Summary
Imprinted genes are marked by parental allele specific DNA methylation and histone modifications which regulate their monoallelic expression. Chromatin immunoprecipitation (ChIP) is the technique of choice to characterize the histones associated with either the maternal or paternal chromosomes. To study allele-specific chromatin composition at imprinted regions, the method has to be efficient to work on limiting amount of starting material, and specific enough to recognize one of the parental alleles. We optimized the commonly used ChIP technique for efficient recovery of one parental allele from small number of cells. We provide examples to show that this ChIP protocol can specifically distinguish between parental alleles in mouse embryo fibroblasts carrying maternal and paternal duplication of mouse distal Chr7 and also in normal mouse embryo fibroblasts carrying single nucleotide polymorphism at imprinted regions.
Keywords: Genomic imprinting, Monoallelic, Differentially methylated regions, Chromatin immunoprecipitation, Histone modifications
1. Introduction
Genomic imprinting is a form of epigenetic regulation whereby the allele inherited from either the mother or father is functional [1, 2]. Imprinted genes often occur in clusters and are often associated with differentially methylated regions (DMRs). DMRs are classified as imprinting control regions (ICRs) when they determine the parental allele-specific expression of the imprinted genes in their respective domain. The promoters of imprinted genes and also the DMRs and ICRs exhibit parental allele specific DNA methylation and post translational histone modifications. Chromatin immunoprecipitation (ChIP) [3] is performed to determine allele specific binding of a given histone modification at the maternal or paternal allele at a DNA sequence in vivo. Histone modifying proteins are crosslinked to DNA with formaldehyde followed by shearing the DNA into smaller fragments by sonication. Antibodies against specific histones are used to precipitate the complex. The crosslinking is reversed and the bound proteins are digested using proteinase K to release the DNA fragments which can be analyzed for enrichment using locus specific PCR primers [4, 5].
One challenge in analyzing allele-specific epigenetic features is to differentiate between the parental alleles. One possible approach is to use cells with uniparental duplications, where two copies of a chromosome segment are inherited from one parent and no copies from the other parent. Chromatin from primary mouse embryo fibroblast (MEFs) carrying maternal and paternal duplication of distal Chr7, MatDup.dist7 and PatDup.dist7, can be used to characterize maternal and paternal allele specific histone modifications, respectively at the H19/Igf2 and Cdkn1c/Kcnq1 domains [1, 2], because these are both located along the duplicated region of Chr7 [6, 7]. The H19-Igf2 region is controlled by an ICR, which is methylated in the paternal chromosome but is associated with the insulator protein CTCF in the maternal chromosome. The Cdkn1c/Kcnq1 domain is controlled by the KvDMR1 that carries DNA methylation in the maternal allele and functions as the promoter for the noncoding regulatory RNA, Kcnq1ot1 in the paternal allele. Another possible approach for allele-specific chromatin analysis utilizes DNA sequence differences, such as single nucleotide polymorphisms (SNPs) between the mother and father to distinguish the parental alleles in normal cells [8, 9].
The other challenge in analyzing imprinted genes is, that the starting material can be limiting. General ChIP protocols suggest using 25 µg of chromatin or 107 cells per reaction, which is hard to obtain when only small embryos or small populations of purified embryonic cells are available. The following ChIP protocol is routinely performed in our laboratory using 4 µg of chromatin or 400000 cells. Using this optimized technique we demonstrate here that an active histone modification mark, H3K4me2, is associated with the unmethylated maternal and paternal allele of H19/Igf2 ICR and KvDMR1, respectively and a repressive mark, H3K9me3, is associated with the reciprocal, methylated alleles at these DMRs. This ChIP protocol has been used in combination with tiling microarrays to reveal chromosome-wide allele-specific features along distal Chr7 and distal Chr15 [7]. It was also used in combination with multiplex single nucleotide primer extension (SNuPE) assays in the sequenom-allelotyping platform to generate histone modification profiles across multiple DMRs in the mouse genome [8, 9]. We also provide an even smaller scale version (*), which gives reproducibly allele-specific results using only 100000 purified mouse embryonic germ cells [10].
2. Materials
All buffers are prepared in tissue culture grade water (Lonza#17-724Q), filter sterilized and stored at appropriate temperature.
DMEM (Dulbecco's modified eagle medium)
FBS (Fetal bovine serum)
Formaldehyde 37%
PBS (phosphate buffered saline without calcium and magnesium)
Sodium dodecyl sulphate 10% (GIBCO-BRL#15553-035 and Roche #11667262001)
Triton X-100 (Sigma# T9284)
Tris-HCl pH 8.0 (Invitrogen # 15568-025)
Sodium Chloride 5 M (USB #75888)
Lithium Chloride (Sigma# L9650)
NP-40 (Biochemika#74385)
Sodium deoxycholate (Sigma#D6750)
EDTA 0.5M (USB# 15694)
Glycine (Fisher scientific#BP381-500)
Sodium Bicarbonate (JT Baker #3506-01)
cØmplete protease inhibitor cocktail tablets, (Roche applied science, Cat.# 1697498) 50×, one tablet dissolved in 1ml water.
ChIP Dilution Buffer: 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8.1 and 167 mM NaCl. Store at RT. Add 4 µl/ml of 50× cØmplete protease inhibitor cocktail before use.
ChIP Lysis Buffer: 1% SDS, 10 mM EDTA and 50mM Tris-HCl, pH 8.1. Store at RT. Add cØmplete protease inhibitor cocktail to 1× concentration before use.
Preblocked A/G Beads: 2ml A/G Beads (Santa cruz# sc-2003), 20 µl herring sperm DNA (Invitrogen# 15634-017), 56 µl 50× cØmplete protease inhibitor cocktail, 280 µl 10mg/ml BSA (PENTEX) and 2ml ChIP dilution Buffer. Preblock the beads by gentle rotation for 2 h in the cold room in 15ml falcon tube. Spin at 250 g for 5 m. Discard supernatant and resuspend the beads in 2 ml of ChIP dilution buffer supplemented with 40µl of 50× complete protease inhibitor cocktail. Store at 4°C for up to a month. (see Notes 1, 2, 3)
Buffer A (Low-salt Buffer): 0.1% SDS, 1.0% Triton X-100, 2.0 mM EDTA, 20mM Tris-HCl, pH 8.1, 150 mM NaCl. Store at 4°C.
Buffer B (High-salt Buffer): 0.1% SDS, 1.0% Triton X-100, 2.0 mM EDTA, 20mM Tris-HCl, pH 8.1, 500 mM NaCl. Store at 4°C.
Buffer C (LiCl Buffer): 0.25 M LiCl, 1.0% Igepal-CA630 (also known as NP-40), 1.0% Sodium deoxycholate), 1.0 mM EDTA, 10 mM Tris-HCl, pH 8.1. Store at 4°C.
ChIP Elution Buffer: 1% SDS, 0.1M NaHCO3
Proteinase K (Roche#03115801001)
TE Buffer: 1.0 mM EDTA, 10 mM Tris-HCl, pH 8.0
3M Sodium Acetate (pH 5.5)
Phenol (equilibrated to pH 8.0)
Chloroform
Linear polyacrylamide [11]
Ethanol
3. Methods
3.1 Cross-linking and cell harvesting
Grow primary mouse embryo fibroblasts (MEFs) close to confluency in a 150 mm dish containing 25 ml of culture medium (DMEM supplemented with 6% fetal bovine serum) to obtain about 2.5×107 cells per plate. Prepare chromatin in two ways, using N-ChIP or X-ChIP conditions, to test out new antibodies. *Trypsinized and flow-sorted cells, such as primordial germ cells, are crosslinked in suspension. (see Notes 4, 5)
3.1.1 Preparation of N-link chromatin
Add formaldehyde directly into the media to a final concentration of 1% (675 µl of 37% formaldehyde per 25 ml medium) inside a fume hood. It is best to tilt the plate and quickly add formaldehyde by touching the side of the plate where medium level is highest and without delay mixing up the medium by swirling. *For formaldehyde crosslinking of 100 K cells, first pellet cells at 2800 rpm for 10 min in an eppendorf tube, then remove medium and add 500 µl of PBS with 13.5 µl of 37% formaldehyde. Suspend cells and mix by pipetting.
Incubate the plate at room temperature with gentle agitation for exactly 2 min on a horizontal shaking platform. *Tubes are placed on a rotating platform for 2 min.
Stop the cross-linking reaction by adding 2.6 ml of 1.25 M glycine solution, mix well immediately. *For 100 K cells, add 25 µl of 1.25 M glycine and spin at 2800 rpm for 10 min.
Remove media inside a fume hood by pouring it from the plate into a glass beaker and blotting the last drop off on a paper towel. Wash cells twice with 20 ml ice-cold PBS. *Remove liquid from 100 K pellet by pipetting. To wash 100 K cells, suspend the cell pellet in 700 µl of cold PBS and spin.
Aspirate PBS completely after the second wash. Place cell culture dish or *eppendorf tube on ice.
Scrape cells off the plate in 5 ml ice-cold PBS with a plastic scraper and collect into a 15 ml falcon tube by pipetting.
Pellet the cells by centrifugation for 5 min at 250 g at 4°C. Aspirate supernatant without touching the cell pellet, keep cell pellet on ice. *For 100 K cells, aspirate the last drop using a pulled pasteur pipette attached to a mouthpiece under a dissecting microscope carefully not to dislodge the cell pellet.
3.1.2 Preparation of X-link chromatin
X-ChIP is the same as N-ChIP, except for step 2.
Incubate at room temperature for exactly 10 min.
3.2 Sonication
Resuspend pelleted cells in 750 µl ChIP Lysis buffer containing cØmplete protease inhibitors. *Add 100 µl of lysis buffer to 100 K cells, mix, snap freeze in liquid N2 and store at −80°C. These small samples are sonicated on the day of use.
Sonication is carried out in 1.7 ml eppendorf tubes using Branson Sonifier cell Disruptor −350 with a micro tip. Pulse 3 to 4 times for 10 seconds at 40 % duty cycle and 4 output control. Keep samples on ice at all times. In our experience, N-ChIP and X-ChIP samples need to be sonicated 3 and 4 times, respectively.
If using Diagenode Bioruptor UCD-200, sonicate at high "H" setting with 15 seconds "ON" 1 min "OFF" for 5 min. Repeat 6 times with 1 min on ice between each cycle. Diagenode Bioruptor is preferred for sonicating 100 K cells in 100 µl volume to prevent frothing and loss of chromatin on the sonicator tip.
Centrifuge the sheared chromatin at 13,000 rpm for 5 min at 4°C. Transfer the supernatant containing the chromatin to a fresh tube. Store chromatin on ice in cold room during quantification/before freezing at −80°C or use directly for ChIP.
3.2.1 Determination of Chromatin DNA Concentration and Fragment Size
To determine the concentration of DNA in the sonicated chromatin, take a small aliquot and reverse cross-link it. For a 25 µl chromatin preparation, add 25 µl of ChIP lysis buffer, add NaCl to a final concentration of 0.3 M (3µl of 5M NaCl) and incubate at 65°C for 4 hrs.
Add 1µl each of 0.5 M EDTA, 1M Tris-HCl, pH 6.5 and 20 mg/ml proteinase K. Incubate at 55°C at 1hr.
Bring the volume to 100 µl by adding 50 µl of TE (pH 8.0). Extract with phenol/chloroform. Add 50 µl of phenol and mix vigorously. Add 50 µl of chloroform and mix well. Centrifuge at 13000 rpm for 10 min. Collect the top, aqueous phase and also the interphase at this point. Add 100 µl of chloroform and mix well. Spin at 13000 rpm. Collect only the top aqueous phase this time and transfer it to a fresh tube.
Precipitate DNA: add 3µl of linear polyacrylamide and 1/10 volume of 3M sodium acetate, pH 5.5 and mix. Add 2.5 volumes of 100 % ethanol, mix and freeze sample at −80°C for 30 min. To collect DNA pellet, centrifuge sample at 13000 rpm for 15 min. Wash DNA pellet with 70% ethanol. Resuspend DNA in 25 µl Tris-EDTA buffer (TE), pH 8.0.
Check DNA concentration using Nanodrop (see Note 6).
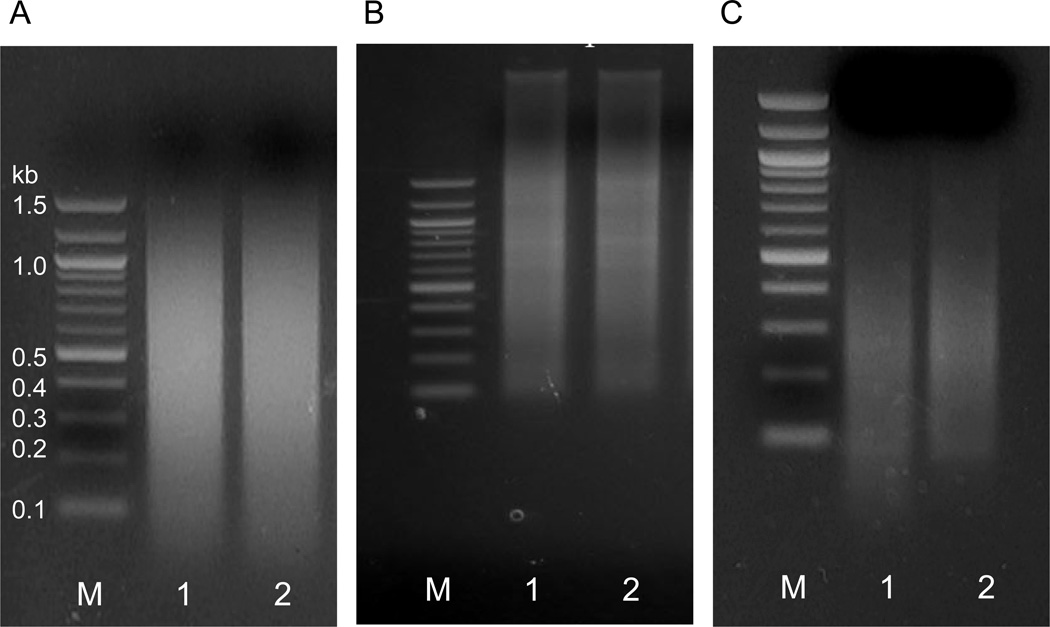
Load 1µg of DNA on a 1.5% agarose gel and run it in 1×TAE (Tris-Acetate-EDTA) buffer to check the size of chromatin fragments. They should range from 0.2 to 1.5 kb. (Figure 1) (see Note 7).
Dilute chromatin preparations (which were stored on ice to this point) to 0.4 µg/µl concentration with ChIP lysis buffer containing 1× cØmplete protease inhibitors. You will use 10 µl (4 µg nucleic acid equivalent chromatin) per ChIP reaction. At this point, chromatin preparations can be used right away or 100 µl aliquots can be snap frozen in liquid nitrogen for long-term storage at −80 °C.
Figure 1. Testing the efficiency of chromatin sonication.
Agarose gel electrophoresis of sonicated DNA to determine the fragment size of the crosslinked chromatin (A) Ideal DNA sonication is shown with fragment size ranging from up to 1200 bp with median fragment length of ~500 bp. The small fraction <200 bp on this gel is RNA and can be eliminated by RNAse treatment. (B) Undersonication. High molecµlar weight unfragmented DNA can be seen on top of the gel. (C) Oversonication. The majority of fragments are below 400 bp. (lane 1, M: 1 kb-ladder, lane 2, MatDup.dist7 and lane 3, PatDup.dist7 DNA)
3.3 Immunoprecipitation
Quickly defrost frozen sheared chromatin sample on the day of ChIP. *100 K cell aliquots can be defrosted, combined and sonicated in a larger batch on the day of ChIP. Sonication efficiency of small aliquots is similar to larger samples. This can be optimized and tested beforehand.
Take out 10 µl sheared chromatin for “input”.
Use 10 µl of chromatin for each IP. Add 90 µl ChIP Lysis Buffer with 1× cØmplete protease inhibitors to the 10 µl chromatin fraction in an eppendorf tube. Dilute 10-fold with ChIP Dilution Buffer containing 4 µl/ml 50× cØmplete. *100 K cells had been already suspended in 100 µl ChIP Dilution Buffer containing 4 µl/ml of 50× cØmplete. Dilute these 10-fold with ChIP dilution buffer containing 4 µl/ml 50× cØmplete.
Pre-clear chromatin with preblocked Protein A/G agarose beads. Add 50 µl Protein A/G agarose into each reaction. Rotate at 4°C for 1 hr. Pellet beads by centrifugation for 5 min at 250 g. Transfer supernatant chromatin to a new tube. (see Note 8)
Add 4 µg of the antibody to the supernatant and incubate overnight at 4 °C on a rotating platform. Set up one sample with normal rabbit IgG antibody as negative control. (see Notes 9, 10, 11)
To collect the antibody-chromatin complexes, the next morning add 50 µl Protein A/G agarose beads to each IP. Rotate for 2 h at 4 °C.
To collect beads centrifuge for 1 min at 1,000 rpm and discard supernatant.
- Wash agarose beads on a rotating platform for 5 min with 1 ml of the ice-cold buffers A, B and C. Collect beads by centrifugation after each wash at 1,000rpm for 1min at 4 °C.
- Buffer A with 2 µl/ml 50 × cØmplete in cold room.
- Buffer B with 2 µl/ml 50 × cØmplete in cold room.
- Buffer C with 2 µl/ml 50 × cØmplete in cold room.
- TE Buffer with 2 µl/ml 50 × cØmplete at room temperature (repeat this wash 2 more times)
Remove as much buffer as possible without disturbing the beads (see Notes 12, 13, 14).
3.4 Elution, Reverse Cross-link and Proteinase K Treatment
Add 150 µl freshly made ChIP Elution buffer to the washed beads (see Note 15). Incubate 15 min at room temperature with gentle agitation.
Pellet beads by centrifugation at 4000 rpm for 2 min. Transfer the ChIP eluate supernatant to a new clean tube.
Repeat elution. Combine the second eluate with the first one.
-
Reverse cross-link the eluted DNA by adding 18 µl 5M NaCl per 300 µl eluate followed by incubation for 4 h at 65°C. (see Note 16)
(Remember to reverse-crosslink the reserved “input” DNA samples also at this point)
Add 6 µl each of 0.5 M EDTA, 1 M Tris-HCl, pH 6.5 and 20 mg/ml proteinase K. Incubate 1 hr at 55°C. (see Note 17)
3.5 DNA Isolation and Purification
Add 1.5 ml of Binding buffer QG (QIAquick Kit, QIAGEN).
Purify DNA according to the manufacturer’s instruction. Wash with PE four times. (see Note 18)
Recover DNA with 100 µl warm elution buffer. Store at −20 °C. Samples are ready for subsequent qPCR assays or sequenom allelotyping (Figures 2 and 3) (see Notes 19, 20, 21, 22. 23).
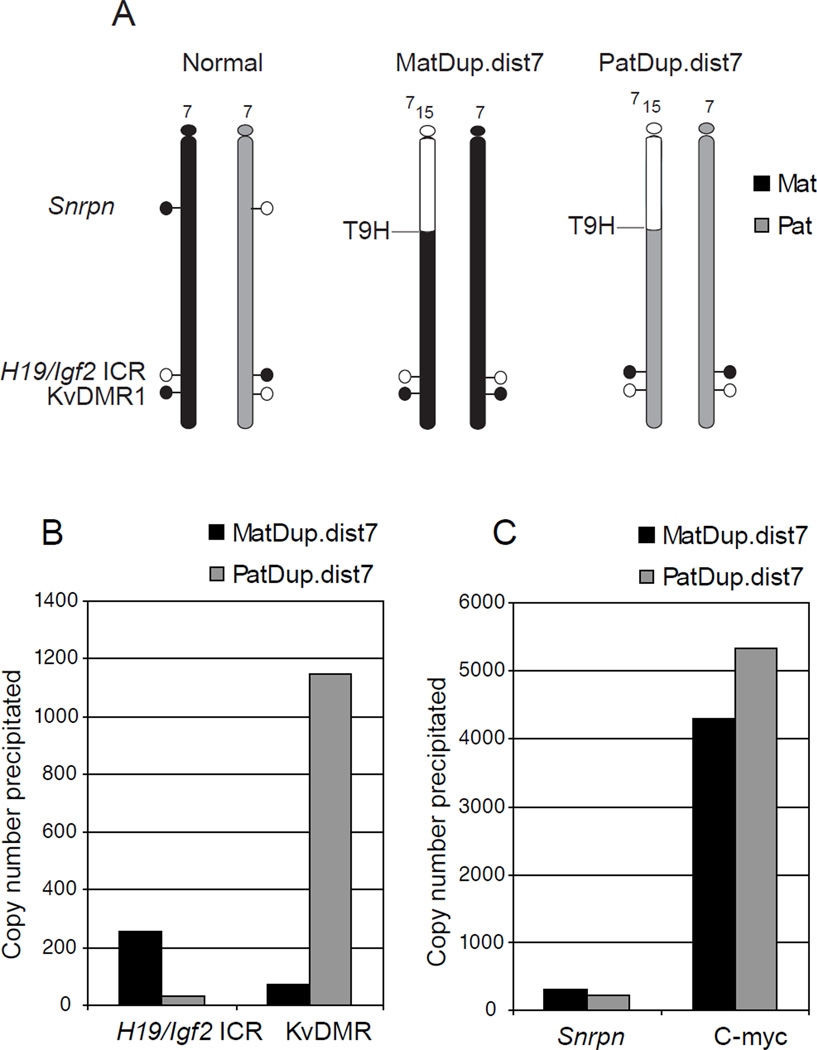
Figure 2. Detecting allele-specific H3K4me2 enrichment at imprinted regions using cells with uniparental duplications of distal Chr7.
(A) MatDup.dist7 and PatDup.dist7 MEFs carry two maternal (black) or two paternal (grey) copies of chromosome 7 regions, located distally to the T9H translocation breakpoint. These cells allow the analysis of allele-specific marks at the H19/Igf2 ICR and at the KvDMR1 along the maternally or paternally duplicated distal chromosome 7 region. Parental allele-specific methylation and hypomethylation of the DMRs is shown by closed and open lollipops, respectively. (B) Real-time PCR was used to quantify an active chromatin mark, H3K4me2, levels at specific loci in MatDup.dist7 and PatDup.dist7 MEFs at two imprinted regions. The paternally methylated H19/Igf2 ICR shows H3K4me2 in the MatDup.dist7 MEFs whereas the maternally methylated KvDMR1 exhibits H3K4me2 enrichment in PatDup.dist7 MEFs. (C) Control regions. The control Snrpn DMR is located outside of the duplicated chromosome region, therefore the allele-specificity of positive H3K4me2 enrichment can’t be discerned at this locus. The control c-myc promoter is constitutively active and enriched in H3K4me2.
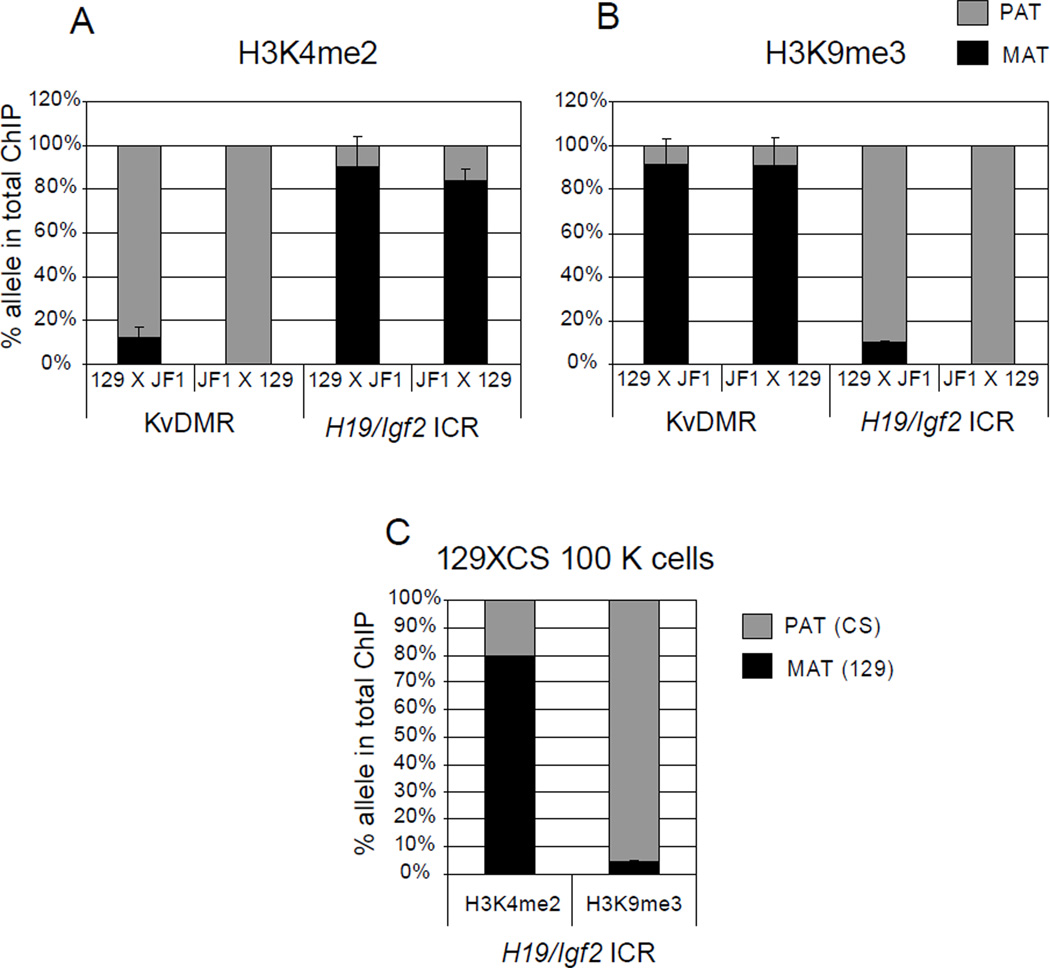
Figure 3. Detecting allele-specific chromatin in normal cells.
Chromatin was prepared from 129XJF1 (129 mother and JF1 father) and JF1X129 (JF1 mother and 129 father) MEFs and was subjected to ChIP using (A) H3K4me2 and (B) H3K9me3 antibodies. Sequenom allelotyping assays were used to measure the percent maternal and paternal component in the total ChIP DNA at the KvDMR1 and at the H19/Igf2 ICR. H3K4me2 is enriched in the unmethylated paternal allele (PAT) at the KvDMR1 and maternal allele (MAT) at the H19/Igf2 ICR. H3K9me3 shows enrichment at the reciprocal, methylated, alleles. (C) ChIP using 100000 MEFs. These MEFs were obtained by mating a 129 mother to a CAST/Ei (CS) father. Sequenom allelotyping shows correct allele specific enrichment in 129XCS MEFs for H3K4me2 and H3K9me3 at the H19/Igf2 ICR similar to ChIP from 4 µg chromatin.
3.6 Multiplex quantitative real-time PCR for allele specific histone modifications
Dissolve all the primers and probes (Table 2) in TE (pH 8.0) to a final concentration of 100 µM. The oligo tubes from IDT are spun briefly before dissolving, as the pellet may dislodge during shipment. Mix equal volumes of the upper and lower (U+L) primers. These can be stored at −20°C (see Note 24).
Prepare MIQ 5× Buffer by mixing equal volumes of 10× iTaq Buffer and 5 mM MgCl2 from the iTaq DNA Polymerase kit (Bio-Rad #170-8875).
-
For amplification standards use a sonicated mouse genomic DNA dilution series: 10 ng/µl, 1 ng/µl, 0.1 ng/µl, 0.01 ng/µl 0.001ng/µl and a no template control. In the iQ5 real-time plate setup menu, define units as copy numbers (10,000; 1000; 100; 10; 1 and 0 copies, respectively. This calculation considers that one diploid mouse genome equals 6 pg of genomic DNA. Set up the multiplex PCR reaction as follows:
X1 (µl) MIQ 5× Buffer 5.0 dNTP mix (25mM each) 0.45 iTaq (Biorad) 5U/µl 0.325 Primer (U+L) (4) 0.2 each pair (up to 5 total pairs) Probes (4) 0.075 each probe (up to 5 total probes) DNA 3 µl of ChIP eluate or 30 ng input or standard genomic DNA H2O up to 25 µl The following PCR parameters are used to run the reactions in Bio-Rad iQ5-Thermal Cycler:95°C 3min 40 cycles 95°C 30sec 55°C 45sec
Table 2.
Primers for multiplex real-time PCR
| DMR | Probe sequence and dye | PCR Primers |
|---|---|---|
| H19-Igf2 ICR-FAM | ACATTCACACGAGCATCCAGGAGGC | CACTTACACCCAGGACTCAAAGG |
| FAM | GCGTATAAACCCCACAACTGATTC | |
| KvDMR1 | CCGCAGTGGCTCCGTATTCGTT | CGGCTGGGCTCCATCTTC |
| TEX | CGACCTCGGGGCTCAAAG | |
| Snrpn promoter | CATGCGTCCCAGGCAATGGCTGC | TCCTTTTGGTAGCTGCCTTTTGG |
| TAMRA | CCGCAATGGCTCAGGTTTGTC | |
| c-myc promoter | CTGCCTCGCTCCACACAATACGCCA | AGATAACTCATTCGTTCGTCCTTCC |
| Cy5 | TGTGTTCTTGCCCTGCGTATATC |
To illustrate the applicability of the of this ChIP protocol to allele-specific analysis, we precipitated chromatin from MatDup.dist7 and PatDup.dist7 MEFs using the H3K4me2 antibody and amplified the H19/Igf2 ICR, the KvDMR, as well-as the control Snrpn promoter and c-myc promoter regions. Four sets of primers and probes (Table 2) were used in a multiplex real-time PCR reaction (Figure 2). We also performed ChIP with H3K4me2 and H3K9me3 antibodies using chromatin from normal 129XJF1 and JF1X129 MEFs and submitted aliquots of the ChIP elution for multiplex sequenom allelotyping as we have done earlier [8, 9]. The percent H3K4me2 and H3K9me3 enrichment in the total immunoprecipitation was measured utilizing SNPs between the parental alleles (Figure 3 A and B). The results for the H3K4me2 antibody were in agreement with the results of the MatDup.dist7-PatDup.dist7 experiment, showing active chromatin-specific H3K4me3 enrichment in the unmethylated DMR alleles. The repressing mark, H3K9me3, however, was present in the CpG-methylated alleles at both DMRs. The ChIP protocol using 100 K 129XCS MEFs also revealed the correct allele specific bias (Figure 3C). These experiments demonstrate that our optimized ChIP protocol efficiently and quantitatively recovers the correct parental alleles of imprinted regions from chromatin even when the cell numbers are limited.
Table 1.
Volumes for preclearing the chromatin
| Chromatin | Lysis Buffer | Dilution Buffer |
Preblocked A/G beads |
|
|---|---|---|---|---|
| Sonicated chromatin from plate | 10 µl (~4 µg) | 90 µl | 900 µl | 50 µl |
| Chromatin from 100 K cells | 100 µl (~1 µg) | 0 µl | 900 µl | 50 µl |
Acknowledgements
We thank Jeff Mann for the MatDup.dist7 and PatDup.dist7 MEFs, Li Han for her work on the initial phases of this project and Diana Tran for her comments on the manuscript. This work was supported by a Public Health Service grant (GM064378) from the National Institute of General Medicine to P.E.S.
Footnotes
Protein A/G agarose beads are preferred over either Protein A or G because they bind nearly all isotypes.
The protein A/G beads are preblocked to reduce non-specific binding.
Gently swirl the beads to uniformly resuspend them before taking an aliquot. Vigorous shaking should be avoided, because the beads may stick to the side of tube and dry. Also, vortexing or high speed centrifugation may break the beads.
For most histone modifications N- linked chromatin works well. For certain non-histone proteins like CTCF insulator and transcription factors X-linking is preferred. The optimal crosslinking strength should be experimentally determined for each antibody by ChIP, real-time PCR and allele-specific protocols.
Mouse embryos/organs can be used for chromatin preparation. The embryo/organ is first suspended in PBS and crosslinked in suspension [9].
Keep in mind that RNAse step is not necessary in this DNA preparation. Therefore, the DNA will have some contaminating RNA (4 µg nucleic acid equivalent chromatin has less than 4 µg DNA; 100 K fetal germ cells contain 600 ng DNA and ~1 µg total nucleic acids).
A <1.2kb smear on the gel is considered optimal shearing with ~500 bp as median fragment size. It is critical to obtain optimal size distribution of sonicated chromatin as fragment size >1.2 kb may result in high background from neighboring chromosomal regions. Fragment size <200bp may result in inadequate labeling yields for downstream ChIP-chip experiments. Fragmentation needs to be optimized with each sonication device and cell types being used for the experiment.
The pre-clearing step inhibits non-specific DNA and protein binding to Protein A/G beads and thereby reduces the background. Preclearing can be done in large amount for multiple ChIPs by multiplying the volumes in Table 1.
Set up your first ever ChIP using 4 µg DNA equivalent chromatin and well-characterized antibodies, such as those in Figure 3, which give reliable allele-specific enrichment at DMRs.
It is a good practice to test new batches of commercial antibodies for specificity in a dot blot before using them in ChIP [9].
A1:1 (4 µg antibody and 4 µg chromatin) ratio usually works for histone modifications. However, it is best to determine optimal concentrations for each antibody. Polyclonal antibodies give better results than monoclonal ones in ChIP.
Washing buffers can be prepared beforehand and stored at 4°C. Add 50× cØmplete protease inhibitor just before use.
1ml pipette tips can be used for washes. Gently dispense washing buffers from the side of the tubes. Avoid touching the beads when taking out supernatant after washes.
Remove buffer leaving ~50 µl behind, spin again and use a 100 µl pipette tip to take out as much buffer as possible.
Prepare elution buffer not more than 10 min before use.
Reverse-crosslinking removes the protein and allows purifying genomic DNA. Proteinase K treatment digests proteins in the DNA-protein mix.
65 °C and 55 °C temperatures should be maintained during reverse crosslinking and proteinase K treatments for best results. Rotating of the samples is not required.
ChIP protocols suggest stopping/freezing after reverse crosslinking or protienase K treatment and continuing the process the next day. In our hands finishing all the steps on the second day gives the best results.
If washes with PE buffer are done less than four times, SDS from the elution buffer may remain and you may see SDS crystals in the eluate.
Repeat freeze-thawing or long-term storage of ChIP-precipitated DNA should be avoided.
The ChIP-precipitated DNA can be extracted using phenol:chloroform and precipitated using linear polyacrylamide or glycogen (small scale ChIP). However, if ChIP DNA is to be amplified for ChIP -on chip, it is best to use Qiagen columns for purification, which removes excess salts.
The copy number of immunprecipitated DNA should be higher for specific antibodies than for nonspecific IgG. Using real-time PCR we generally measure less than 10 copies for nonspecific IgG and above 20 copies, up to 10000 copies for specific antibodies from a 3 µl ChIP eluate (Figure 2) [9]. The downstream allele-specific measurement is more accurate with higher copy numbers for the specific region in the ChIP elution.
ChIPs and downstream allele-specific allelotyping assays should be done in duplicates or triplicates. Small standard deviation in these tests (Figure 3) indicates that the precipitation worked and there is real enrichment with a specific antibody at a specific site. High standard deviation is a warning signal suggesting that the PCR randomly amplified background precipitation.
Reciprocal mouse crosses substantiate the allele-specific findings (Figure 3).
Use the Beacon Designer software for four-color real time qPCR probe and primer design for other regions or consult our paper for more DMR sets [9]. Multiplexing in the real-time PCR provides internal controls, saves time and importantly, saves the majority of the ChIP elution for downstream processes, such as sequenom-allelotyping or amplification for microarray hybridization.
References
- 1.Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647(1–2):77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koerner MV, Barlow DP. Genomic imprinting-an epigenetic gene-regulatory model. Curr Opin Genet Dev. 2010;20(2):164–170. doi: 10.1016/j.gde.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53(6):937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 4.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11(2):205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 5.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. Embo J. 1988;7(5):1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLaughlin KJ, et al. Mouse embryos with paternal duplication of an imprinted chromosome 7 region die at midgestation and lack placental spongiotrophoblast. Development. 1996;122(1):265–270. doi: 10.1242/dev.122.1.265. [DOI] [PubMed] [Google Scholar]
- 7.Singh P, et al. Chromosome-wide analysis of parental allele-specific chromatin and DNA methylation. Mol Cell Biol. 2011;31(8):1757–1770. doi: 10.1128/MCB.00961-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh P, et al. Allele-specific H3K79 Di- versus trimethylation distinguishes opposite parental alleles at imprinted regions. Mol Cell Biol. 2010;30(11):2693–2707. doi: 10.1128/MCB.01537-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh P, et al. Coordinated allele-specific histone acetylation at the differentially methylated regions of imprinted genes. Nucleic Acids Res. 2010;38(22):7974–7990. doi: 10.1093/nar/gkq680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee DH, et al. CTCF-dependent chromatin bias constitutes transient epigenetic memory of the mother at the H19-Igf2 imprinting control region in prospermatogonia. PLoS Genet. 2010;6(11):e1001224. doi: 10.1371/journal.pgen.1001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaillard C, Strauss F. Ethanol precipitation of DNA with linear polyacrylamide as carrier. Nucleic Acids Res. 1990;18(2):378. doi: 10.1093/nar/18.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]