Abstract
Aromatic amines and heterocyclic aromatic amines (HAAs) are a class of structurally related carcinogens that are formed during the combustion of tobacco or during the high temperature cooking of meats. These procarcinogens undergo metabolic activation by N-oxidation of the exocyclic amine group to produce N-hydroxylated metabolites, which are critical intermediates implicated in toxicity and DNA damage. The arylhydroxylamines and their oxidized arylnitroso derivatives can also react with cysteine (Cys) residues of glutathione or proteins to form, respectively, sulfenamide and sulfinamide adducts. However, sulfur-nitrogen linked adducted proteins are often difficult to detect because they are unstable and undergo hydrolysis during proteolytic digestion. Synthetic N-oxidized intermediates of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a carcinogenic HAA produced in cooked meats, and 4-aminobiphenyl, a carcinogenic aromatic amine present in tobacco smoke were reacted with human serum albumin (SA) and formed labile sulfenamide or sulfinamide adducts at the Cys34 residue. Oxidation of the carcinogen-modified SA with m-chloroperoxybenzoic acid (m-CPBA) produced the arylsulfonamide adducts, which were stable to heat and the chemical reduction conditions employed to denature SA. The sulfonamide adducts of PhIP and 4-ABP were identified, by liquid chromatography/mass spectrometry, in proteolytic digests of denatured SA. Thus, selective oxidation of arylamine-modified SA produces stable arylsulfonamide-SA adducts, which may serve as biomarkers of these tobacco and dietary carcinogens.
Introduction
Aromatic amines and heterocyclic aromatic amines (HAAs) are a class of structurally related carcinogens that are formed during the combustion of tobacco or during the high temperature cooking of meats.1,2 Certain aromatic amines, such as 4-aminobiphenyl (4-ABP) and 2-naphthylamine (2-NA) are classified as human carcinogens (Group 1), and several prevalent HAAs, including 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), which is present in tobacco smoke and well-done cooked meats, have been listed as probable or possible human carcinogens (Group 2A or 2B), on the basis of toxicity data reviewed by the International Agency for Research on Cancer.3,4 Thus, there is much concern about the health risk associated with the exposure to these structurally related classes of chemicals.
There is a critical need to establish long-term biomarkers that can be implemented in molecular epidemiology studies to evaluate the health risk of HAAs and aromatic amines. DNA and protein adducts are a measure of the biologically effective dose and have been used for human risk assessment of environmental and dietary genotoxicants.5,6 However, DNA adduct measurements are often precluded by the unavailability of biopsy samples in large scale human studies. Protein carcinogen adducts of hemoglobin (Hb) and serum albumin (SA) have been used as an alternative to DNA adducts for biomarkers of several different classes of genotoxicants.6,7 Stable carcinogen protein adducts are expected to accumulate and follow the kinetics of the lifetime of Hb or half-life of SA, during chronic exposure, and augment the sensitivity of adduct detection.8
Arylnitroso compounds are reactive metabolites of arylamines that bind to Cys residues of glutathione and proteins to form arylsulfinamide adducts.6,9,10 The occurrence of arylsulfinamide adducts in intact proteins has been shown by mass spectrometric analysis.11,12 However, arylsulfinamide adducts undergo hydrolysis to form the arylamines and cysteine sulfinic acid during the heat and chemical reduction conditions employed to denature proteins,13,14 or during proteolysis (Scheme 1).6,12 The site of carcinogen adduction is inferred by tandem mass spectrometry (MS/MS) sequencing and identification of the sulfinic acid residue in peptides from protease digests.11,13,14 In the case of 4-ABP, the location and identity of the sulfinamide adduct at the human Hb-Cys93β chain was proven by X-ray crystallography.15 Following that hallmark study, the sulfinamide adducts of several arylamines were measured indirectly as the parent amines following mild acid treatment of Hb.16 However, covalent binding of N-oxidized metabolites of HAAs to Hb is low and alternative protein carcinogen adduct biomarkers have been sought.2
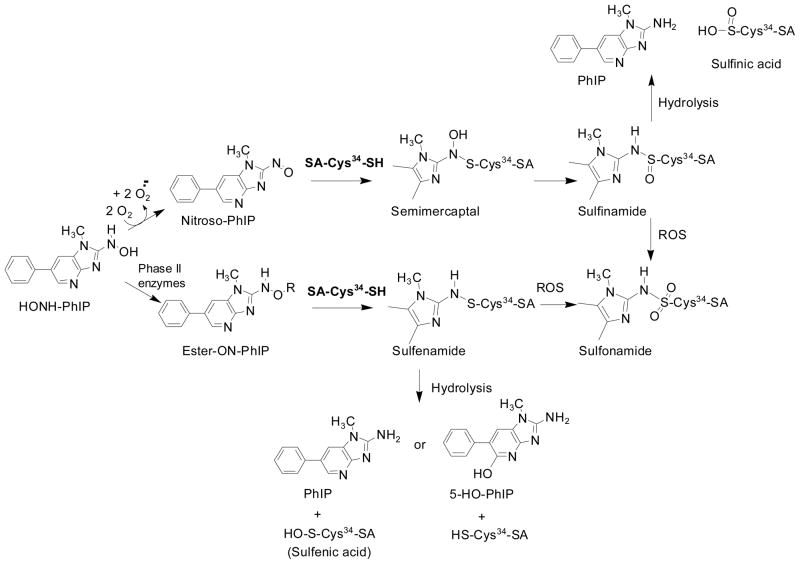
Scheme 1.
Formation of SA-Cys34 sulfenamide, sulfinamide, and sulfonamide adducts with N-oxidized metabolites of PhIP. The genotoxic metabolite HONH-PhIP undergoes oxidation to form Nitroso-PhIP, which forms a sulfinamide adduct, following rearrangement of the semimercaptal. HONH-PhIP forms reactive sulfate or acetate esters following conjugation reactions with phase II enzymes. These esters of HONH-PhIP react with SA to form the sulfenamide adduct at Cys34. The superoxide generated during the oxidation of HONH-PhIP to Nitroso-PhIP, or other reactive oxygen species (ROS), can convert the labile sulfenamide and sulfinamide adducts of SA-Cys34-PhIP to the stable sulfonamide linkage.
Many genotoxicants also bind covalently to human SA.6–8,17 PhIP was reported to bind to human SA in vivo, but structures of the PhIP-SA adduct(s) are unknown.18 We and others have shown that N-oxidation products of PhIP covalently bind to human SA in vitro.14,19–21 The genotoxic metabolites 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (HONH-PhIP) and 2-nitroso-1-methyl-6-phenylimidazo[4,5-b]pyridine (NO-PhIP) were reacted with SA in vitro to produce the N2-[cystein-S-yl-PhIP]-S-oxide at Cys34.14 This sulfinamide adduct is also labile toward Pronase or even weak acid, and some portion of the PhIP bound to SA in vivo may exist as the sulfinamide.18 N-(Acetyloxy)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-acetoxy-PhIP) a penultimate metabolite of PhIP that forms covalent adducts with DNA, did not form stable adducts with human SA. Instead, PhIP and 2-amino-1-methyl-6-(5-hydroxy)-phenylimidazo[4,5-b]pyridine (5-HO-PhIP), an aqueous reaction product of the proposed nitrenium ion of PhIP, were identified in the proteolytic digest of modified SA.21 We surmise that N-acetoxy-PhIP reacted with SA to from an unstable sulfenamide adduct at Cys34, because the pretreatment of SA with thiol specific-reagents prior to reaction with N-acetoxy-PhIP greatly decreased the recovery of PhIP and 5-HO-PhIP. (Scheme 1).19–21
During the course of characterizing SA adduction products with PhIP, we discovered that a portion of the sulfinamide adduct at Cys34 underwent oxidation to the sulfonamide, N2-[cystein-S-yl-PhIP]-S-dioxide.14,21 In contrast to the instability of many arylsulfenamide and arylsulfinamide linkages, the sulfur-nitrogen bond of the SA-PhIP sulfonamide adduct is stable toward acid, heat, and reducing agents (Peng, L., unpublished observations). We discovered that 4-hydroxaminobiphenyl (HONH-4-ABP), the carcinogenic metabolite of 4-ABP,22 also reacts with SA to form a sulfinamide linkage and undergoes oxidation to produce the sulfonamide. Thus, selective chemical oxidation of SA may allow us to biomonitor labile Cys34 adducts of HAAs and arylamines through these newly discovered sulfonamide adducts.
Experimental Section
Caution: PhIP and 4-ABP are carcinogens and should be handled in a well-ventilated fume hood with the appropriate protective clothing
Chemicals and Materials
PhIP and 2-amino-1-[2H3]-methyl-6-phenylimidazo[4,5-b]pyridine ([2H3]-PhIP, 99% isotopic purity) were purchased from Toronto Research Chemicals (Toronto, ON, Canada). 2-Amino-1-methyl-6-[2H5]-phenylimidazo[4,5-b]pyridine ([2H5]-PhIP, 99% isotopic purity) was a gift from Dr. Mark Knize and Dr. Kristen Kulp, (Lawrence Livermore National Laboratory, Livermore, CA). Human SA, β-mercaptoethanol, iodoacetamide (IAA), dithiothreitol (DTT), m-chloroperoxybenzoic acid (m-CPBA), and 4-nitrobiphenyl were obtained from Sigma-Aldrich (St. Louis, MO). LQQCPF was obtained from Biosynthesis (Lewisville, TX). Sequencing grade trypsin and chymotrypsin were purchased from Promega (Madison, WI). Pronase E, leucine aminopeptidase, and prolidase were purchased from Sigma (St. Louis, MO). All solvents were high-purity B & J Brand from Honeywell Burdick and Jackson (Muskegon, MI). Isolute C18 SPE columns (25 mg) were from Biotage (Charlotte, NC). Spire PEP tips were a gift from Thermo Scientific (Bellefonte, PA).
Synthesis of HONH-PhIP, 2-Nitroso-1-methyl-6-phenylimidazo[4,5-b]pyridine (NO-PhIP), N-Acetoxy-PhIP, 5-HO-PhIP, and HONH-4-ABP
HONH-PhIP and HONH-[2H5]-PhIP, HONH-4-ABP were prepared by reduction of their respective nitro derivatives with hydrazine, using Pd/C as a catalyst.23 NO-PhIP was obtained by oxidation of HNOH-PhIP with K3Fe(CN)6.14 N-Acetoxy-PhIP was prepared by reaction of HONH-PhIP with acetic anhydride,21 and 5-HO-PhIP was obtained by solvolysis of N-acetoxy-PhIP.21,24
Synthesis of PhIP Sulfinamide and Sulfonamide Adducts with LQQCPF
LQQCPF was reacted with NO-PhIP in H2O to form the PhIP sulfinamide LQQC*PF (C-[S=O]-PhIP).14 The sulfonic acid of LQQCPF and the sulfonamide adduct of PhIP were obtained by oxidation of LQQCPF or LQQC*PF (C-[S=O]-PhIP) (1 nmol) with m-CPBA (1.5 nmol) in 0.1 M phosphate buffer (pH 7.4) at room temperature for 30 min. Another 1.5 nmol of m-CPBA was added and the reaction continued for 1 h. Thereafter, the peptides were enriched by SPE and LQQC*PF(C-[SO2H]) and PhIP sulfonamide LQQC*PF (C-[SO2]-PhIP) were purified by HPLC.14 The identities of the adducts were confirmed by MS/MS (vide infra).
Modification of Human SA with N-Oxidized Derivatives of PhIP and 4-ABP
Human SA was pretreated with β-mercaptoethanol to reduce mixed disulfides formed at Cys34.13 Following PD-10 gel filtration in 0.1 M potassium phosphate buffer (pH 7.4), the sulfhydryl content of SA was determined using Ellman’s reagent.25 The SA reduced with s-mercaptoethanol contained a sulfhydryl content of 0.95 mol SH/mol SA. SA (1 mg, 15 nmol) was reacted with HONH-PhIP, N-acetoxy-PhIP or HONH-4-ABP (15 nmol, 40 μL C2H5OH) in 10 mM potassium phosphate buffer (1 mL, pH 7.4). The reactions of HONH-PhIP and HONH-ABP with SA were performed at 37 °C for 18 h, whereas the reaction of N-acetoxy-PhIP with SA was performed at 37 °C for 2 h. Thereafter, unbound carcinogen was removed with ethyl acetate, and the modified SA was run through a PD-10 gel filtration column in 0.1 M phosphate buffer (pH 7.4).
Oxidation of PhIP-Modified Human SA with m-CPBA
Unmodified or PhIP-modified SA (0.5 mg, 7.5 nmol) was oxidized with m-CPBA for 4 h in 0.1 M phosphate buffer (pH 7.4, 1 mL) with a protein/m-CPBA molar ratio of 1:10, 1:50, 1:100, or 1:200. The samples were then incubated for 1 h with a Branson 2510 model sonicator. Thereafter, the SA was run through a PD-10 gel filtration column in 50 mM ammonium bicarbonate (pH 8.5). The oxidation of PhIP-modified SA was also examined as a function of time with m-CPBA (protein/m-CPBA molar ratio = 1:50) at room temperature for 1, 3, 5, or 17 h, followed by sonication for 1 h. Protein samples were processed by gel filtration to remove the oxidant.
Proteolytic Digestion
Enzymes were prepared as described previously.14 Two different proteolytic conditions were employed to digest SA adducts modified by HONH-PhIP and HONH-4-ABP.
Trypsin/chymotrypsin digestion: The PhIP- or 4-ABP-modified SA (10 μg, 150 pmol, in 50 μL 50 mM ammonium bicarbonate buffer (pH 8.5) containing CaCl2 (1 mM) was digested with trypsin at a protease:protein ratio of 1:50 (w/w) and chymotrypsin at a protease:protein ratio of 1:12.5 (w/w) at 37° C for 20 h, followed by adduct enrichment with SPE.14,21 In some instances, the PhIP-modified SA (0.5 mg) was denatured prior to enzyme digestion by heating the protein in 6 M guanidine–HCl (pH 7.4, 0.5 mL) with DTT (20 mM) at 55 °C for 1 h, followed by alkylation with excess IAA.21 The SA was then subjected to gel filtration chromatography with the PD-10 column in 50 mM ammonium bicarbonate buffer (pH 8.5).
Pronase E/leucine aminopeptidase/prolidase digestion: PhIP- or 4-ABP-modified SA (10 μg, 150 pmol) in 50 mM ammonium bicarbonate buffer (pH 8.5) containing 1 mM MnCl2 was treated with Pronase E at a protease:protein ratio of 1:2 (w/w), leucine aminopeptidase at a protease:protein ratio of 1:30 (w/w), and prolidase at a protease:protein ratio of 1:8 (w/w). The digestion was conducted at 37° C for 20 h, followed by adduct enrichment with SPE.14,21
Estimates PhIP-Cys34 SA Adducts
LQQC*PF (C-[SO2]-[2H5]-PhIP) sulfonamide (7.9 pmol), LQQC*PF (C-[S=O]-[2H5]-PhIP) sulfinamide (12.3 pmol), and [2H3]-PhIP (3.95 pmol) were added as internal standards to PhIP-modified SA (10 μg protein) prior to proteolysis. The major adducts were identified as the missed cleavage adducts LQQC*PFEDHVK (vide infra). The ionization efficiencies of the mixed cleavage peptide adducts were assumed to be comparable to the ionization efficiencies of the internal standards of [2H5]-PhIP-modified LQQC*PF.
Ultraperformance Liquid Chromatography-Electrospray Ionization/Multistage Mass Spectrometry (UPLC-ESI/MSn)
A NanoAcquity UPLC system (Waters Corp., Milford, MA) equipped with a Magic C18AQ column (0.3 × 150 mm, Michrom Bioresource Inc., Auburn, CA) was employed for chromatography. The peptides were resolved with a linear gradient starting from 90% A solvent (0.01% HCO2H) to 100% B solvent (95% CH3CN containing, 4.99% H2O, and 0.01% HCO2H) over 20 min, at a flow rate of 5 μL/min. MS spectra were acquired with a linear quadrupole ion trap mass spectrometer (LTQ, Thermo Fisher, San Jose, CA). Analyses were conducted in the positive ionization mode and employed an Advance CaptiveSpray™ source from Michrom Bioresource Inc. The temperature of capillary tube was set at 200 °C; the spray voltage was 1.5 kV; and the in-source fragmentation was 10 V. There was no sheath or auxiliary gas. The automatic gain control (AGC) settings were full MS target 30,000 and MSn target 10,000; the maximum injection time was 10 ms, and one microscan was used for data acquisition. Xcalibur version 2.07 software was used for data manipulations. The tandem mass spectra of peptides were interpreted manually and facilitated by online software (Protein Prospector, Univ. of California, San Francisco, http://us.expasy.org/proteomics).
Results and Discussion
The Cys34 residue of human SA is an important adduction site for electrophilic N-oxidized metabolites of PhIP.14,21 Treatment of SA or plasma with HNOH-PhIP produced the N2-cysteinesulfinyl-PhIP adduct: greater than 90% of the HONH-PhIP bound to the Cys34 of SA.14 The absence of other PhIP-SA adducts signifies that the reactivity of HONH-PhIP with SA is poor and adduct formation only occurs following the oxidation of HONH-PhIP to NO-PhIP. The sulfhydryl SH of Cys34 reacts with the N=O bond of NO-PhIP to form a semimercaptal, which undergoes rearrangement to form the more stable sulfinamide structure (Scheme 1). During the course of analysis, the sulfonamide adduct of PhIP was also identified.21 The mechanism of formation of this adduct is not known. The superoxide anion slowly generated over the time, by HONH-PhIP,26 may have oxidized a portion of the existing N2-[cystein-S-yl-PhIP]-S-oxide linkage to the N2-[cystein-S-yl-PhIP]-S-dioxide adduct (Scheme 1). Alternatively, a portion of the N2-[cystein-S-yl-PhIP]-S-oxide adduct may have undergone oxidation during enzymatic digestion of SA.
Oxidation of LQQCPF and N2-Cysteinesulfinyl-PhIP Peptide LQQC*PF (C-[S=O-PhIP])
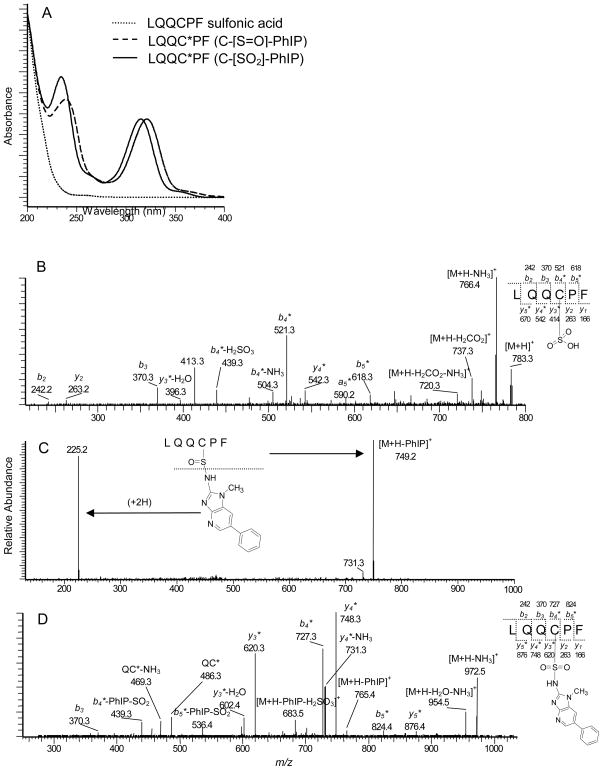
The goal was to oxidize the N2-cysteinesulfinyl-PhIP adduct of SA to form the chemically stable sulfonamide linkage in high yield. Initially, LQQCPF, a target peptide recovered from human SA digested with trypsin and chymotrypsin,14,27 was treated with several different oxidants. The oxidation of LQQCPF [MH+ observed at m/z 735.3] with m-CPBA produced the sulfonic acid LQQC*PF (C-[SO2H]) [MH+ observed at m/z 783.3] in higher yields (80%) than the oxidation reactions conducted with Fenton reagents (Fe2+/H2O2) or NaOCl (Peng, L., unpublished observations).28 Thereafter, the oxidation of PhIP sulfinamide LQQC*PF (C-[S=O]-PhIP) peptide was examined: treatment with m-CPBA converted LQQC*PF (C-[S=O]-PhIP) to the sulfonamide LQQC*PF (C-[SO2]-PhIP) adduct [MH+ observed at m/z 989.5]. The UV and product ion spectra of the oxidized sulfonic acid peptide LQQC*PF (C-[SO2H]), and the sulfinamide and sulfonamide adducts of PhIP formed with LQQC*PF are shown in Figure 1.
Figure 1.
(A) UV spectra of modified LQQC*PF peptides and ESI/MS product ion spectra of (B) LQQC*PF sulfonic acid ([M + H]+ at m/z 783.3), (C) LQQC*PF (C-[S=O]-PhIP) sulfinamide ([M+H]+ at m/z 973.3), and (D) LQQC*PF (C-[SO2]-PhIP) sulfonamide ([M+H]+ at m/z 989.5).
Oxidation of N2-cysteinesulfinyl-PhIP at Cys34 of Human SA, by m-CPBA, to Form the N2-Cysteinesulfonamide-PhIP Adduct
The condition of oxidation of the SA-N2-cysteinesulfinyl-PhIP adduct with m-CPBA was optimized to produce the arylsulfonamide linkage in high yield. A 50-fold mole excess of m-CPBA over protein produced the highest amount of the sulfonamide adduct within a period of 6 h (Supporting Information, Figure S-1).
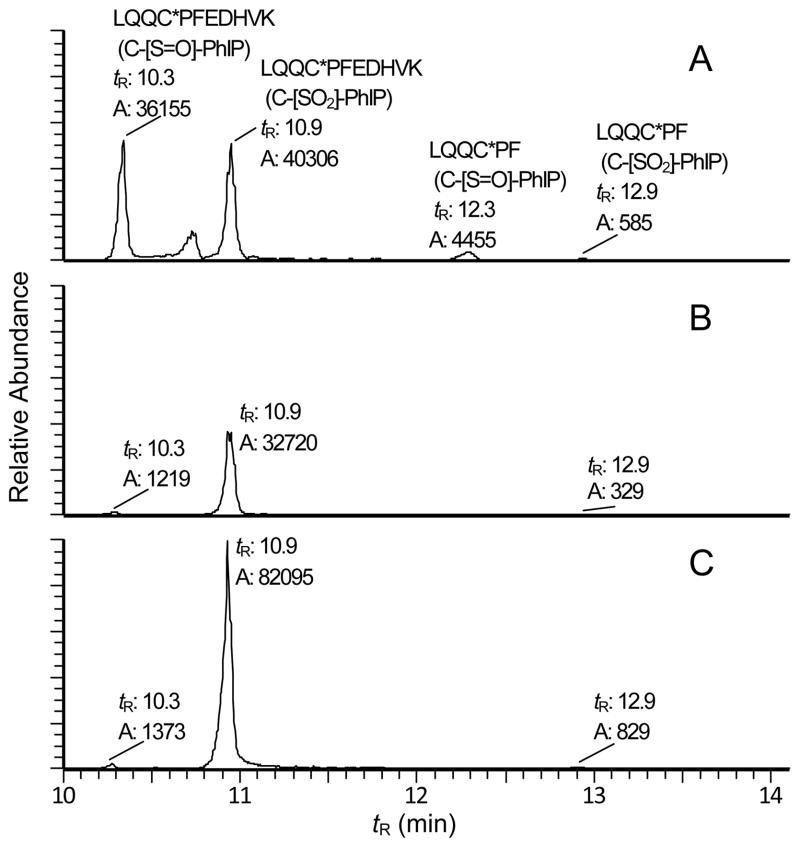
The stability of the sulfinamide and sulfonamide adducts of HOHN-PhIP-modified SA under the conditions of denaturation, which included heat and chemical reduction of SA disulfide bonds with DTT, followed by alkylation of the unfolded SA with IAA, were assessed. The mass chromatograms of the Cys34 peptide adducts recovered from the trypsin/chymotrypsin digests of SA are shown in Figure 2. The sulfinamide LQQC*PF (C-[S=O]-PhIP) and sulfonamide LQQC*PF (C-[SO2]-PhIP) adducts, as well as the missed cleavage adducts LQQC*PFEDHVK (C-[S=O]-PhIP) and LQQC*PFEDHVK (C-[SO2]-PhIP), are detected in the nonoxidized, nondenatured SA sample (Figure 2A). However, the sulfinamide adduct underwent hydrolysis during protein denaturation, and only the sulfonamide peptides LQQC*PF (C-[SO2]-PhIP) and LQQC*PFEDHVK (C-[SO2]-PhIP) were identified in the digest of denatured SA, either without (Figure 2B) or with m-CPBA treatment (Figure 2C). The missed cleavage adducts LQQC*PFEDHVK were the predominant peptides identified in both nondenatured and denatured samples. IAA-modified LQQC*PFEDHVK was also recovered in higher amount than IAA-modified LQQC*PF in denatured SA, and the sulfonic acid peptide LQQC*PFEDHVK (C-[SO2H]) was recovered in higher yield than LQQC*PF (C-[SO2H]) in m-CPBA-oxidized SA, irrespective of denaturation of the protein (Supporting Information, Figure S-2). Thus, the proline residue and not the PhIP-Cys adduct in LQQC*PFEDHVK, impeded the efficacy of digestion by chymotrypsin. The employment of larger quantities of chymotrypsin overcame some of this missed cleavage peptide (Peng, L, unpublished observations).
Figure 2.
UPLC-ESI/MS2 chromatograms of Cys34 peptide adducts recovered from 100 ng of SA digest of (A) HONH-PhIP-modified SA, following trypsin/chymotrypsin digestion; (B) HONH-PhIP-modified SA, which was denatured by heating the SA in 6 M guanidine–HCl with DTT, followed by alkylation with IAA, prior to trypsin/chymotrypsin digestion; and (C) HONH-PhIP-modified SA was oxidized with m-CPBA prior to denaturation and digestion with trypsin/chymotrypsin. Targeted UPLC-ESI/MS2 was done on the following peptides: LQQC*PFEDHVK (C-[S=O]-PhIP) as triply charged species ([M+3H]3+ at m/z 527.9); LQQC*PFEDHVK (C-[SO2]-PhIP) ([M+3H]3+ at m/z 533.2); LQQC*PF (C-[S=O]-PhIP) ([M+H] + at m/z 973.3); and LQQC*PF (C-[SO2]-PhIP) ([M+H]+ at m/z 989.4). The scale of the signals for all 3 treatments were normalized to the response of LQQC*PFEDHVK (C-[SO2]-PhIP) in panel C.
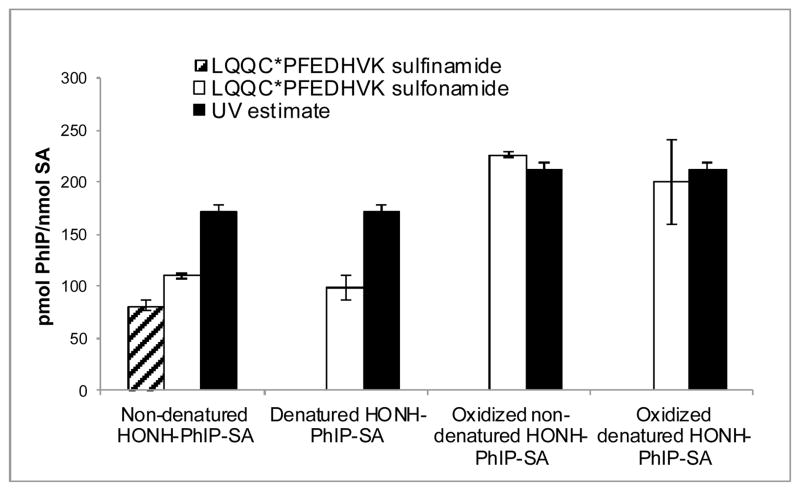
The estimates of LQQC*PFEDHVK (C-[S=O]-PhIP) and LQQC*PFEDHVK (C-[SO2]-PhIP) adduct formation in HONH-PhIP-modified SA, either without or with oxidation with m-CPBA and without or with denaturation of SA prior to trypsin/chymotryspin digestion, are depicted in Figure 3. The treatment of HONH-PhIP-modified SA with m-CPBA efficiently oxidized the sulfinamide LQQC*PFEDHVK (C-[S=O]-PhIP) to the sulfonamide LQQC*PFEDHVK (C-[SO2]-PhIP). Comparable amounts of LQQC*PFEDHVK (C-[SO2]-PhIP) were recovered from m-CPBA-oxidized SA for the denatured and non-denatured SA samples, signifying that the PhIP sulfonamide adduct is stable under the conditions employed for denaturation. The level of LQQC*PFEDHVK (C-[SO2]-PhIP) recovered from HONH-PhIP-modified SA accounted for ≥95% of the PhIP bound to SA on the basis of UV measurement of PhIP-modified SA at 320 nm.14 A comparable estimate of the sulfonamide adduct was obtained by measurement of tripeptide C*PF (C-[SO2]-PhIP) in m-CPBA oxidized PhIP-modified SA, following digestion with Pronase E/leucine aminopeptidase/prolidase (Supporting Information, Figure S-3).
Figure 3.
Estimates of LQQC*PFEDHVK adducts recovered from trypsin/chymotrypsin digestion of m-CPBA-oxidized or nonoxidized HONH-PhIP-modified SA, without or with denaturation. Adduct formation was estimated by UPLC-ESI/MS2, employing LQQC*PF (C-[SO2]-[2H5]-PhIP) sulfonamide and LQQC*PF (C-[S=O]-[2H5]-PhIP) sulfinamide as internal standards. The ionization efficiencies of the mixed cleavage peptide adducts LQQC*PFEDHVK were assumed to be comparable to the ionization efficiencies of the internal standards of [2H5]-PhIP-modified LQQC*PF. Total PhIP bound to SA was determined by UV measurement at 320 nm.14
Trapping Labile Putative N2-Cysteinesulfenamide Adduct of PhIP at Cys34 of Human SA, by m-CPBA Oxidation, to Form the N2-Cysteinesulfonamide Adduct
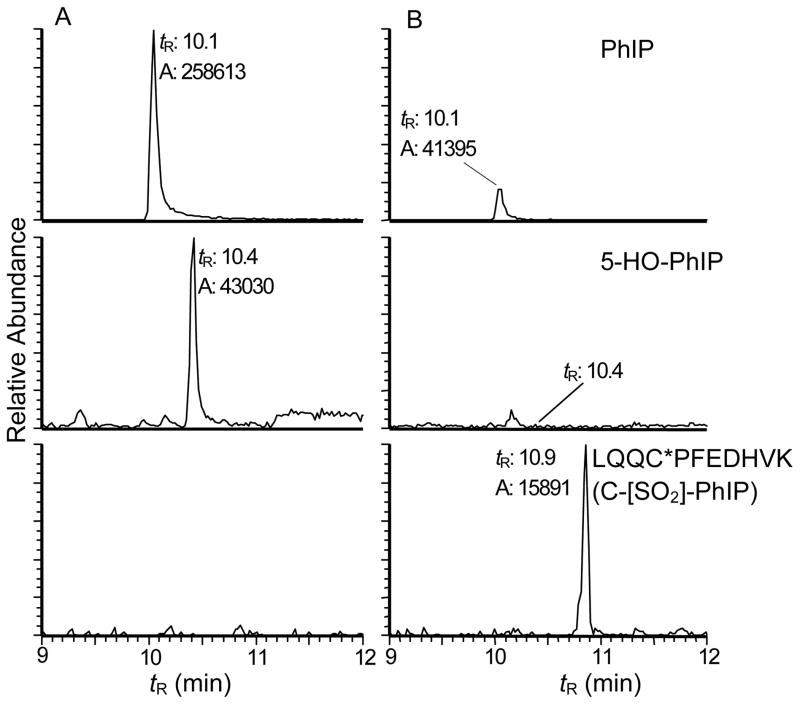
The mass chromatograms of the tryptic/chymotryptic digest of N-acetoxy-PhIP-modified SA either without or with oxidation by m-CPBA are shown in Figure 4. There was no evidence for sulfur-nitrogen PhIP adducts (sulfenamide, sulfinamide or sulfonamide adducts of LQQC*PFEDHVK) in the non-oxidized SA sample, whereas the sulfonamide LQQC*PFEDHVK (C-[SO2]-PhIP) was identified in the m-CPBA-treated SA sample. Moreover, the amount of PhIP and 5-HO-PhIP recovered in the m-CPBA treated SA sample was ~80% lower than those levels present in the non-oxidized SA sample. These findings suggest that a labile sulfenamide adduct had formed at Cys34 in N-acetoxy-PhIP modified SA (Scheme 1): the treatment of the modified SA with m-CPBA oxidized the sulfur-nitrogen linkage to the stable PhIP sulfonamide adduct.
Figure 4.
Reconstructed ion chromatograms of N-acetoxy-PhIP-modified SA monitoring: PhIP ([M+H]+, m/z 225.1 → 210.1); 5-HO-PhIP ([M+H]+, m/z 241.1 → 210.1); and LQQC*PFEDHVK (C-[SO2]-PhIP) ([M+3H]3+, m/z 533.2 → 225.2, 521.8, 670.5) in (A) Non-oxidized SA and (B) m-CPBA-oxidized SA, digested with trypsin/chymotrypsin. The scales of the signals were fixed to the maximum response of the respective analytes between the nonoxidized and oxidized samples.
Sulfinamide and Sulfonamide Adduct Formation of HONH-4-ABP at Cys34 of Human SA
The reactivity of SA with HONH-4-ABP, the carcinogenic metabolite of 4-ABP,22 was examined. Similar to the results obtained with HONH-PhIP-modified SA, the missed cleavage sulfinamide (C-[S=O]-4-ABP) and sulfonamide (C-[SO2]-4-ABP) adducts of LQQC*PFEDHVK were recovered in higher yields than the LQQC*PF adducts in the trypsin/chymotrypsin digest of SA (non-oxidized and non-denatured), as depicted in Figure 5. Following oxidation with the m-CPBA, the total ion counts of sulfonamide adduct (C-[SO2]-4-ABP) was 10-fold greater than the ion counts of the sulfinamide linked adduct (C-[S=O]-4-ABP), demonstrating that the oxidation of the sulfur-nitrogen linkage, by m-CPBA, was efficient (data not shown).
Figure 5.
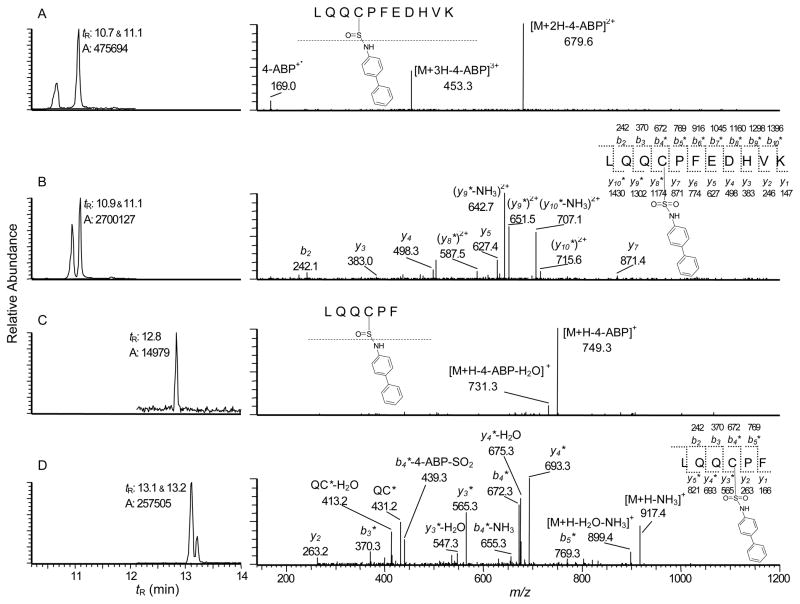
UPLC-ESI/MS2 chromatograms of Cys34 peptide adducts recovered from HONH-4-ABP-modified SA following digestion with trypsin/chymotrypsin (left panel) and corresponding product ion mass spectra of (A) LQQC*PFEDHVK (C-[S=O]-4-ABP) sulfinamide ([M+3H]3+ at m/z 509.6), (B) LQQC*PFEDHVK (C-[SO2]-4-ABP) sulfonamide ([M+3H]3+ at m/z 514.9), (C) LQQC*PF (C-[S=O]-4-ABP) sulfinamide ([M+H]+ at m/z 918.4, and (D) LQQC*PF (C-[SO2]-4-ABP) sulfonamide ([M+H]+ at m/z 934.4).
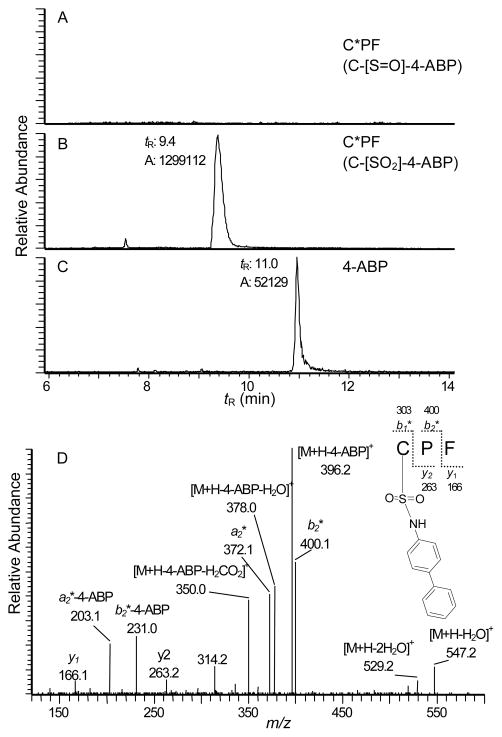
Pronase alone or in combination with leucine aminopeptidase and prolidase has been employed to digest a number of carcinogens adducted at the Cys34 of rat or human SA. For many carcinogen adducts, including the N2-cysteinsulfinyl adduct of the HAA, 2-amino-3-methylimidazo[4,5-f]quinoline formed with rat SA,13 and Cys34 adducts of human SA derived from the reactive N-acetyl-p-benzoquinoneimine metabolite of acetaminophen,29 sulfur mustard,30 and acrylamide31 do not undergo digestion beyond the tripeptide stage of rat (C*PY) or human (C*PF) SA treated with Pronase. Consistent with those findings, the digestion of HONH-4-ABP-modified human SA (nonoxidized and nondenatured) with Pronase E/leucine aminopeptidase/prolidase also produced a prominent tripeptide adduct at CP*F. The UPLC-ESI/MS2 chromatogram (Figure 6) of the digest shows the presence of the sulfonamide CP*F (C-[SO2]-4-ABP) along with 4-ABP, but the sulfinamide (C-[S=O]-4-ABP), which is labile, underwent complete hydrolysis during enzyme digestion with Pronase E.
Figure 6.
UPLC-ESI/MS2 chromatograms of (A) a putative C*PF (C-[S=O]-4-ABP) sulfinamide adducts ([M+H]+ at m/z 549.2), (B) C*PF (C-[SO2]-4-ABP) sulfonamide ([M+H]+ at m/z 565.2), and (C) 4-ABP ([M+H]+, m/z 170.1 → 153.1) recovered from HONH-4-ABP-modified SA digested with Pronase E, leucine aminopeptidase, and prolidase. The scale of the signal of the putative C-[S=O]-4-ABP adduct, which was not detected, was normalized to the scale of response of (C-[SO2]-4-ABP). D) Product ion mass spectrum of C*PF (C-[SO2]-4-ABP) sulfonamide adduct.
Conclusions
Many arylsulfinamide protein adducts undergo extensive hydrolysis during protein denaturation or proteolytic digestion to form the arylamines and cysteine sulfinic acid of the protein. As a consequence, the analysis of arylsulfinamide peptide adducts is a challenging analytical task. The sulfinamide adducts of several one- and two-ring aromatic amines formed at the Hb-Cys93β chain have been measured indirectly as their liberated parent amines, following mild acid treatment of Hb (Scheme 1).16 However, aromatic amines and HAAs having a larger ring system do not form Hb sulfinamide adducts in vivo, perhaps because alternative metabolic pathways predominate or because N-oxidized metabolites do not efficiently penetrate the erythrocyte or do not react with the heme moiety to produce the reactive nitrosoarene intermediates.6
The nucleophilic Cys34 residue of human SA reacts with many genotoxicants and toxic electrophiles to form covalent adducts,17,32 and several putative labile arylsulfinamide adducts have been detected.6,13,14,33 The formation of arylamine adducts with SA does not require the transport of the arylhydroxylamine or the arylnitroso metabolite to the blood since SA is synthesized in the hepatocyte,34 the cell where the arylhydroxylamine metabolites are formed by cytochrome P450 enzymes.2 The Cys34 of SA is also a major scavenger of reactive oxygen species (ROS) in plasma.35,36 The Cys34 sulfenic acid of SA is a proposed transient intermediate that undergoes further oxidation to form stable Cys34 sulfinic (Cys-SO2H) and sulfonic (Cys-SO3H) acids.36,37 Moreover, the Cys34 sulfenic acid is thought to be the intermediate of SA that is involved in the formation of mixed disulfides with low molecular weight thiols, such as Cys or glutathione: these mixed disulfides account for about 30% of the circulating SA in plasma.34,35 The high level of oxidation of Cys34 of SA by ROS suggests that arylsulfinamide adducts of SA also may undergo oxidation in vivo. The ability to trap labile sulfur-nitrogen arylamine adducts at Cys34 of SA by chemical oxidation to form stable arylsulfonamide adducts may allow us to simultaneously biomonitor exposures to many carcinogenic aromatic amines and HAAs that was not previously possible.
Supplementary Material
Acknowledgments
Funding
This research was supported by Grant R01 CA122320 (L.P. and R.J.T.)
Footnotes
Supporting Information Available: Additional information as noted in text (Figure S-1-Figure S-3). This material is available free of charge via the Internet at http://pubs.acs.org.
Reference List
- 1.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turesky RJ, Le Marchand L. Chem Res Toxicol. 2011;24:1169–1214. doi: 10.1021/tx200135s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Tobacco smoking. International Agency for Research on Cancer; Lyon, France: 1986. [Google Scholar]
- 4.Some naturally occurring substances: food items and constituents, heterocyclic aromatic amines and mycotoxins. International Agency for Research on Cancer; Lyon, France: 1993. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. [Google Scholar]
- 5.Jarabek AM, Pottenger LH, Andrews LS, Casciano D, Embry MR, Kim JH, Preston RJ, Reddy MV, Schoeny R, Shuker D, Skare J, Swenberg J, Williams GM, Zeiger E. Crit Rev Toxicol. 2009;39:659–678. doi: 10.1080/10408440903164155. [DOI] [PubMed] [Google Scholar]
- 6.Skipper PL, Tannenbaum SR. Carcinogenesis. 1990;11:507–518. doi: 10.1093/carcin/11.4.507. [DOI] [PubMed] [Google Scholar]
- 7.Liebler DC. Environ Health Perspect. 2002;110(Suppl 1):3–9. doi: 10.1289/ehp.02110s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tornqvist M, Fred C, Haglund J, Helleberg H, Paulsson B, Rydberg P. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;778:279–308. doi: 10.1016/s1570-0232(02)00172-1. [DOI] [PubMed] [Google Scholar]
- 9.Dolle B, Topner W, Neumann HG. Xenobiotica. 1980;10:527–536. doi: 10.3109/00498258009033787. [DOI] [PubMed] [Google Scholar]
- 10.Kiese M, Taeger K. Naunyn Schmiedebergs Arch Pharmacol. 1976;292:59–66. doi: 10.1007/BF00506490. [DOI] [PubMed] [Google Scholar]
- 11.Guo Z, Wagner CR, Hanna PE. Chem Res Toxicol. 2004;17:275–286. doi: 10.1021/tx030045o. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Wagner CR, Hanna PE. Chem Res Toxicol. 2008;21:2005–2016. doi: 10.1021/tx800215h. [DOI] [PubMed] [Google Scholar]
- 13.Turesky RJ, Skipper PL, Tannenbaum SR. Carcinogenesis. 1987;8:1537–1542. doi: 10.1093/carcin/8.10.1537. [DOI] [PubMed] [Google Scholar]
- 14.Peng L, Turesky RJ. Chem Res Toxicol. 2011;24:2004–2017. doi: 10.1021/tx2003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringe D, Turesky RJ, Skipper PL, Tannenbaum SR. Chem Res Toxicol. 1988;1:22–24. doi: 10.1021/tx00001a003. [DOI] [PubMed] [Google Scholar]
- 16.Bryant MS, Vineis P, Skipper PL, Tannenbaum SR. Proc Natl Acad Sci U S A. 1988;85:9788–9791. doi: 10.1073/pnas.85.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubino FM, Pitton M, Di FD, Colombi A. Mass Spectrom Rev. 2009;28:725–784. doi: 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- 18.Dingley KH, Curtis KD, Nowell S, Felton JS, Lang NP, Turteltaub KW. Cancer Epidemiol Biomarkers Prev. 1999;8:507–512. [PubMed] [Google Scholar]
- 19.Reistad R, Frandsen H, Grivas S, Alexander J. Carcinogenesis. 1994;15:2547–2552. doi: 10.1093/carcin/15.11.2547. [DOI] [PubMed] [Google Scholar]
- 20.Chepanoske CL, Brown K, Turteltaub KW, Dingley KH. Food Chem Toxicol. 2004;42:1367–1372. doi: 10.1016/j.fct.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Peng L, Dasari S, Tabb DL, Turesky RJ. Chem Res Toxicol. 2012;25:2179–2193. doi: 10.1021/tx300253j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beland FA, Beranek DT, Dooley KL, Heflich RH, Kadlubar FF. Environ Health Perspect. 1983;49:125–134. doi: 10.1289/ehp.8349125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turesky RJ, Lang NP, Butler MA, Teitel CH, Kadlubar FF. Carcinogenesis. 1991;12:1839–1845. doi: 10.1093/carcin/12.10.1839. [DOI] [PubMed] [Google Scholar]
- 24.Langouët S, Paehler A, Welti DH, Kerriguy N, Guillouzo A, Turesky RJ. Carcinogenesis. 2002;23:115–122. doi: 10.1093/carcin/23.1.115. [DOI] [PubMed] [Google Scholar]
- 25.Riener CK, Kada G, Gruber HJ. Anal Bioanal Chem. 2002;373:266–276. doi: 10.1007/s00216-002-1347-2. [DOI] [PubMed] [Google Scholar]
- 26.Hiramoto K, Negishi K, Namba T, Katsu T, Hayatsu H. Carcinogenesis. 1988;9:2003–2008. doi: 10.1093/carcin/9.11.2003. [DOI] [PubMed] [Google Scholar]
- 27.Aldini G, Regazzoni L, Orioli M, Rimoldi I, Facino RM, Carini M. J Mass Spectrom. 2008;43:1470–1481. doi: 10.1002/jms.1419. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Vivekananda S, Men L, Zhang Q. J Am Soc Mass Spectrom. 2004;15:697–702. doi: 10.1016/j.jasms.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Damsten MC, Commandeur JN, Fidder A, Hulst AG, Touw D, Noort D, Vermeulen NP. Drug Metab Dispos. 2007;35:1408–1417. doi: 10.1124/dmd.106.014233. [DOI] [PubMed] [Google Scholar]
- 30.Noort D, Hulst AG, de Jong LP, Benschop HP. Chem Res Toxicol. 1999;12:715–721. doi: 10.1021/tx9900369. [DOI] [PubMed] [Google Scholar]
- 31.Noort D, Fidder A, Hulst AG. Arch Toxicol. 2003;77:543–545. doi: 10.1007/s00204-003-0484-5. [DOI] [PubMed] [Google Scholar]
- 32.Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER. Toxicol Lett. 2012;213:83–90. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynch AM, Murray S, Zhao K, Gooderham NJ, Boobis AR, Davies DS. Carcinogenesis. 1993;14:191–194. doi: 10.1093/carcin/14.2.191. [DOI] [PubMed] [Google Scholar]
- 34.Peters T., Jr Adv Clin Chem. 1970;13:37–111. doi: 10.1016/s0065-2423(08)60385-6. [DOI] [PubMed] [Google Scholar]
- 35.Turell L, Botti H, Carballal S, Ferrer-Sueta G, Souza JM, Duran R, Freeman BA, Radi R, Alvarez B. Biochemistry. 2008;47:358–367. doi: 10.1021/bi701520y. [DOI] [PubMed] [Google Scholar]
- 36.Turell L, Botti H, Carballal S, Radi R, Alvarez B. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3384–3392. doi: 10.1016/j.jchromb.2009.03.035. [DOI] [PubMed] [Google Scholar]
- 37.Grigoryan H, Li H, Iavarone AT, Williams ER, Rappaport SM. Chem Res Toxicol. 2012;25:1633–1642. doi: 10.1021/tx300096a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.