, growth, and survival, and is manipulated by many herpesviruses at some stage of their life cycle. Interestingly, several hCMV proteins directly alter the activities of mTOR signaling molecules (reviewed in 14). Therefore, the modulation of cellular miRNA expression may allow hCMV further control over a key cell signaling pathway.
VIRAL SUPPRESSION OF CELLULAR MICRORNA EXPRESSION
Defining the boundary between cellular miRNAs actively induced or repressed by viral factors and those miRNAs altered by host responses can be difficult. Indeed, in the absence of phenotypic data, this difference is essentially semantic. For instance, two cellular miRNAs, miR-17-5p and miR-20a, are suppressed by HIV-1 infection. These miRNAs target PCAF, a cellular histone acetylase and proposed cofactor of the HIV-1 Tat transactivator (92). Similar to hCMV suppression of miR-100 and miR-101, it is unclear whether these miRNAs are actively inhibited by viral factors or whether their downregulation is due to host responses.
One interesting question is whether viruses might directly target cellular miRNA biogenesis to interfere with antiviral defenses. The adenovirus VA RNAs, for example, compete with cellular pre-miRNAs for Exportin-5 in the nucleus and inhibit Dicer function in the cytoplasm, thereby interfering with miRNA biogenesis (2, 59, 105). Viruses may further disrupt global cellular miRNA expression by overexpressing their own viral miRNAs. Both KSHV and mCMV miRNAs dominate the miRNA profile during infection (31, 94). Furthermore, viruses may express proteins or RNAs that selectively inhibit specific miRNA function. One recent example is the rapid downregulation of miR-27a activity following mCMV infection (15). Pri-miRNA processing and miR-27a* levels are unaffected, indicating that the mature miRNA itself is being targeted by mCMV. Interestingly, overexpression of miR-27a using miRNA mimics decreases viral titers, suggesting that miR-27a and/or miR-27a-regulated genes inhibit some aspect(s) of the mCMV life cycle (15). Hence, virus-induced suppression of specific cellular miRNAs can favor virus replication. The question then arises, can cellular miRNAs inhibit virus replication as part of the innate antiviral immune response?
ANTI-VIRAL IMMUNITY AND RNAI
The role of RNA interference (RNAi) in antiviral immunity is well established for plant and invertebrate systems. RNA virus infection can stimulate production of viral dsRNA-derived siRNAs to specifically target viral genomes and mRNAs for degradation (reviewed in 30, 93). Examination of small RNAs in mammalian cells infected with several RNA viruses, however, has not revealed any viral-derived siRNAs (56, 72; R.L. Skalsky, J.L. Umbach, and B.R. Cullen, unpublished). Thus, siRNA-directed silencing as a method of combating viral infection appears not to occur in mammalian somatic cells (30, 93). Rather, the equivalent of the innate RNAi antiviral responses in mammalian cells is the more sophisticated protein-based interferon system, which is induced following the activation of intracellular dsRNA sensors such as PKR, RIG-I, and MDA-5.
miRNA expression has been linked to type I interferon stimulation. Interferon β, for example, is upregulated during HCV infection and rapidly induces miR-196, miR-296, miR-351, miR-431, and miR-448, among others (71). These miRNAs can attenuate HCV replication, and may directly target HCV genomes to downregulate viral accumulation (71). However, the idea that an RNA virus such as HCV would retain cellular miRNA binding sites presents a paradox. Why would HCV, a virus with a highly error prone RNA polymerase and therefore, a high mutation rate, retain such sites unless the virus were under selective pressure to do so? Mahajan et al. proposed that robust interferon activation by HCV might result in the loss of infected cells (61). Thus, HCV may exploit IFN-stimulated cellular miRNAs to downregulate its own replication to a level where persistent infection can be achieved.
To add to this intriguing interplay between cellular miRNAs and RNA viruses, miR-122 has been shown To Whom It May Concern:To Whom It May Concern: bind to two adjacent sites in the 5′ non-coding region of HCV genomes to, surprisingly, positively regulate HCV RNA accumulation (51, 52). As HCV is the leading cause of liver disease worldwide, the importance of the miR-122 interaction with HCV has recently been evaluated in vivo. Encouragingly, treatment of chronically HCV-infected chimpanzees with locked-nucleic acid (LNA)-modified antisense oligonucleotides directed against miR-122 reduced HCV levels, IFN activation, and importantly, liver pathology, demonstrating that miRNA inhibition can be a promising new strategy for the treatment of virus-induced diseases (54).
MICRORNA TARGETING OF RNA VIRUSES
Is host miRNA targeting of viral RNAs truly a bona fide mechanism of innate antiviral immunity? Given that the fate of miRNA-bound mRNAs is translational repression and/or degradation, one would predict that RNA viruses would evolve to avoid miRNA-targeting altogether. Yet, other examples of miRNA:RNA virus interactions have been reported. For example, primate foamy virus type 1 (PFV-1) is restricted by miR-32 in 293T cells, while miR-24 and miR-93 have been reported to target vesicular stomatitis virus (VSV) RNAs in mice (55, 69). However, until these findings have been validated in the context of their natural target species and/or tissues, it is unclear whether they are physiologically relevant.
Similarly, several cellular miRNAs were reported to target HIV-1 RNAs in cultivated resting CD4+ T cells to maintain HIV-1 latency (47). While the latent viral reservoir is certainly relevant in HIV-1 individuals undergoing highly active anti-retroviral therapy (HAART), HIV-1 latency is thought to occur inadvertently (45). Thus, in the absence of HAART, under normal physiological conditions, there is no obvious reason why HIV-1 should retain these host miRNA target sites.
On the other hand, knockdown of Dicer results in hypersensitivity to VSV infection (69). Additionally, knockdown of P body and miRNA biogenesis components increases HIV-1 virus production (23, 67, 92), indicating a role for miRNA pathways in viral replication. Intriguingly, HIV-1 RNAs were recently shown to be targeted to P bodies through direct interactions with miR-29a, which consequently limits virus accumulation (67).
However, miRNA-mediated entry into P bodies is not necessarily a dead-end for mRNAs and can be reversible (34). CAT-1 mRNA, for example, can escape translational inhibition by miR-122 during amino acid starvation as it is released from P bodies and recruited back to polysomes (11). Recent studies indicate that P bodies may actually function as sites of virus replication and/or assembly (reviewed in 8). In the case of both HCV and HIV-1, the RNA genome serves as a template for viral protein translation and is also packaged into virion particles, while, in the case of HCV, the same genome also functions as the template for viral RNA replication. Thus, it is possible that these RNA viruses exploit cellular miRNAs to target viral RNAs away from the translational machinery so that viral RNA synthesis and/or assembly can occur.
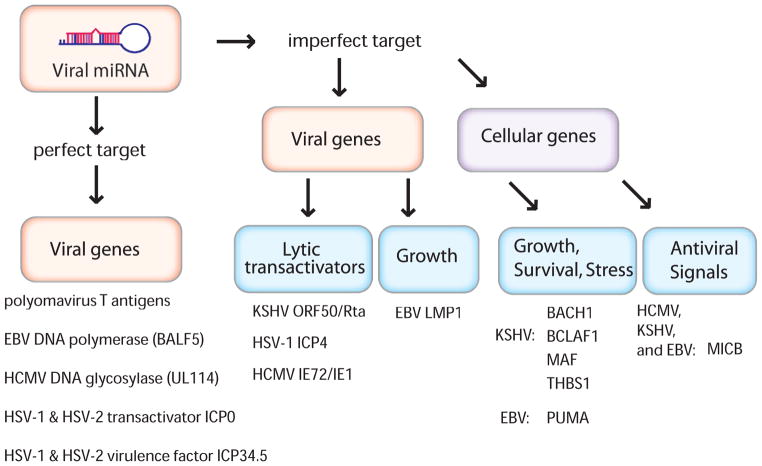
Figure 3. Viral miRNA targets.
Viral miRNAs target perfectly complementary viral mRNAs as well as imperfectly complementary viral and/or cellular mRNAs. Viral miRNA targets can be further divided into functional categories. Some targets have roles in lytic transactivation while others are involved in cellular growth, stress, or immune activation.
SUMMARY POINTS.
Viral miRNAs downregulate selected viral and cellular mRNAs to establish a host environment conducive to completion of the viral life cycle. Many of these mRNAs encode factors involved in the latent-lytic switch (i.e., lytic viral transactivators) or immune activation (i.e., viral antigens or cellular signaling molecules)
Viruses can mimic cellular miRNAs or affect cellular miRNA expression to control existing regulatory pathways
Cellular miRNAs can directly influence viral replication, and some miRNAs can directly target mammalian RNA virus genomes.
FUTURE ISSUES.
What are the mRNA targets of viral miRNAs and how do these facilitate the various stages of the viral life cycle, e.g., lytic versus latent?
Is targeting of RNA viruses, such as HCV or HIV-1, by cellular miRNAs a bona fide mammalian antiviral immune response? Or have target sites for cellular miRNAs been retained by viruses to benefit the viral life cycle in some manner?
Are viral transcripts expressed by vertebrate DNA viruses also targeted by cellular miRNAs?
What is the contribution of viral and/or cellular miRNAs to viral pathogenesis? Can these viral miRNA:host or host miRNA:virus interactions be strategically targeted for anti-viral therapy?
Acknowledgments
Research in the Cullen laboratory was supported by R01-AI-067968 from the National Institutes of Health. RLS was supported by NIH T32-CA009111.
Key Terms and Acronyms
- miRNA
microRNA, a ~19–24 nt non-coding RNA
- pre-miRNA
precursor microRNA, the stem-loop structure from which a miRNA is cleaved
- RISC
RNA-induced silencing complex, a cytoplasmic complex containing at minimum a mature miRNA and Argonaute protein
- P body
Processing body
- RNAi
RNA interference
- Herpesviruses
large, nuclear, dsDNA viruses that establish long-term infections
Acronyms list
- EBV
Epstein-Barr Virus
- KSHV
Kaposi’s sarcoma-associated herpesvirus
- HSV-1 and HSV-2
Herpes simplex virus 1 and 2
- LAT
latency associated transcript
- hCMV
Human Cytomegalovirus
- HCV
Hepatitis C Virus
- HIV-1
Human immunodeficiency virus 1
- IFN
interferon
Contributor Information
Rebecca L. Skalsky, Email: rebecca.skalsky@duke.edu.
Bryan R. Cullen, Email: bryan.cullen@duke.edu.
LITERATURE CITED
- 1.Anastasiadou E, Boccellato F, Vincenti S, Rosato P, Bozzoni I, et al. Epstein-Barr virus encoded LMP1 downregulates TCL1 oncogene through miR-29b. Oncogene. 2009 doi: 10.1038/onc.2009.439. [DOI] [PubMed] [Google Scholar]
- 2.Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–65. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol. 2006;80:1376–84. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–85. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. This review describes our current knowledge of miRNA-based targeting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–75. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckham CJ, Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–12. doi: 10.1016/j.chom.2008.03.004. This review describes the relationship of P body components to viral life cycles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6:570–5. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian miRtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–24. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol Cell. 2010;37:135–42. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolisetty MT, Dy G, Tam W, Beemon KL. Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J Virol. 2009;83:12009–17. doi: 10.1128/JVI.01182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchkovich NJ, Yu Y, Zampieri CA, Alwine JC. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat Rev Microbiol. 2008;6:266–75. doi: 10.1038/nrmicro1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buck AH, Perot J, Chisholm MA, Kumar DS, Tuddenham L, et al. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA. 2010 doi: 10.1261/rna.1819210. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. Discrete clusters of virus-encoded microRNAs are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol. 2007;81:13761–70. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burnside J, Bernberg E, Anderson A, Lu C, Meyers BC, et al. Marek’s disease virus encodes microRNAs that map to meq and the latency-associated transcript. J Virol. 2006;80:8778–86. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X, Li G, Laimins LA, Cullen BR. Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. J Virol. 2006;80:10890–3. doi: 10.1128/JVI.01175-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570–5. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, et al. Epstein-Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82:1946–58. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantalupo P, Doering A, Sullivan CS, Pal A, Peden KW, et al. Complete nucleotide sequence of polyomavirus SA12. J Virol. 2005;79:13094–104. doi: 10.1128/JVI.79.20.13094-13104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chable-Bessia C, Meziane O, Latreille D, Triboulet R, Zamborlini A, et al. Suppression of HIV-1 replication by microRNA effectors. Retrovirology. 2009;6:26. doi: 10.1186/1742-4690-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–60. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui C, Griffiths A, Li G, Silva LM, Kramer MF, et al. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80:5499–508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–5. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009;457:421–5. doi: 10.1038/nature07757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diebel KW, Smith AL, van Dyk LF. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA. 16:170–85. doi: 10.1261/rna.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–26. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolken L, Perot J, Cognat V, Alioua A, John M, et al. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J Virol. 2007;81:13771–82. doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn W, Trang P, Zhong Q, Yang E, van Belle C, Liu F. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 2005;7:1684–95. doi: 10.1111/j.1462-5822.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- 33.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–28. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 34.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 35.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furnari FB, Adams MD, Pagano JS. Unconventional processing of the 3′ termini of the Epstein-Barr virus DNA polymerase mRNA. Proc Natl Acad Sci U S A. 1993;90:378–82. doi: 10.1073/pnas.90.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gandy SZ, Linnstaedt SD, Muralidhar S, Cashman KA, Rosenthal LJ, Casey JL. RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication. J Virol. 2007;81:13544–51. doi: 10.1128/JVI.01521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottwein E, Cai X, Cullen BR. Expression and function of microRNAs encoded by Kaposi’s sarcoma-associated herpesvirus. Cold Spring Harb Symp Quant Biol. 2006;71:357–64. doi: 10.1101/sqb.2006.71.004. [DOI] [PubMed] [Google Scholar]
- 39.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–9. doi: 10.1038/nature05992. One of two studies demonstrating a virally encoded ortholog of cellular miR-155 that shares seed sequence homology, and consequently can target, an overlapping set of cellular transcripts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, et al. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. 2005;79:12095–9. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grey F, Nelson J. Identification and function of human cytomegalovirus microRNAs. J Clin Virol. 2008;41:186–91. doi: 10.1016/j.jcv.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–50. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol. 2007;5:95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- 46.Hansen A, Henderson S, Lagos D, Nikitenko L, Coulter E, Roberts S, Gratrix F, Plaisance K, Renne R, Bower M, Kellam P, Boshoff C. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes & Development. 2010;24:195–205. doi: 10.1101/gad.553410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J, Wang F, Argyris E, Chen K, Liang Z, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241–7. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 48.Hussain M, Asgari S. Functional analysis of a cellular microRNA in insect host-ascovirus interaction. J Virol. 84:612–20. doi: 10.1128/JVI.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hussain M, Taft RJ, Asgari S. An insect virus-encoded microRNA regulates viral replication. J Virol. 2008;82:9164–70. doi: 10.1128/JVI.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 51.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. This study describes the first direct interaction of a cellular miRNA with an RNA virus genome that positively regulates viral RNA accumulation. [DOI] [PubMed] [Google Scholar]
- 53.Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, et al. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. This study demonstrates the potential for therapeutic targeting of miRNAs involved in viral infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–60. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 56.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218–26. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 58.Lo AK, To KF, Lo KW, Lung RW, Hui JW, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104:16164–9. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–76. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–43. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 61.Mahajan VS, Drake A, Chen J. Virus-specific host miRNAs: antiviral defenses or promoters of persistent infection? Trends Immunol. 2009;30:1–7. doi: 10.1016/j.it.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–82. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McAllister SC, Hansen SG, Ruhl RA, Raggo CM, DeFilippis VR, et al. Kaposi sarcoma-associated herpesvirus (KSHV) induces heme oxygenase-1 expression and activity in KSHV-infected endothelial cells. Blood. 2004;103:3465–73. doi: 10.1182/blood-2003-08-2781. [DOI] [PubMed] [Google Scholar]
- 64.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 65.Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci U S A. 2008;105:5453–8. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–85. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 67.Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okamura K, Chung WJ, Lai EC. The long and short of inverted repeat genes in animals: microRNAs, mirtrons and hairpin RNAs. Cell Cycle. 2008;7:2840–5. doi: 10.4161/cc.7.18.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otsuka M, Jing Q, Georgel P, New L, Chen J, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–34. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 70.Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, et al. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–65. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–22. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–76. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 73.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–6. doi: 10.1126/science.1096781. This study provides the first evidence of virally-encoded miRNAs. [DOI] [PubMed] [Google Scholar]
- 74.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79:9301–5. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, et al. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sano M, Kato Y, Taira K. Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett. 2006;580:1553–64. doi: 10.1016/j.febslet.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 79.Schafer A, Cai X, Bilello JP, Desrosiers RC, Cullen BR. Cloning and analysis of microRNAs encoded by the primate gamma-herpesvirus rhesus monkey rhadinovirus. Virology. 2007;364:21–7. doi: 10.1016/j.virol.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 81.Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383:183–7. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 82.Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J Virol. 2008;82:9823–8. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, et al. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–45. doi: 10.1128/JVI.01804-07. One of two studies demonstrating a virally encoded ortholog of cellular miR-155 that shares seed sequence homology, and consequently can target, an overlapping set of cellular transcripts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–81. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stern-Ginossar N, Saleh N, Goldberg MD, Prichard M, Wolf DG, Mandelboim O. Analysis of human cytomegalovirus-encoded microRNA activity during infection. J Virol. 2009;83:10684–93. doi: 10.1128/JVI.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–6. doi: 10.1038/nature03576. This study validated the first target of a viral miRNA. [DOI] [PubMed] [Google Scholar]
- 87.Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, et al. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology. 2009;387:157–67. doi: 10.1016/j.virol.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci U S A. 2008;105:10931–6. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol. 2009;83:1433–42. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 91.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–82. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 93.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–64. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Umbach JL, Cullen BR. In-depth analysis of Kaposi’s sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J Virol. 2010;84:695–703. doi: 10.1128/JVI.02013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–3. doi: 10.1038/nature07103. This paper reports evidence indicating that viral miRNAs can act to stabilize viral latency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol. 2009;83:10677–83. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Umbach JL, Wang K, Tang S, Krause PR, Mont EK, et al. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J Virol. 2010;84:1189–92. doi: 10.1128/JVI.01712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Waidner LA, Morgan RW, Anderson AS, Bernberg EL, Kamboj S, et al. MicroRNAs of Gallid and Meleagrid herpesviruses show generally conserved genomic locations and are virus-specific. Virology. 2009;388:128–36. doi: 10.1016/j.virol.2009.02.043. [DOI] [PubMed] [Google Scholar]
- 99.Walz N, Christalla T, Tessmer U, Grundhoff A. A global analysis of evolutionary conservation among known and predicted gammaherpesvirus microRNAs. J Virol. 84:716–28. doi: 10.1128/JVI.01302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol. 2008;82:9065–74. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang X, Wang HK, McCoy JP, Banerjee NS, Rader JS, et al. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637–47. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–42. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 104.Xing L, Kieff E. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J Virol. 2007;81:9967–75. doi: 10.1128/JVI.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu N, Segerman B, Zhou X, Akusjarvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the RNA-induced silencing complex and associate with polyribosomes. J Virol. 2007;81:10540–9. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu Y, Rodriguez-Huete A, Pari GS. Evaluation of the lytic origins of replication of Kaposi’s sarcoma-associated virus/human herpesvirus 8 in the context of the viral genome. J Virol. 2006;80:9905–9. doi: 10.1128/JVI.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, et al. Marek’s disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J Virol. 2007;81:7164–70. doi: 10.1128/JVI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, et al. MicroRNA profile of Marek’s disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J Virol. 2008;82:4007–15. doi: 10.1128/JVI.02659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yin Q, McBride J, Fewell C, Lacey M, Wang X, et al. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82:5295–306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 111.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, et al. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83:3333–41. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41:130–4. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]