Abstract
The tail of the enzyme RNA polymerase II is responsible for integrating the diverse events of gene expression in eukaryotes and is indispensable for life in yeast, fruit flies and mice. The tail features a C-terminal domain (CTD), which is comprised of tandemly repeated Y1-S2-P3-T4-S5-P6-S7 amino acid heptads that are highly conserved across evolutionary lineages, with all mammalian polymerases featuring 52 identical heptad repeats. However, the composition and function of protozoan CTDs remain less well understood. We find that malaria parasites (genus Plasmodium) display an unprecedented plasticity within the length and composition of their CTDs. The CTD in malaria parasites which infect human and non-human primates has expanded compared to closely related species that infect rodents or birds. In addition, this variability extends to different isolates within a single species, such as isolates of the human malaria parasite, Plasmodium falciparum. Our results indicate that expanded CTD heptads in malaria parasites correlates with parasitism of primates and provide the first demonstration of polymorphism of the RNA polymerase II CTD within a single species. The expanded set of CTD heptads feature lysine in the seventh position (Y1-S2-P3-T4-S5-P6-K7), a sequence only otherwise seen in the distal portion of mammalian polymerases. These observations raise new questions for the radiation of malaria parasites into diverse hosts and for the molecular evolution of the RNA polymerase II.
Keywords: protozoa, Plasmodium, Apicomplexa, transcription
Introduction
Malaria is responsible for a significant burden on human health and economic productivity in the developing world. The malaria parasite Plasmodium falciparum has co-evolved with humans for millennia and displays sophisticated patterns of gene expression that enable the parasite to multiply in several distinct morphological stages in remarkably different environments, including clonal replication in the liver and circulating red blood cells of its mammalian host as well as both sexual and asexual replication within its mosquito vector. Despite these major changes in gene expression, analyses of the completed sequence of the P. falciparum genome have resulted in the report of a surprising “paucity” of identifiable transcription factors (Aravind, Iyer et al. 2003) or well-characterized domains that are found in transcription factors shared from budding yeast to humans (Iyer, Anantharaman et al. 2008). Nonetheless, the basic aspects of transcription in Plasmodium, mediated by RNA polymerase II, appear to be similar to those described for metazoans, including 5′ capping, polyadenylation, splicing and chromatin modifications (Aravind, Iyer, Wellems, and Miller 2003). Hence, attention is rapidly turning to the study of the role of RNA polymerase II in transcriptional regulation to more fully understand the basic biology of malaria parasites.
RNA polymerase II is a central component of a complex apparatus responsible for controlling the basic biochemical steps required for mRNA production, including transcription initiation, capping, elongation, splicing, and polyadenylation (Howe, 2002;Zorio and Bentley 2004;Proudfoot, Furger et al. 2002). These diverse processes are integrated by the tail of the molecule, referred to as the C-terminal domain (CTD) of the protein (Carty and Greenleaf 2002). The CTD, essential for gene expression in animals and fungi, is comprised of a tandem array of heptapeptide repeats, featuring a signature amino acid sequence: Y1-S2-P3-T4-S5-P6-S7 (Corden, 1990;Allison, Moyle et al. 1985). In yeast, rodents and humans, these repeats coordinate transcriptional events by undergoing reversible phosphorylation at the 2nd and 5th serines of the heptad (Corden, 2007;Schroeder, Schwer et al. 2000;Licatalosi, Geiger et al. 2002), with more recent data implicating the 7th serine as critical for transcriptional elongation (Chapman, Heidemann et al. 2007;Egloff, O’Reilly et al. 2007). The combinatorial phosphorylation patterns create novel docking platforms for the sequential recruitment of specific transcription factors involved in initiation, capping, splicing and termination.
The length of the CTD varies in different organisms and roughly tracks with genome size. For example, the CTD of the microsporidian Encephalitozoon cuniculi contains 15 repeats while a CTD containing 52 repeats is present in all mammalian species thus far described (Chapman, Heidemann, Albert, Mailhammer, Flatley, Meisterernst, Kremmer, and Eick 2007;Egloff, O’Reilly, Chapman, Taylor, Tanzhaus, Pitts, Eick, and Murphy 2007). The perfect conservation of the mammalian CTD demonstrates the strict conservation of this portion of RNA pol II within broad evolutionary lineages. The length of the CTD is essential for RNA polymerase II stability, transcriptional efficiency and ultimately cell viability. For example, the CTD of the budding yeast Sacharomyces cerevisiae consists of 26 heptad repeats and truncation to 17, 14, 13 or 11 repeats leads to respective reductions in specific transcript levels to 58%, 8%, 5% and 2% of that found in wild type cells (Liao, Taylor et al. 1991). Further reduction of the repeat length to 8–10 repeats yields cells that are temperature-sensitive, and truncation to less than eight repeats is lethal. Similarly, 30 of the 52 hepatads are necessary for viability in mice (Meininghaus, Chapman et al. 2000) and mutants with 31–39 heptads exhibit growth defects (Bartolomei, Halden et al. 1988;Litingtung, Lawler et al. 1999). The importance of CTD length is reflected in the fact that this region of the protein is strictly conserved in closely related organisms, and no variability within an individual species in the number of heptad repeats has been observed.
Throughout evolution, the 7th position of the conserved heptad repeat displays the most heterogeneity in higher eukaryotes (Guo and Stiller 2005;Liu, Greenleaf et al. 2008). In yeast the serine in this position is non-essential, with substitution of alanine tolerated (Stiller, McConaughy et al. 2000). Intriguingly, mammals, including rodents and primates, feature seven to nine non-consensus Y1-S2-P3-T4-S5-P6-K7 heptads near the distal end of the CTD (Barron-Casella and Corden 1992;Guo and Stiller 2005). In mammals, while 20 of the first 25 heptads contain a serine in the 7th position, only 6 of the last 27 heptads do, and 8 of the last 17 heptads all contain a lysine in this position. In human cell lines, however, these lysine-containing repeats are dispensable, with polymerases consisting of only consensus repeats supporting normal growth and viability of cells (Chapman, Conrad et al. 2005). A further study by Chapman et al. indicated that polymerases featuring an excess of lysine heptads fail to support cell viability (Chapman, Heidemann, Albert, Mailhammer, Flatley, Meisterernst, Kremmer, and Eick 2007). Thus the role of the lysine heptads in mammalian polymerases remains an outstanding question in RNA polymerase II biology and awaits functional, biochemical and regulatory characterization.
The completion and release of the genome sequences from several Plasmodium species has revealed that this genus – alone outside of mammalian genera-- extensively utilize lysine-containing repeats within their RNA pol II CTDs. This observation raised the question of what role these repeats might play CTD function, and led us to study the composition of the CTD across the Plasmodium genus. We find that the number of heptad repeats in Plasmodium is remarkably small, with the rodent and bird malaria parasites possessing only eight heptad repeats within their CTDs. Surprisingly, unlike other evolutionary lineages in which heptad numbers are tightly conserved, closely related Plasmodium species display very different numbers of heptad repeats. In particular, primate parasites posses an expanded CTD of up to nine additional lysine containing repeats, a motif otherwise only found in the distal half of mammalian polymerases. The expansion appears to have occurred at least twice in evolution, once in a lineage giving rise to P. falciparum and independently in the lineage giving rise to P. vivax and other non-human primate parasites. Strikingly, the plasticity appears to extend to individual species of Plasmodium. Different geographical isolates of the human parasites P. falciparum and P. vivax and the primate parasite P. cynomolgi also exhibit significant flexibility in repeat numbers. These findings provide the first examples of polymorphism of the RNA polymerase II CTD within a single species. Taken together, our observations raise new questions for both the evolution of malaria parasites and the RNA polymerase II molecule as well as malaria parasitism of diverse hosts. In addition, as P. falciparum and P. berghei are genetically tractable systems, this work opens the possibility to elucidating the hitherto cryptic function of lysine heptads in eukaryotes.
Materials and Methods
Amplification of Plasmodium CTDs
Genomic DNA for P. falciparum, P. vivax and P. simium strains were obtained from the Malaria Research and Reference Reagent Resource Center (MR4; www.mr4.org) and Dr. X. Su of the National Institute of Allergy and Infectious Disease. Genomic DNA from P. ovale was a gift by J. Carlton (New York University, New York City) and genomic DNA for isolates and sub-species of rodent malarias (P. berghei, P. yoelii and P. vinckei) were from stocks held at the University of Edinburgh by R. Carter (Perkins, Sarkar et al. 2007). Oligonucleotide primers complementary to sequences flanking the first repeat in the CTD and the carboxy-terminus of the largest subunit of RNA polymerase II (Rbp1) gene were used to PCR-amplify the CTD. All PCR reactions were carried out on a PTC-2000® Peltier thermal cycler using Taq polymerase® (Invitrogen) under the following conditions: 95 °C for 5 min followed by 35–38 cycles of 94 °C for 30 s, 56 °C for 60 s, 60 °C for 1 min, and a final extension step of 65 °C for 5 min. Reaction products were purified and directly sent for automated sequencing by the university core facility. The forward primer that was used to amplify the RNA polymerase II CTD from P. falciparum isolates was 5′-CCTAAACCTCAAATTAATCATAATATTTATTCA-3′ and the reverse primer was 5′-CATATTTTCCTTCATTTCGTCCTCGTATAT-3′. The forward primer for P. vivax and P. fragile RNA polymerase II CTD was 5′ – TCCCC(A/C)TT(C/T)TCTCC(A/T/C)TTTGAT-3′ and the reverse primer was: 5′-CATTTCGTCCTC(C/G)TC(C/T)ATGTTGTA-3′ The forward primer for P. berghei RNA polymerase II CTD was 5′-CCAAAACCTCAGATGCAAAATAATATATATTCT-3′ and the reverse primer was: 5′-TTATTCTTCCTGCATCTCCTCTTCATCCAT-3′; forward primer for P. yoelii yoelii RNA polymerase II CTD was 5′-CCAAAACCTCAGATGCAAAATAATATATATTCT-3′ and the reverse primer was as above for P. berghei.
Tabulation of Repeat Numbers
Repeats were counted manually and defined conventionally, as heptads featuring a serine in the second and fifth position (S2 and S5) (Liu, Greenleaf, and Stiller 2008). The lone exception to this rule was made for the repeat YAIASPK in the non-human primate parasite Plasmodium fragile as it aligned with the repeat YSITSPK in rodent and human parasites. We note that this definition differs from recent work of Chapman et al. (Liu, Greenleaf, and Stiller 2008) who include phasing in their definition. This group identifies five repeats in P. yoelii; whereas we identify eight. Diheptads or heptad pairs refers to two heptad repeats found in tandem with no intervening amino acid residues. A single heptad can be counted as part of two diheptads. For example, three consecutive heptad repeats would constitute two diheptads.
Results
Plasticity of the CTD within the Genus Plasmodium
Previously, two laboratories described a greater number of CTD heptad repeats in P. falciparum when compared to the rodent malarias P. berghei and P. yoelii (Chapman, Heidemann et al. 2008;Egloff and Murphy 2008;Giesecke, Barale et al. 1991), suggesting an unusual variability in CTD structure between these closely related organisms. Since the number of CTD heptad repeats is generally stringently conserved between even distantly related genera, we wondered if the differences between the human and rodent parasites reflected a general plasticity of CTD heptads in the genus Plasmodium. To obtain a broad perspective of CTD sequences from throughout the Plasmodium genus, we collected sequences from the largest subunit of RNA polymerase II (Rbp1) from parasites infecting humans (P. falciparum, P. vivax and P. ovale), non-human primates (P. reichenowi, P. knowlesi and P. fragile), rodents (P. berghei and P. yoellii) and birds (P. gallinaceum). The sequences were either obtained from genome sequence databases at the NCBI or Sanger Institute, or directly amplified from parasitized blood obtained from field samples. This collection of sequences was then aligned using CLUSTALW, adjusted through visual inspection and repeats tabulated as described in the Materials and Methods.
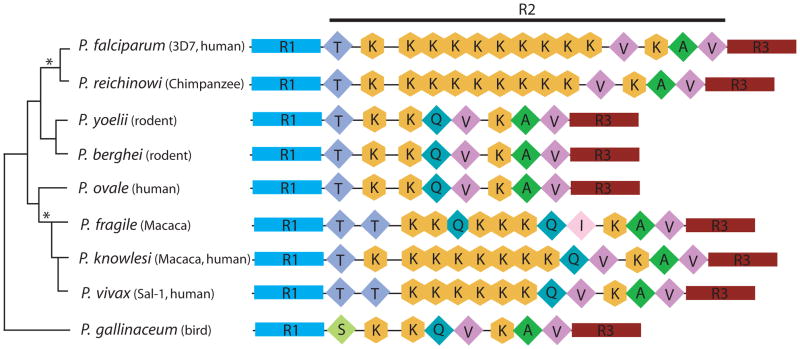
Analysis of the amino acid sequences from the various species showed that they all maintained the typical, conserved CTD structure consisting of a linker region (R1), the heptad repeat-containing region (R2), and a tail region following the last repeat (R3) (Chapman, Heidemann, Hintermair, and Eick 2008). However, while all of the species possessed the heptad repeat structure typical of CTDs from higher eukaryotes, there was a remarkable degree of variability in the number of repeats in region R2 as well as in the amino acid found in the 7th position of each heptad (Figure 1). This is in stark contrast to the extreme conservation of heptad sequence observed in many other evolutionary lineages. For example, mammals from different orders exhibit no variability in either heptad structure or number of repeats, with all species thus far examined displaying nearly indistinguishable (one amino acid difference between mice and humans) CTDs consisting of 52 identical repeats (Barron-Casella and Corden 1992). Second, the Plasmodium CTDs are remarkably short, consisting of only 8 repeats in the rodent and bird parasites, P. berghei, P. yoellii, and P. gallinaceum, and reaching a maximum of 13–15 repeats in the primate parasites P. knowlesi, P. vivax, P. reichenowi and P. falciparum. The exception to this trend is P. ovale which possesses only 8 repeats within the CTD. In addition, the different Plasmodium species all displayed a propensity toward a non-serine amino acid in the 7th position, and in particular the primate parasites exhibited an expanded set of tandemly arrayed repeats containing lysine in this position. All of the species that display an expanded CTD infect monkeys, apes or humans, thus correlating increased heptad number with parasitism of primates.
Figure 1. Plasticity of the heptads in the RNA polymerase II CTD across the Plasmodium genus.
Schematic depiction of the CTD heptads from eight species of Plasmodium. The species names are listed at the left of each line along with a tree indicating their evolutionary relationships, constructed using sequences from the three mitochondrial protein-coding genes of the parasites as described in Perkins (2008). R1 is the linker domain, R2 is the heptad-bearing region and R3 is the acidic tail region following the last repeat of the CTD. Diamonds and hexagons symbolize individual heptads, with the amino acid in the 7th position indicated. The plasticity of the CTD is restricted to heptad-containing region, R2. Rodent malaria parasites (P. yoelii and P. berghei) and the bird parasite (P. gallinaceum) exhibit eight repeats with a common stalk (Q,V,K,A,V). Primate malaria parasites feature an expanded set of tandemly arrayed heptads all containing lysine in position 7 of heptads immediately preceding this stalk. Two separate, independent expansions are proposed to have occurred (indicated by asterisks). Sequences of the CTDs drawn from PlasmoDB (www.plasmodb.org) and contigs at the Sanger Institute: P. berghei: ANK,PB000038.00.0; P. yoelii17xNL: PY03187; P. vivax Sal-1: PV095320; P. knowlesi Strain H: PKH_082310; P. falciparum 3D7: Pfc0805w. P. gallinaceum: EU840284 and P. reichinowi: EU850397, P. fragile: EU840282 and P. ovale: EU887536.
When phylogenetic analyses between the various species of Plasmodium are overlaid on the CTD data, there is support for two separate expansions of the lysine heptads (denoted by asterisk in Figure 1) in primate parasites. One expansion occurred after P. fragile, P. knowlesi and P. vivax diverged from P. ovale and the other expansion occurred in the lineage giving rise to P. falciparum and P. reichinowi. Codon usage data supports this argument; P. falciparum and P. reichinowi share single expansion of a codon for the lysine repeats (AAA), whereas the other primate parasites that have undergone expansion (P. vivax, P. knowlesi and P. fragile) feature lysine codon AAG as the common or predominant codon in the repeats. Moreover, the rest of the RNA polymerase II molecule of these parasites is more closely related to the polymerase of the rodent parasite P. berghei, which lacks the expanded motifs, than to the primate parasite P. falciparum, which features them (data not shown). The separate expansions imply that the increased CTD length provides a selective advantage for parasites infecting primate hosts.
Plasticity of the CTD within a single species of Plasmodium
Such extensive plasticity in CTD heptad repeat numbers and composition between closely related species of a single genus has not been described before. We therefore questioned whether the CTD structure might be rapidly diverging within the Plasmodium genus, and consequently whether such plasticity might extend to an individual species. For this analysis we took advantage of recent efforts to sequence the genomes of several independent isolates of P. falciparum obtained from infected individuals from different malarious regions around the world. The sequence of RNA polymerase II from an isolate from Honduras ((Li, Bzik et al. 1989); NCBI database) and one from Papua New Guinea (D10; Broad Institute database) were analyzed for CTD sequence and structure. Notably, both of these isolates exhibited fewer heptad repeats (Honduras: 14 repeats; D10: 13 repeats) than 3D7 (15 repeats), the reference strain used for the initial genome sequencing project. To determine if the change in repeat number was an artifact of genome sequence assembly, we obtained genomic DNA from both 3D7 and D10 (gift from Dr. X. Su, NIAID), and directly amplified the CTD encoding region of the gene using species-specific PCR primers and sequenced the purified amplicons. We found that both sequences precisely matched the sequences reported in the initial genome nucleotide sequence databases, thus verifying the observed polymorphisms.
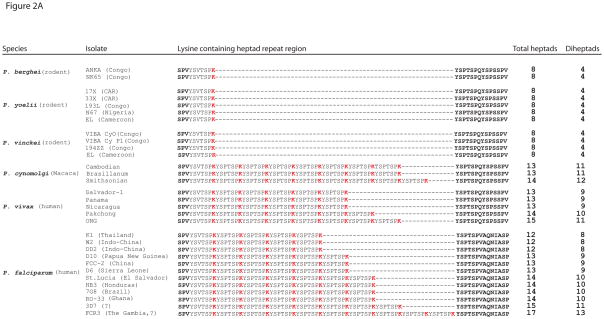
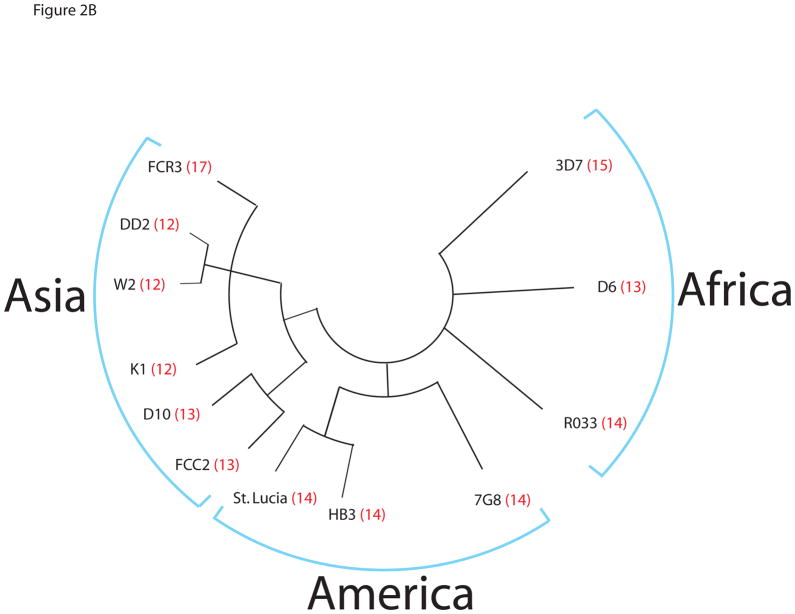
To determine the degree of repeat number polymorphism within P. falciparum, we extended the analysis to additional isolates from other geographically diverse locations, including 4 African, 5 Asian and 3 American isolates of P. falciparum from the Malaria Research and Reference Reagent Resource Center (MR4) of the American Type Culture Collection (ATCC). We amplified the CTD from genomic DNA and directly sequenced the purified amplicons. The results displayed an even greater degree of polymorphism than initially observed, with isolates exhibiting CTD lengths ranging from as few as 12 to as many as 17 repeats (Figure 2A). Both heptad repeats and diheptad repeats (repeats in tandem) were tabulated. The variations appear to weakly cluster geographically. Asian isolates (e.g. DD2, K1 and D10) tend to be shorter (12–13 repeats) while American isolates (St. Lucia, HB3 and 7G8) all have 14 repeats. African isolates tend to be the longest with 15 in 3D7, 17 in FCR3 and 14 in RO33; with the exception of the African isolate D6, which has only 13 repeats. Figure 2B shows a phylogenetic tree of the various P. falciparum isolates based on 137 SNPs (Volkman, Sabeti et al. 2007), which indicates the CTD polymorphism tracks with continental Asian and American clades. The one exception is FCR3, which was reportedly isolated from the Gambia, but genetic analysis has suggested might actually be of Asian origin (Volkman, Sabeti, DeCaprio, Neafsey, Schaffner, Milner, Jr., Daily, Sarr, Ndiaye, Ndir, Mboup, Duraisingh, Lukens, Derr, Stange-Thomann, Waggoner, Onofrio, Ziaugra, Mauceli, Gnerre, Jaffe, Zainoun, Wiegand, Birren, Hartl, Galagan, Lander, and Wirth 2007).
Figure 2. Plasticity of the heptads in the RNA polymerase II CTD across Plasmodium species.
A. Amino acid sequences of the R2 domain of 2 isolates of P. berghei, 5 isolates of P. yoelii and 4 isolates of P. vinckei (all rodent parasites), 3 isolates of P. cynomolgi, 5 isolates of P. vivax and 12 geographical isolates of P. falciparum. The lysine in the 7th position of each heptad repeat is shown in red. Numbers of total heptads and diheptads are tabulated at the right. B. Phylogenetic tree of various P. falciparum isolates based on 137 SNPs and arranged according to geographical origin. The number of heptad repeats found in the CTD of each isolate is shown in red. Adapted from (Volkman, Sabeti, DeCaprio, Neafsey, Schaffner, Milner, Jr., Daily, Sarr, Ndiaye, Ndir, Mboup, Duraisingh, Lukens, Derr, Stange-Thomann, Waggoner, Onofrio, Ziaugra, Mauceli, Gnerre, Jaffe, Zainoun, Wiegand, Birren, Hartl, Galagan, Lander, and Wirth 2007).
We next asked whether two other primate parasites, P. vivax and P. cynomolgi, exhibit plasticity in CTD repeat numbers similar to P. falciparum. As done for the P. falciparum samples, using known database sequence and genomic DNA from MR4, we amplified the CTD from several DNA samples and directly sequenced purified amplicons. Similar to P. falciparum, we found plasticity of the CTD specific to the lysine-containing, tandemly arrayed heptad repeats (Figure 2A). There is also a geographic correlation to the repeat number with Asian isolates of P. vivax (ONG and Pakchong) exhibiting more repeats (15 and 14), respectively than the American (Nicaragua, Panama and Salvador-1) isolates (13 repeats). The Smithsonian isolate of P. cynomolgi features one more repeat than the Bastianelli and Cambodian isolates.
To determine if the intra-species polymorphisms in CTD length extend to other Plasmodium species, we took advantage of 11 rare samples of genomic DNA obtained from various isolates and sub-species of the rodent malaria parasites P. berghei, P. yoelii and P. vinckei as described in Perkins et al., 2007 (Perkins, Sarkar, and Carter 2007). These rodent parasite species are restricted to West and Central Africa. We observed no differences in repeat numbers either across or within the P. berghei, P. yoelii or P. vinckei species or subspecies (Figure 2A). The number of repeats was also identical to the number reported for the bird parasite, P. gallinaceum. These data suggest that the diversity of heptad repeat numbers may be specific to the primate malaria parasites P. falciparum, P. vivax and P. cynomolgi. In addition, the polymorphisms do not appear to be an artifact of laboratory culture, as seen in budding yeast (Nonet, Sweetser et al. 1987), because fresh uncultured clinical isolates also display plasticity of repeats (unpublished data).
Evolution of the CTD in protozoa
The heptad repeat structure of the CTD is a ubiquitous feature of RNA pol II of metazoa, and it is indispensable for transcriptional activity in these organisms. Among the unicellular eukaryotes, however, some organisms possess RNA pol II enzymes that contain CTDs with recognizable heptads, while others do not, suggesting that the CTD evolved within an ancient organism near this point in the eukaryotic lineage. In light of the remarkable diversity of CTD heptads within the Plasmodium genus and within different isolates of P. falciparum and P. vivax, we investigated whether closely related protozoa possess conserved CTD heptads. In particular we were interested in other apicomplexans, as well as other alveolates such as dinoflagellates and ciliates. Our examination of CTDs from protozoa using conventional genome sequence databases indicated that CTD heptads are also present in the apicomplexans Babesia bovis; Theileria spp; Toxoplasma gondii and Cryptosporidium spp as well as in the early branching dinoflagellate Perkinsus marinus. The apicomplexans and dinoflagellates examined possessed heptad repeats that varied in size from nine in Toxoplasma to as many as 24 in Cryptosporidium. The 7th position of the heptads varies across organisms with an alanine in Babesia and Perkinsus and a histidine in Cryptosporidium. The two closely related parasites, Cryptosporidium parvum and Cryptosporidium hominis, possess the same heptad repeat composition and number throughout their CTDs; whereas the murine parasite Cryptosporidium muris displays the same number of heptads but utilizes a different heptad composition that relies on histidine and arginine in the 7th position. Thus the remarkable variability observed in repeat numbers in Plasmodium may be unique to this genus.
DISCUSSION
The extreme plasticity that we observed for the CTD heptad repeats in Plasmodium has not been described for any other eukaryotic species. Moreover, the intra-species plasticity is not observed across any of the rodent parasites we studied. It is possible that the repetitive nature of the DNA sequence encoding the CTD simply leads to frequent duplications and deletions, and in this regard Plasmodium might have a propensity toward replication errors in the absence of purifying selection regulating CTD length. However, the lack of tandem repeats in the ancestral Plasmodium CTD suggest that the amplification observed in primate parasites was not simply the result of random replication errors (i.e. unequal crossover) and instead likely arose from positive selection. Rather than displaying random variation in length and composition, the primate malarias appear to have a specific expansion of a particular heptad, leading to a longer CTD. Further, both phylogenetic analyses and codon-usage bias support the hypothesis that the expansion of the CTD occurred twice in primate parasites – once in the line giving rise to P. falciparum and once in a line giving rise to P. vivax, and not any non-primate parasites. The fact that these expansion events appear to have occurred independently and in parallel suggests a strong functional basis for the longer CTD. The nature of this selective force in primates remains cryptic and no available data correlates CTD length in human parasites with virulence, drug resistance or any other obvious phenotype.
We did not observe variation in the CTDs of any of the rodent parasites, and their length was identical to that of P. gallinaceum, a parasite of birds, suggesting that the non-primate parasites might be fixed in their repeat numbers at eight heptads. In addition, the eight specific heptads found in bird and rodent parasites are also conserved in the CTDs of primate parasites (Figure 1), indicating that this represents the ancestral structure of the CTD prior to the expansion observed in primate parasites. Interestingly, eight repeats is consistent with the minimal number of heptads required for viability in yeast and the minimal number in the mouse RNA pol II to form a secondary structure resembling a full length CTD (Bienkiewicz, Moon et al. 2000;Liao, Taylor, Kingston, and Young 1991). We therefore surmise that the 8 repeats might be the minimal number required for a functional RNA polymerase II, and thus this represents the shortest CTD possible within this genus. However, in yeast the functional unit of the CTD was shown to be pairs of tandemly arranged heptad repeats, and that at least 7 such “diheptads” were required for viability (Stiller and Cook 2004). Thus, it appears that both the number and tandem arrangement of repeats is important for function. It is not yet clear which specific RNA polymerase functions require the tandem arrangement of the heptads, though the tandem arrangement is present in all eukaryotes from yeast to mammals. Notably, the rodent and bird parasites do not possess the minimal number of diheptad repeats required for viability in yeast, while the heptad expansion found in the CTD of primate parasites is specific for tandemly arranged repeats. This expansion greatly increases the number of diheptads (Figure 2A) resulting in CTDs that meet or exceed what is required in higher eukaryotes. In contrast to metazoan CTDs, in which the tandemly repeated structure of the CTD is strongly conserved, heptads are often found in isolation in protists. Hence, the expanded set of perfect tandemly arranged repeats in several Plasmodium species suggests that the CTD-related proteins of these parasites might share functional requirements with those from higher eukaryotes.
Interestingly, the expanded repeats all feature lysine in the seventh position, a motif only otherwise found in mammalian polymerases. The expansion of lysine containing heptads in primate parasites highlights the potential significance of repeats that deviate from the consensus sequence YSPTSPS, particularly in the 7th position. The 7th position of the conserved heptad repeat displays the most heterogeneity in higher eukaryotes (Guo and Stiller 2005;Liu, Greenleaf, and Stiller 2008) and in yeast, the serine in this position is non-essential (Stiller, McConaughy, and Hall 2000). Thus the role of the lysine-enrichment in the distal end of the RNA polymerase II remains an outstanding question in CTD biology (Egloff and Murphy 2008;Phatnani and Greenleaf 2006). It has been hypothesized that lysine containing heptads might confer additional specificity in recruitment of specific proteins to the transcription complex (Phatnani and Greenleaf 2006). Naturally, this raises the question of why plasmodia species utilize lysine containing repeats within their CTDs, particularly as our analysis indicates that the extensive reliance on lysine containing heptads is unique to Plasmodium and not present in other organisms, including other apicomplexan parasites. In the case of P. falciparum the lysine (codon: AAA or AAG) enrichment could simply be a natural consequence of selective pressures of its A/T-rich (80%) genome. However, P. vivax also displays a similar number of lysine containing repeats but is nearly balanced in its A/T content (57%), implying functional significance for these heptads. In light of recent data concerning lysine modifications on non-histone targets involved in gene regulation, it is possible that the lysines in the CTD might be differentially acetylated, particularly by adjoining transcription factors (e.g. TAF1 subunit of TFIID) (Mizzen, Yang et al. 1996).
Why Plasmodium should evolve a longer CTD in just a few and not all of its mammalian host species is intriguing. It is likely that the expanded lysine heptads reflect individual or small sets of interactions that have been incrementally added to the transcriptional repertoires of primate parasites. One could imagine that the primate-infecting parasites have co-opted functional requirements for transcription shared with higher eukaryotes during co-evolution with host transcription systems. It is less likely, however, that the expanded lysine heptads are themselves interacting with mammalian CTD-related proteins, at least during the erythrocytic stages, as this portion of the parasite life cycle transpires in the aenucleated red blood cell environment.
This work also sheds light on the evolution of the CTD. We find that all Apicomplexa studied possess heptad repeats. By contrast, ciliates lack CTD heptads altogether despite their grouping with apicomplexans within Alveolata. Interestingly, the RNA polymerases of ciliates possess serine-proline (SP) motifs that are enriched within the C-terminus but not elsewhere in RNA polymerase II (Figure 3). A similar pattern of “SP” richness is also seen in the eukaryotic human pathogen Trichomonas (Dacks, Marinets et al. 2002), but is not seen in Giardia or the kinetoplastids (Trypanosoma cruzi, Trypansoma brucei and Leishmania major). The “SP” motifs are reminiscent of Y1-S2-P3-T4-S5-P6-X7 heptad motifs found in metazoan and apicomplexan CTDs, suggesting the possibility that they are either a direct result of degeneracy or serve as early, primitive repeats. The ciliate Stylonchia mytilus, for instance, features regular, repeating SP motifs that are often preceded by tyrosine residues (e.g. YSP motifs). These observations suggest that that the SP motifs have evolved as specific sites for phosphorylation of RNA pol II. However, the absence of SP motifs or heptads does not necessarily imply the absence of phosphorylation of the RNA pol II; for example, the CTD of T. brucei is phosphorylated (Chapman and Agabian 1994).
Figure 3. Enrichment of “SP” motifs in the tails of RNA Pol II from eight different protozoan species.
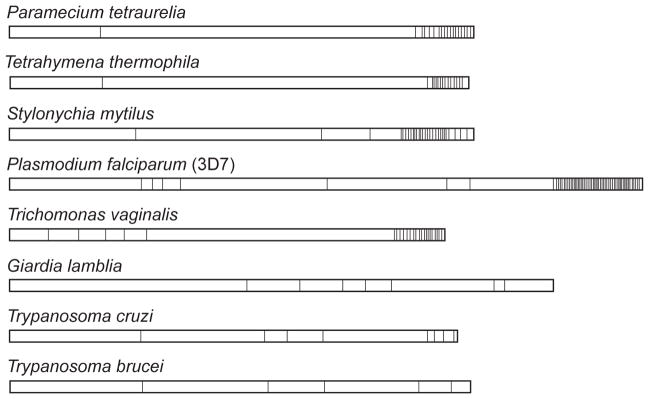
Serine-proline (SP) motifs that represent potential phosphorylation sites are marked with vertical lines (|). Sequence accession numbers are: Stylonchia mytilus: AAK00313.1; Paramecium tetraurelia: CAI39063.1; Tetrahymena thermophila: GI accession: 118348890; Trypanasoma brucei: P17545; Trypanasoma cruzi: XP_812569; Trichomonas vaginalis: TVU20501; Giardia lamblia: XP_001704218
The study of RNA polymerase II CTDs in protozoa, particularly with the ever expanding number of genome sequences that are becoming available, is providing valuable insights into the evolution of this important protein domain. The fact that different species of Plasmodium naturally display variable CTDs, and that both P. falciparum and P. berghei, haploid single-celled protists, are genetically tractable systems, means that it might be possible to use these organisms to test many of the current hypotheses regarding the roles of heptad repeats in CTD function. Thus Plasmodium, an organism of great importance to human health and economic development, could also serve as a model system for understanding the evolution of transcription in eukaryotic biology.
Acknowledgments
This work was supported by grant AI 52390 from the National Institutes of Health to KWD and the Medical Scientist Training Program grant GM07739 and the Paul and Daisy Soros Fellowship for SPK. We thank John Stiller for his critical reading of the manuscript. We thank the Malaria Research and Reference Reagent Resource Center (MR4), Dr. X Su of the NIAID for P. falciparum isolate gDNAs and Mr. Bryan Falk of the American Museum of Natural History for extracting the P. yoelii, P. berghei and P. vinckei species and sub-species gDNAs.
Abbreviations
- CTD
carboxy-terminal domain or C-terminal domain
Footnotes
Data deposition footnote:
Sequences generated by this study: RNA Polymerase II CTD (regions R2 and R3)
| Accession Code: | Source | |
|---|---|---|
| P. gallinaceum RNA polymerase II (whole) | EU840284 | Sanger |
| P. berghei isolate ANKA | EU827171 | Edinburgh |
| P. berghei isolate NK65 | EU827172 | Edinburgh |
| P. yoelii yoelii 17X | EU827173 | Edinburgh |
| P. yoelii yoelii 33X | EU827174 | Edinburgh |
| P. yoelii killicki | EU827175 | Edinburgh |
| P. yoelii nigeriensis | EU827176 | Edinburgh |
| P. yoelii (new subspecies) | EU827177 | Edinburgh |
| P. vinckei VIBA CyO | EU827178 | Edinburgh |
| P. vinckei VIBA Cy P1 | EU827179 | Edinburgh |
| P. vinckei lentum 194ZZ | EU827180 | Edinburgh |
| P. vinckei (new subspecies) | EU827181 | Edinburgh |
| P. reichinowi | EU850397 | Sanger |
| P. vivax isolate ONG | EU840276 | MRA-341G |
| P. vivax isolate Pakchong | EU840277 | MRA-342G |
| P. vivax isolate Nicaragua | EU840278 | MRA-343G |
| P. vivax isolate Panama | EU840283 | MRA-340G |
| P. cynomolgi isolate Smithsonian | EU840279 | MRA-351G |
| P. cynomolgi isolate Cambodian | EU840280 | MRA-579G |
| P. cynomolgi isolate Bastianelli | EU840281 | MRA-350G |
| P. fragile | EU840282 | MRA-352G |
| P. falciparum isolate FCR3 | EU827182 | MRA-731 |
| P. falciparum isolate D6 | EU827183 | MRA-285 |
| P. falciparum isolate RO33 | EU827184 | MRA-200 |
| P. falciparum isolate 7G8 | EU827185 | NIH |
| P. falciparum isolate HB3 | EU827186 | MRA-155 |
| P. falciparum isolate Santa (St.) Lucia | EU827187 | MRA-331 |
| P. falciparum isolate FCC-2 | EU827188 | MRA-733 |
| P. falciparum isolate D10 | EU827189 | NIH |
| P. falciparum siolate K1 | EU827190 | MRA-159 |
| P. falciparum isolate W2 | EU827191 | MRA-157 |
| P. falciparum isolate DD2 | EU827192 | MRA-156 |
| P. ovale | EU887536 | NYU |
Reference List
- Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: Genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- Barron-Casella E, Corden JL. Conservation of the mammalian RNA polymerase II largest-subunit C-terminal domain. J Mol Evol. 1992;35:405–410. doi: 10.1007/BF00171818. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Halden NF, Cullen CR, Corden JL. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1988;8:330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkiewicz EA, Moon WA, Woody RW. Conformation of the RNA polymerase II C-terminal domain: circular dichroism of long and short fragments. J Mol Biol. 2000;297:119–133. doi: 10.1006/jmbi.2000.3545. [DOI] [PubMed] [Google Scholar]
- Carty SM, Greenleaf AL. Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and RNA processing. Mol Cell Proteomics. 2002;1:598–610. doi: 10.1074/mcp.m200029-mcp200. [DOI] [PubMed] [Google Scholar]
- Chapman AB, Agabian N. Trypanosoma brucei RNA polymerase II is phosphorylated in the absence of carboxyl-terminal domain heptapeptide repeats. J Biol Chem. 1994;269:4754–4760. [PubMed] [Google Scholar]
- Chapman RD, Conrad M, Eick D. Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol Cell Biol. 2005;25:7665–7674. doi: 10.1128/MCB.25.17.7665-7674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- Corden JL. Transcription. Seven ups the code. Science. 2007;318:1735–1736. doi: 10.1126/science.1152624. [DOI] [PubMed] [Google Scholar]
- Dacks JB, Marinets A, Ford DW, Cavalier-Smith T, Logsdon JM., Jr Analyses of RNA Polymerase II genes from free-living protists: phylogeny, long branch attraction, and the eukaryotic big bang. Mol Biol Evol. 2002;19:830–840. doi: 10.1093/oxfordjournals.molbev.a004140. [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Egloff S, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesecke H, Barale JC, Langsley G, Cornelissen AW. The C-terminal domain of RNA polymerase II of the malaria parasite Plasmodium berghei. Biochem Biophys Res Commun. 1991;180:1350–1355. doi: 10.1016/s0006-291x(05)81344-0. [DOI] [PubMed] [Google Scholar]
- Guo Z, Stiller JW. Comparative genomics and evolution of proteins associated with RNA polymerase II C-terminal domain. Mol Biol Evol. 2005;22:2166–2178. doi: 10.1093/molbev/msi215. [DOI] [PubMed] [Google Scholar]
- Howe KJ. RNA polymerase II conducts a symphony of pre-mRNA processing activities. Biochim Biophys Acta. 2002;1577:308–324. doi: 10.1016/s0167-4781(02)00460-8. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Li WB, Bzik DJ, Gu HM, Tanaka M, Fox BA, Inselburg J. An enlarged largest subunit of Plasmodium falciparum RNA polymerase II defines conserved and variable RNA polymerase domains. Nucleic Acids Res. 1989;17:9621–9636. doi: 10.1093/nar/17.23.9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao SM, Taylor IC, Kingston RE, Young RA. RNA polymerase II carboxy-terminal domain contributes to the response to multiple acidic activators in vitro. Genes Dev. 1991;5:2431–2440. doi: 10.1101/gad.5.12b.2431. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, et al. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Litingtung Y, et al. Growth retardation and neonatal lethality in mice with a homozygous deletion in the C-terminal domain of RNA polymerase II. Mol Gen Genet. 1999;261:100–105. doi: 10.1007/s004380050946. [DOI] [PubMed] [Google Scholar]
- Liu P, Greenleaf AL, Stiller JW. The essential sequence elements required for RNAP II carboxyl-terminal domain function in yeast and their evolutionary conservation. Mol Biol Evol. 2008;25:719–727. doi: 10.1093/molbev/msn017. [DOI] [PubMed] [Google Scholar]
- Meininghaus M, Chapman RD, Horndasch M, Eick D. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J Biol Chem. 2000;275:24375–24382. doi: 10.1074/jbc.M001883200. [DOI] [PubMed] [Google Scholar]
- Mizzen CA, et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- Nonet M, Sweetser D, Young RA. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Perkins SL. Molecular systematics of the three protein-coding genes of malaria parasites: corrobative and new evidence for the origins of human malaria. Mitochondrial DNA. 2008;19:471–478. doi: 10.1080/19401730802570926. [DOI] [PubMed] [Google Scholar]
- Perkins SL, Sarkar IN, Carter R. The phylogeny of rodent malaria parasites: simultaneous analysis across three genomes. Infect Genet Evol. 2007;7:74–83. doi: 10.1016/j.meegid.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller JW, Cook MS. Functional unit of the RNA polymerase II C-terminal domain lies within heptapeptide pairs. Eukaryot Cell. 2004;3:735–740. doi: 10.1128/EC.3.3.735-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller JW, McConaughy BL, Hall BD. Evolutionary complementation for polymerase II CTD function. Yeast. 2000;16:57–64. doi: 10.1002/(SICI)1097-0061(20000115)16:1<57::AID-YEA509>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Volkman SK, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- Zorio DA, Bentley DL. The link between mRNA processing and transcription: communication works both ways. Exp Cell Res. 2004;296:91–97. doi: 10.1016/j.yexcr.2004.03.019. [DOI] [PubMed] [Google Scholar]