Abstract
Microorganisms play key roles in biogeochemical and nutrient cycling in all ecosystems on Earth, yet little is known about the processes controlling their biogeographic distributions. Here we report an investigation of magnetotactic bacteria (MTB) designed to evaluate the roles of niche-based process and spatial process in explaining variation in bacterial communities across large spatial scales. Our results show that both environmental heterogeneity and geographic distance play significant roles in shaping dominant populations of MTB community composition. At the spatial scale in this study, the biogeography of MTB is relatively more influenced by environmental factors than geographic distance, suggesting that local conditions override the effects of dispersal history on structuring MTB community. Of note, we found that the strength of geomagnetic field may influence the biogeography of MTB. We argue that MTB have the potential to serve as a model group to uncover the underlying processes that influence microbial biogeography.
A major challenge in biogeography is to identify the factors that regulate diversity and distribution of organisms on Earth. Although biogeographic patterns for animals and plants are well documented, the diversity and distribution of microorganisms, which play key roles in all ecosystems, are poorly understood. Whether microorganisms represent cosmopolitan or ecologically restricted distribution is a contentious and hotly debated topic1. It was previously assumed that microorganisms have a random and cosmopolitan distribution because of their large population numbers, small sizes, short generation times, and high dispersal capabilities2. However, with the advent of molecular techniques, a rapidly growing body of evidence suggests that microorganisms may exhibit biogeographic patterns1,3,4. Niche-based process and spatial process are two alternative strategies proposed to generate and maintain microbial diversity1. The former emphasizes the importance of local environmental conditions and assumes that same environments should support similar microbial communities regardless of geographic distances, the so called “everything is everywhere, but, the environment selects” situation. In contrast, the latter strategy emphasizes the dependence of geographic distances rather than environmental gradients, which is the similar in concept to Hubbell's neutral theory for macroorganisms that stochastic processes and dispersal limitation affect variation in species composition5.
Magnetotactic bacteria (MTB) are diverse microbes united by the ability to form intracellular magnetic crystals of magnetite and/or greigite usually arranged into one or more linear chains6. These magnetic inclusions called magnetosomes help these bacteria to sense and swim along the Earth's magnetic field lines (a behavior known as magnetotaxis)6. All known MTB are found within the Alphaproteobacteria, Deltaproteobacteria, Gammaproteobacteria, phylum Nitrospirae, or the candidate division OP37,8,9,10. MTB are able to accumulate up to 2–3% iron per cell by dry weight, which is several orders of magnitude higher than iron in Escherichia coli11. Considering their wide distribution in diverse aquatic and sedimentary ecosystems and high intracellular iron content, MTB may have global significance in iron cycling12,13 as well as bulk magnetization of sediments14,15.
Despite of their remarkable magnetic abilities and proposed ecological functions, our understanding of MTB biogeography remains very poor. Although several studies have found that some environmental factors, such as salinity16,17, temperature18, nitrate19, or sulfur compounds20, could explain MTB abundance or community differences at local or regional scales, little information is available concerning the biogeography of MTB across large spatial scales21. In this study we have compared the diversity and distribution of MTB communities from different aquatic ecosystems ranging over a large spatial scale (Fig. 1 and Supplementary Table S1 online). The goals of the present study were to (i) describe the large-scale biogeographic pattern of MTB communities, (ii) identify environmental factors that may contribute to the distribution of MTB, and (iii) quantify the relative abundances of niche-based and spatial processes involved with the structuring of MTB communities. These results may provide a starting point for understanding the underlying mechanism(s) leading to the biogeography of these, and perhaps other, microorganisms.
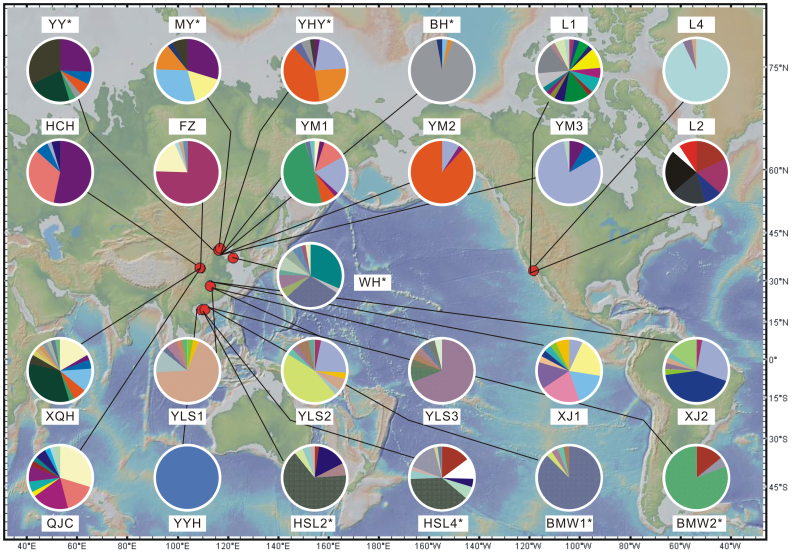
Figure 1. The locations of 16 sampling sites in this study and 9 locations (those with *) from previous study21 that are compared here.
Distribution pies of distinct OTUs (98% similarity threshold) are shown for each site. The sampling sites are described in more detail in Supplementary Table S1 online. The map was generated using GeoMapApp version 2 (http://www.geomapapp.org/).
Results
Magnetic enrichment of MTB and their phylogenetic diversity
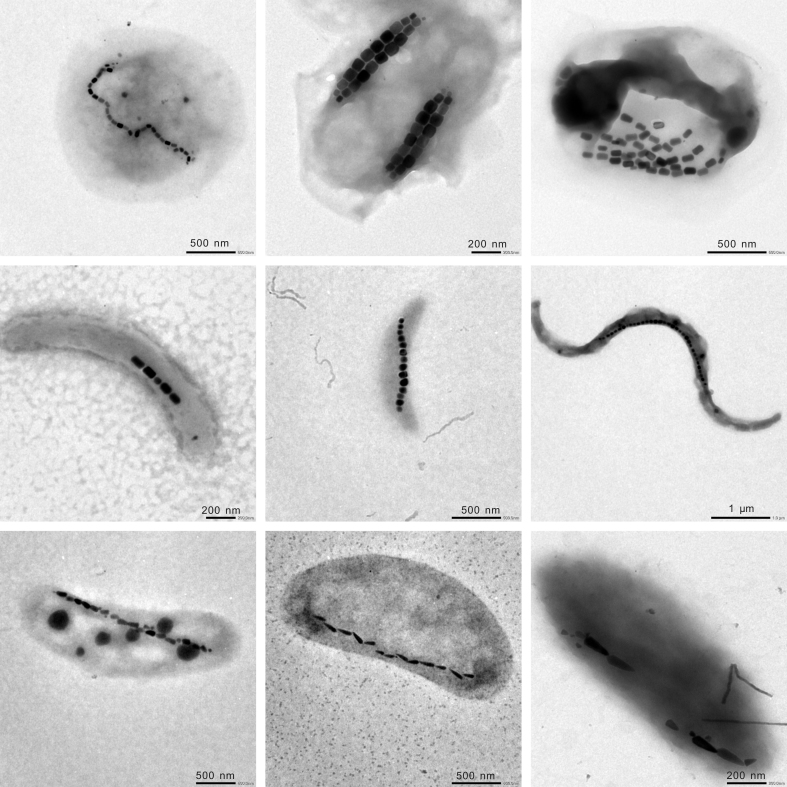
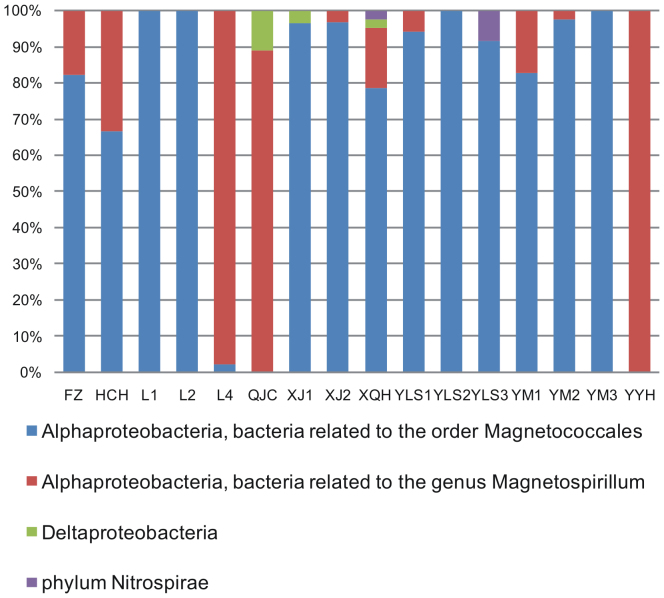
MTB were discovered from all 16 locations across various ecosystems (Supplementary Table S1 online). Different morphologies of MTB cells were identified, such as cocci, rods, vibrios, and spirilla (Fig. 2). Living MTB cells were concentrated and enriched by taking advantage of their motility and magnetotaxis through the “MTB trap” method22. The diversity of enriched bacterial samples was assessed by comparison of 16S rRNA genes. Nearly 700 sequences were retrieved after removing sequences of insufficient quality or potential chimeras. The most highly represented taxa were members of the phylum Proteobacteria (> 90%). Other sequences were identified to belong to the phyla Nitrospirae, Bacteroidetes, TM7, OD1, Actinobacteria, Firmicutes, or unclassified Bacteria. It was noted that some fast-swimming non-magnetotactic bacteria could be collected during the magnetic enrichment23. In order to remove these potential contaminations, sequences most similar to non-magnetotactic organisms were arbitrarily attributed to contaminations and were removed from further analyses. We ended up with a total of 580 sequences, in which bacteria related to the order Magnetococcales and the genus Magnetospirillum in the Alphaproteobacteria were the most dominant groups, representing 72% and 26% of all sequences, respectively. Consistent with previous studies7, bacteria related to the order Magnetococcales dominated the MTB communities in most sampling locations, while bacteria related to the genus Magnetospirillum were the major group in a few locations (e.g., L4, QJC and YYH) (Fig. 3). Sequences belonging to the Deltaproteobacteria and the phylum Nitrospirae were also detected (Figs. 3 and 4). Sequences in the Deltaproteobacteria were identified to affiliate with the orders Desulfobacterales and Desulfovibrionales, while Nitrospirae sequences identified here were related to MTB sequences belonging to groups 1 and 3 as reported previously9.
Figure 2. Representative transmission electron micrographs of MTB cells retrieved in this study.
Figure 3. Taxonomic classification of MTB sequences retrieved from 16 locations in this study.
Refer to Supplementary Table S1 online for detailed sample information.
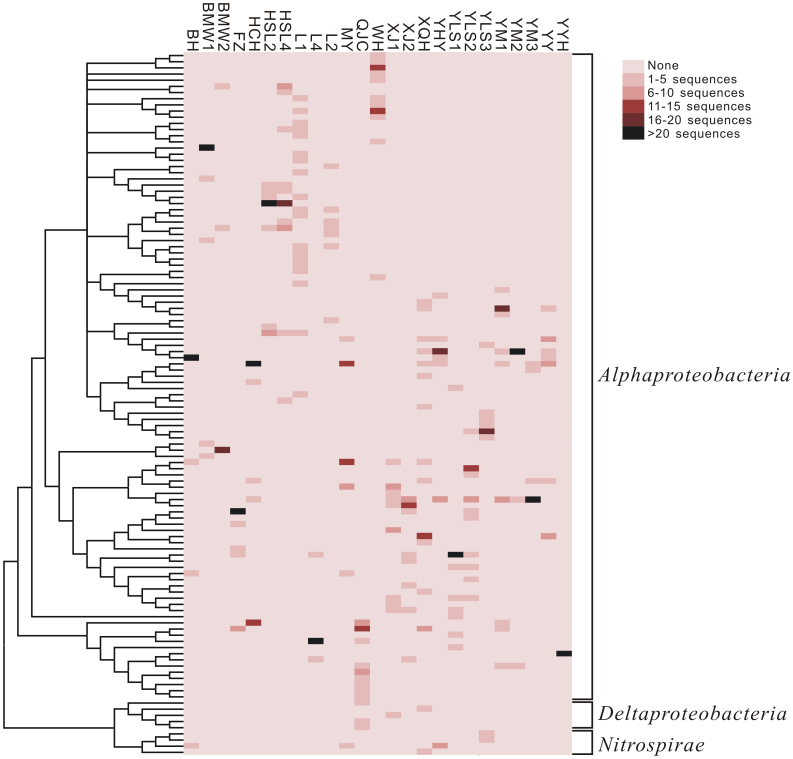
Figure 4. Heatmap showing the abundance and distribution of operational taxonomic units (OTUs at 98% threshold similarity) for 25 16S rRNA gene clone libraries of MTB communities that are compared in this study.
The abundance of each OTU in each library is indicated by different colors. On the left-hand side, a neighbor-joining phylogenetic tree shows the phylogenetic relationship between OTUs.
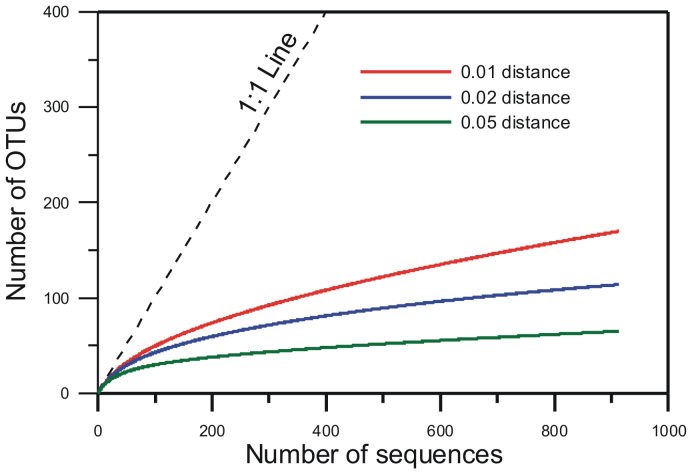
In addition to sequence data generated from 16 locations in this study, we included our previously described data set of MTB communities from 9 locations across northern and southern China21, and compared all these MTB communities together (Fig. 1). It is appropriate to combine these two data sets because of similar sampling, enrichment, and experimental approaches performed in these studies. Together, a total of more than 900 MTB sequences from 25 locations were analyzed (Fig. 1). These sequences can be clustered into 170, 114 and 65 operational taxonomic units (OTUs) at 99%, 98% and 95% similarity cutoffs, respectively (Figs. 4 and 5). Rarefaction curves for all samples nearly reached an asymptote, indicating that we successfully captured the major extent of MTB diversity (Fig. 5).
Figure 5. Rarefaction curves for sequences at 99%, 98%, and 95% sequence similarity levels, respectively.
Analyzing the biogeographic pattern of the MTB
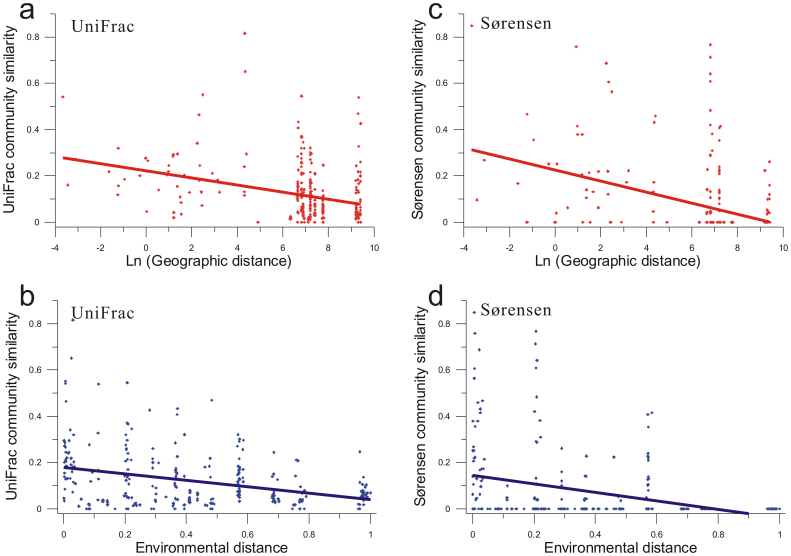
To investigate the biogeography of MTB across studied locations, we used two distinct approaches to determine pairwise community similarities between samples: the Sørensen index and the UniFrac index. The Sørensen index is a taxonomy-based approach that assesses community differences at a single level of taxonomic resolution by defining OTUs at an arbitrary sequence similarity level (e.g., 98% in this study)24. While, the phylogeny-based UniFrac index measures the overall degree of phylogenetic divergence between sets of communities, which allows us to compare community phylogenies in a more integrated manner than the taxonomy-based approach25.
It has been demonstrated that similarities between MTB communities significantly decreased with increasing geographic distance (Fig. 6a and c, P < 0.001), reflecting the distance-decay relationship26. Thus, geographic distance plays a role in controlling MTB distribution similar to that seen in other microorganisms27,28. Changes of MTB community also significantly depend on environmental distance between sites (Fig. 6b and d, P < 0.001), indicating that environmental conditions influence MTB species composition as well. In addition, we noted that although a few OTUs are shared by up to 8 locations, nearly 70% of OTUs are endemic, i.e., found at a unique sample location (Fig. 4). Taken together, these results provide strong evidence that the dominant populations of MTB communities at scales used in this study represent restricted distribution, and both local environment and dispersal history influence their biogeographic pattern. The patterns are similar irrespective of which methods (phylogeny-based UniFrac index or taxonomy-based Sørensen index) are used (Fig. 6).
Figure 6. Correlations of MTB community similarity with geographic and environmental distances.
Both phylogeny-based UniFrac index and taxonomy-based Sørensen index were used as community similarity. Environmental distances are normalized. All correlations are statistically significant (P < 0.001).
Factors influencing MTB biogeography
Permutation-based multiple regression on distance matrices (MRM) was performed to determine environmental variables that significantly contributed to explain the observed variation in dominant MTB communities (Table 1). When the UniFrac index was considered, the variables that significantly explained MTB patterns were salinity, Eh, sulfate, temperature, and strength of geomagnetic field. When the Sørensen index was used, in addition to the above-mentioned factors, total iron was also found to significantly contribute to variance in MTB communities (Table 1).
Table 1. Permutation-based multiple regression on distance matrices of MTB community distances, based on the UniFrac index or the Sørensen index, with geographic distance and environmental factors between sampling sites.
| Variable | UniFrac index | Sørensen index |
|---|---|---|
| R2 | R2 | |
| Ln-transformed geographic distance | 0.110*** | 0.175*** |
| Geomagnetic field strength | 0.106*** | 0.128*** |
| Salinity | 0.301*** | 0.123*** |
| Sulfate | 0.179*** | 0.099*** |
| Temperature | 0.042** | 0.035* |
| Eh | 0.062*** | 0.026* |
| Iron | 0.015 (n.s.) | 0.045* |
| Nitrate | 0.010 (n.s.) | 0.011 (n.s.) |
| Phosphate | 0.007 (n.s.) | 0.000 (n.s.) |
| Nitrite | 0.024 (n.s.) | 0.021 (n.s.) |
| pH | 0.001 (n.s.) | 0.002 (n.s.) |
Abbreviation: n.s., not significant; * P < 0.05; ** P ≤ 0.01;*** P ≤ 0.001.
Quantification of the relative roles of niche-based and spatial processes
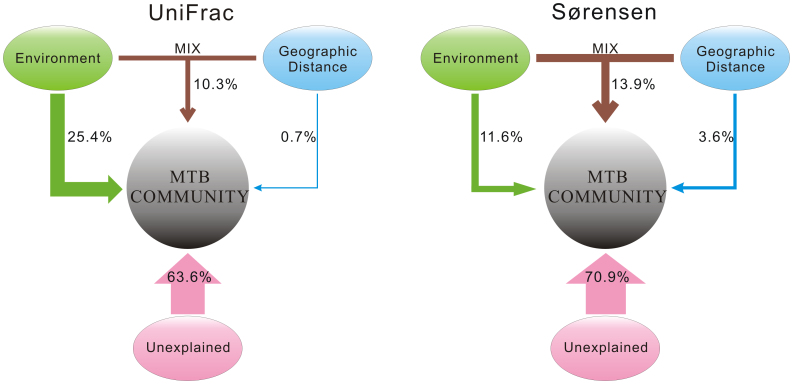
Regressing community similarity against selected environmental factors and geographic distance is an effective approach to quantify the relative roles of niche-based process and spatial process in control of community composition, and is widely applied in biogeographic studies of macroorganisms29. When this approach was used, it was possible to partition the variation in MTB community distance into four components. As shown in Figure 7, most of the explanatory power was pure environment (25.4% for UniFrac index) or both environmental heterogeneity and geographic distance (MIX) (13.9% for Sørensen index). Pure geographic distance alone explained only a minor portion of the variation in MTB communities (0.7% for UniFrac index and 3.6% for Sørensen index). Not surprisingly, more than half of the total variation (63.6–70.9%) remained unexplained by either measured environmental factors or spatial distances. Such poorly explained variance appears to be a common pattern for microorganisms, which may be either due to non-measured environmental variables or accounted for ecosystem productivity, biological interactions, historical events, and other factors that are not considered here27,30.
Figure 7. Relative importance of different variables in explaining variation in MTB communities between sites.
The diagrams are based on multiple regression on distance matrices to partition the variation into four components. MTB community distances were based on either the UniFrac index or the Sørensen index.
Discussion
In this analysis, both environmental heterogeneity and geographic distance are found to play significant roles in shaping dominant populations of MTB community composition. This observation on MTB is in line with results from tropical forests31,32,33 and terrestrial vertebrates34, suggesting that biogeographic patterns between dominant MTB communities and macroorganisms may not be fundamentally different35. However, it must be considered that microorganisms are significantly different from macroorganisms in many aspects, such as body sizes, generation times, dispersal capabilities, and reproduction modes. One of the primary differences between them is the population abundance, i.e., the number of microorganisms on Earth is many orders-of-magnitude larger than that of macroorganisms. For microorganisms, low-abundance populations are normally difficult to detect due to masking by dominant species, which may lead to underestimation of low-abundance cosmopolitan microbes36. We are aware that some MTB strains with slow motility may not be collected using magnetic enrichment approach in this study. Therefore, at this stage, our results only represent the distribution patterns of dominant MTB populations in the studied locations. Additional information will be necessary to fully assess the biogeography of low-abundance MTB with regard to their true ecological nature.
For environmental factors characterized here, salinity was found to contribute to a large part of regression coefficient (R2 = 0.123–0.301, P < 0.001; Table 1). Salinity has been identified as a key determinant of overall microbial communities37,38 as well as MTB abundance16,17 and biogeography21. Salinity is believed to directly affect microbial community structure by selecting groups adapted to a particular salt concentration39. Alternatively, competitors or predators of MTB may change across locations with different salinity, which could influence the diversity and distribution of MTB as well. Therefore, the freshwater-saline boundary may be a difficult barrier for the MTB to cross. Temperature was another noteworthy significant factor in driving biogeography of MTB (Table 1). It extends the recent microcosm-based experiment that revealed community structure of MTB changed with elevated temperature18 to natural habitats, implying that climate changes may influence the diversity and distribution of MTB in nature.
One striking finding in this study was the significant correlation of MTB community with the gradient of the Earth's magnetic field strength (approximately 44000–55000 nT) across the large spatial scale considered here (P < 0.001, Table 1). Since strength of geomagnetic field varies with latitude and temperature, the correlation between geomagnetic field strength and MTB community could be a result of co-variation with latitude or temperature. However, it was noted that geomagnetic field explains more variability (R2 = 0.106–0.128) in MTB communities than latitude (R2 = 0.073–0.089) and temperature (R2 = 0.035–0.042). Moreover, the significant correlation between geomagnetic field and MTB community was not affected by removing effects of latitude or temperature using partial Mantel test (P ≤ 0.01). All these results indicate that geomagnetic field is probably an important geophysical factor that may influence diversity and/or activities of MTB in the studied locations. There are several possible mechanisms that may account for the influence of geomagnetic field. The strength of geomagnetic field may directly affect the growth, metabolism, swimming behavior or biomineralization of MTB40,41 and thus plays a role in regulating their community composition. In addition, for life on Earth, the geomagnetic field acts as an important protective barrier against cosmic radiation42. Regions of relatively weak geomagnetic field strength are likely to experience an increased influence of cosmic radiation at the Earth's surface, which may affect biological processes of MTB communities. Since our studied samples are all from the Northern Hemisphere, it is necessary in future studies to analyze and compare MTB communities from the Southern Hemisphere, as well as higher latitude regions, and to confirm whether variations of geomagnetic field would affect the global distribution of MTB. In addition, further experimental analyses in lab are also necessary to address the underlying mechanisms of magnetic field effects on MTB activity and/or diversity. To our knowledge, this is the first report of potential effects of the Earth's magnetic field on the biogeography of microorganisms in nature, which may improve our understanding links between variation of the Earth's magnetic field and evolution of life on Earth.
This study on MTB contributes to the current debate in microbial biogeography about the relative roles of niche-based process and spatial process in structuring microbial communities. Some studies have found that environmental heterogeneity, like pH43 or salinity38, is a primary factor influencing microbial distribution, while others have suggested that the distribution of microbial communities is largely controlled by geographic distance44,45,46. In the present study, MRM-based variation partitioning analyses have quantitatively revealed that pure environmental factors (for UniFrac indext) or MIX (for Sørensen index) explain more of the variation in community similarity of MTB than do pure geographic distances (Fig. 7). The fraction of MIX is a consequence of co-variation of environmental and spatial variables in nature, and can be interpreted as a spatially structured environmental condition47. Thus a high fraction of MIX suggests that environmental heterogeneity could be of great importance in shaping community composition48. It thus appears that the niche-based process has stronger influence on MTB community distribution than the spatial process of dispersal history, which is consistent with several studies that emphasize the importance of local environmental conditions in structuring microbial communities1,4.
It is important to recognize that in this study geographic distance plays a minor but significant role that should not be ignored. This result indicates that while microbes are thought to have high dispersal capabilities (e.g., transport by migrating animals or water currents)3, spatial process of dispersal limitation and/or historical events still play a role in MTB distribution over the spatial scale considered in this study. A number of studies on macroorganisms have concluded that the relative importance of environment and geographic distance is spatial scale dependent, and a similar conclusion was recently reached for microorganisms as well28. Taken together, our study highlights the importance of integrating both niche-based and spatial processes in investigations of microbial biogeography.
One should be aware that the data presented here are based on magnetic enrichment of MTB cells followed by comparison and classification of 16S rRNA genes. This may introduce some potential biases. For example, some slow motile MTB may not be captured through magnetic enrichment, or those sequences not similar to any known MTB populations that were discounted in this study may be from totally novel MTB strains not yet described. Therefore, further culturing efforts, fluorescence in situ hybridization or single-cell analyses will be necessary to better understand the overall diversity of MTB in nature. Nevertheless, in spite of these potential biases, this study represents one of the largest cross-site surveys of MTB biogeography conducted to date, and our results have revealed clear biogeographic patterns of studied MTB communities across different locations, suggesting that 16S rRNA gene analysis of magnetically enriched MTB is still an effective approach to compare the general diversity of dominant MTB communities in nature21,49. In addition, identical approach was performed for all samples in this study, which would minimize the potential bias of experimental procedures.
Despite recent progress, describing biogeography of microbial communities and ascertaining the relative importance of different factors that account for these trends remains very difficult. One of the challenges is the enormous diversity of microorganisms in natural environments that is difficult to be fully addressed even using the most advanced sequencing technologies. Hence, our knowledge of the fundamental principles influencing microbial biogeography remains limited. Our results presented here suggest that MTB provide an opportunity to test microbial biogeographic theories. Advantages for choosing MTB for microbial biogeography analysis are: (i) MTB are free-living bacteria that are ubiquitous in diverse sedimentary ecosystems; (ii) living MTB cells can be easily enriched through their active magnetotactic behavior, which should better reflect contemporary diversity because the enriched bacteria are free of dead cells and/or ancient DNA; and (iii) the sequence diversity of MTB communities in nature, compared with the whole bacterial community, is moderate, and therefore is easily handled and can be addressed at a high degree of taxonomic resolution. Therefore, MTB have the potential to serve as a model group to uncover the underlying processes that influence microbial biogeography.
In summary, our results show that major populations of MTB do not randomly distribute at large spatial scales but represent an ecologically restricted distribution. Both environmental heterogeneity and geographic distance contribute to this distribution, indicating that the biogeography of MTB is controlled by a combination of niche-based and spatial processes. Environmental heterogeneity (with or without spatial structure) is found to explain more variation in MTB than pure geographic distance, indicating that contemporary environmental condition is one of major factors in structuring MTB community composition in nature. The community similarity of MTB significantly correlates with strength of the Earth's magnetic field, which suggests that geomagnetic field may affect the diversity and biogeography of MTB. This study will form the basis of more detailed studies to further define the global biogeography and ecological functions of MTB communities.
Methods
Site sampling, MTB enrichment, and microscopic observation
Surface sediment samples from sixteen locations were collected across different ecosystems in China and USA (Supplementary Table S1 online). Geographic distances between sampling sites ranged from 0.026 km to 12,240 km. At each sampling site, surface sediments from the top 5–20 cm were collected. The existence of MTB in sediment samples was checked through the “hanging-drop” method50. MTB were magnetically enriched using the “MTB trap” method as described previously19,22. For TEM observation, 20 μl of MTB enrichments were deposited on Formvar-carbon-coated copper grids and were imaged using a JEM-1400 microscope operating at 80 kV (JEOL Corporation, Japan). The rest of the enrichments were frozen at −20°C prior to molecular analysis.
Environmental factors analysis
Several environmental factors of bulk surface sediments were measured. Salinity and pH were measured using a HQ40d salinity meter (HACH, Loveland, Colorado, USA) and a Mettler Toledo Delta 320 pH meter (Mettler-Toledo, Greifensee, Switzerland), respectively. Nitrate, nitrite, sulfate, phosphate, and total iron in pore water were also analyzed spectroscopically using a DR2800 Spectrophotometers (HACH, Loveland, Colorado, USA) and powder pillows detection kits (HACH, Loveland, Colorado, USA) based on the cadmium reduction method, diazotization method, SulfaVer 4 method, ascorbic acid method, and the FerroMo method, respectively, by following the manufacturer's instructions. Redox potential (Eh) was measured using a Metrohm 842 titrando Eh meter (Metrohm, Herisau, Switzerland). The geomagnetic field intensity of each sampling site was acquired from NOAA's National Geophysical Data Center using the model IGRF 11 (the 11th International Geomagnetic Reference Field). We also included five-year mean land surface temperature (2007–2011) of each site as a climatic factor. The temperature data set was from MODIS Land Product Subsets (http://daac.ornl.gov/MODIS/MODIS-menu/).
16S rRNA gene sequences amplification and analysis
16S rRNA genes were directly amplified from the magnetically enriched MTB using bacterial universal primers 27F (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492R (5'-GGTTACCTTGTTACGACTT-3') as previously described51. Each 20 μl PCR mixture contained 1 μl of template, 10 μl of DreamTaq PCR Master Mix (MBI Fermentas, Vilnius, Lithuania), and 8 pmol of each primer. PCR was performed using a T-Gradient thermocycler (Whatman Biometra, Göttingen, Germany). The PCR amplification program consisted of 95°C for 5 min, 30 cycles of 92°C for 1.5 min, 50°C for 1 min, and 72°C for 2 min, and a final 10-min extension at 72°C. To avoid potential sample biases, triplicate PCR products for each sample were pooled and purified by 0.8% (w/v) agarose gel electrophoresis. Purified PCR products were cloned into the pMD19-T vector (TaKaRa, Dalian, China) and chemically DH5α competent cells (Tiangen, Beijing, China) by following the manufacture's instructions. Randomly selected clones were sequenced using the 27F primer (Beijing Genomics Institute, Beijing, China).
After removing vector contaminations and low-quality sequences, the rest were screened for chimeras using the Greengenes chimera-check tool (Bellerophon server)52. Those sequences which were most similar to non-MTB bacteria but unrelated to known MTB sequences were attributed to potential contaminations by non-magnetotactic microorganisms and were removed from further analyses. In this way, a total of 580 MTB sequences were retrieved. These sequence data have been submitted to the GenBank database under accession nos. JX294995-JX295574. The lengths of sequences were about 310–500 bp, covering V1 to V3 hypervariable regions53. We compared the MTB communities retrieved in this study with our previously described dataset of MTB communities from 9 locations across northern and southern China (Genbank accession nos. HQ437323-HQ437656)21. The latter dataset (334 MTB sequences) was combined with sequences acquired here, resulting in a total of 914 sequences from 25 locations. Sequences were aligned and clustered into OTUs at 95%, 98%, and 99% similarities, respectively, and rarefaction curves were then calculated using the RDP's Pyrosequencing Pipeline54. Representative sequences of OTUs at 98% threshold similarity were aligned using the NAST aligner at the Greengenes web site and were then taxonomically classified according to the best match with the Greengenes reference database52. A phylogenetic tree was constructed using MEGA version 5.0 through the neighbor-joining method55.
Statistical analyses
Statistical analyses in this study were based on resemblance matrices. Similarities between MTB communities were determined using two distinct approaches: phylogeny-based UniFrac matrix25 and taxonomy-based Sørensen matrix24. Environmental resemblance matrices were computed using Euclidean distances. A geographic distance matrix was calculated using latitudinal and longitudinal coordinates and the ‘Haversine’ formula56.
The plots of community similarity (both unweighted UniFrac matrix and Sørensen matrix) versus environmental distance and geographic distance were described, respectively. In these analyses, Euclidean distance of environment was obtained by using one climate variable (5-year mean temperature) and nine environmental factors (pH, Eh, salinity, nitrate, nitrite, sulfate, phosphate, total iron, and strength of geomagnetic field). Geographic distances were ln-transformed as suggested by Martiny et al28. Linear regressions of community similarity against geographic and environmental distances were calculated, respectively.
We used permutation-based multiple regression on matrices or MRM to quantify the relative contributions of measured environmental factors and geographic distance on the biogeography of MTB communities. In brief, the community similarity was partitioned into four components by MRM as suggested by Duivenvoorden et al31 and Jones et al33: (i) variation explained by pure environmental heterogeneity, (ii) variation explained by pure geographic distance, (iii) variation explained by both environmental heterogeneity and distance (MIX), and (iv) unexplained variation.
For MRM analyses, we first identify those environmental factors that significantly contribute to the variation in MTB community similarity. To do so, MRM was performed using each environmental factor as independent matrix. Those factors with significant contribution were selected for further analysis (as shown in Table 1). Then, the R-squares of geographic distance matrix as an independent matrix (R2G), selected environmental factors as independent matrices (R2E), and both geographic distance and selected environmental factors as independent matrices (R2T) were used to calculate the four components of variation of MTB community as suggested by Jones et al33: (i) pure environmental heterogeneity = R2T − R2G, (ii) pure geographic distance = R2T − R2E, (iii) MIX = R2G + R2E − R2T, and (iv) unexplained variation = 1 − R2T. The regression analyses were carried out using Ucinet version 6 with 999 permutations57.
To remove the effects of potential co-variables (latitude and temperature) with geomagnetic field, partial Mantel test was carried out to assessed how much the correlation between strength of geomagnetic field and MTB community decreased when the effects of latitude or temperature were partialled out. Partial Mantel tests were carried out using the program PASSaGE 2 and assessed by 999 permutations58. For all statistical analyses, a value of P < 0.05 was considered significant.
Author Contributions
W.L., Y.P. and K.N. designed research; W.L., Y.W. and Y.G. performed field work; W.L. and Y.W. performed research; W.L. analyzed data; W.L., Y.G., Y.P. and K.N. wrote the paper.
Supplementary Material
Table S1
Acknowledgments
We thank Andrea Cheung, Yuelong Huang, Wayne Harris, Haitao Chen, and Youliang Su for assistance with sample collection; and Greig Paterson for helpful discussions about data analysis. This work was supported by the CAS/SAFEA International Partnership Program for Creative Research Teams (KZCX2-YW-T10), the CAS project, and the NSFC grants 40821091 and 41104041. Wei Lin is grateful to the NASA Planetary Biology Internship for supporting his study in University of Southern California in Los Angeles, CA.
References
- Martiny J. B. H. et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112 (2006). [DOI] [PubMed] [Google Scholar]
- Finlay B. J. Global dispersal of free-living microbial eukaryote species. Science 296, 1061–1063 (2002). [DOI] [PubMed] [Google Scholar]
- Fontaneto D. Biogeography of microscopic organisms: is everything small everywhere? Vol. 79 (Cambridge University Press, 2011). [Google Scholar]
- Hanson C. A., Fuhrman J. A., Horner-Devine M. C. & Martiny J. B. H. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506 (2012). [DOI] [PubMed] [Google Scholar]
- Hubbell S. P. The unified theory of biodiversity and biogeography. (Princeton University Press, 2001). [Google Scholar]
- Bazylinski D. A. & Frankel R. B. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2, 217–230 (2004). [DOI] [PubMed] [Google Scholar]
- Amann R., Peplies J. & Schüler D. in Magnetoreception and Magnetosomes in Bacteria (ed D Schüler) 25–36 (Springer, 2007). [Google Scholar]
- Kolinko S. et al. Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ. Microbiol. 14, 1709–1721 (2012). [DOI] [PubMed] [Google Scholar]
- Lin W., Li J. & Pan Y. Newly isolated but uncultivated magnetotactic bacterium of the phylum Nitrospirae from Beijing, China. Appl. Environ. Microbiol. 78, 668–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre C. T. et al. A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science 334, 1720–1723 (2011). [DOI] [PubMed] [Google Scholar]
- Heyen U. & Schüler D. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Biochem. Biotechnol. 61, 536–544 (2003). [DOI] [PubMed] [Google Scholar]
- Faivre D. & Schüler D. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108, 4875–4898 (2008). [DOI] [PubMed] [Google Scholar]
- Roberts A. P. et al. Magnetotactic bacterial abundance in pelagic marine environments is limited by organic carbon flux and availability of dissolved iron. Earth Planet. Sci. Lett. 310, 441–452 (2011). [Google Scholar]
- Pan Y. et al. The detection of bacterial magnetite in recent sediments of Lake Chiemsee (southern Germany). Earth Planet. Sci. Lett. 232, 109–123 (2005). [Google Scholar]
- Larrasoaña J. C. et al. Magnetotactic bacterial response to Antarctic dust supply during the Palaeocene–Eocene thermal maximum. Earth Planet. Sci. Lett. 333–334, 122–133 (2012). [Google Scholar]
- Martins J. L., Silveira T. S., Silva K. T. & Lins U. Salinity dependence of the distribution of multicellular magnetotactic prokaryotes in a hypersaline lagoon. Int. Microbiol. 12, 193–201 (2009). [PubMed] [Google Scholar]
- Martins J. L. et al. Spatiotemporal distribution of the magnetotactic multicellular prokaryote Candidatus Magnetoglobus multicellularis in a Brazilian hypersaline lagoon and in microcosms. Int. Microbiol. 15, 141–149 (2012). [DOI] [PubMed] [Google Scholar]
- Lin W., Wan Y. & Pan Y. Short-term effects of temperature on the abundance and diversity of magnetotactic cocci. MicrobiologyOpen 1, 53–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. & Pan Y. Temporal variation of magnetotactic bacterial communities in two freshwater sediment microcosms. FEMS Microbiol. Lett. 302, 85–92 (2010). [DOI] [PubMed] [Google Scholar]
- Postec A. et al. Magnetotactic bacteria in microcosms originating from the French Mediterranean coast subjected to oil industry activities. Microb. Ecol. 63, 1–11 (2012). [DOI] [PubMed] [Google Scholar]
- Lin W., Wang Y., Li B. & Pan Y. A biogeographic distribution of magnetotactic bacteria influenced by salinity. ISME J. 6, 475–479 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler C. et al. Towards cloning the magnetotactic metagenome: identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from different aquatic sediments. Appl. Environ. Microbiol. 75, 3972–3979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons S. L., Sievert S. M., Frankel R. B., Bazylinski D. A. & Edwards K. J. Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond. Appl. Environ. Microbiol. 70, 6230–6239 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magurran A. E. Measuring Biological Diversity. (Blackwell, Oxford, UK, 2004). [Google Scholar]
- Lozupone C. A. & Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekola J. C. & White P. S. The distance decay of similarity in biogeography and ecology. J. Biogeogr. 26, 867–878 (1999). [Google Scholar]
- Ramette A. & Tiedje J. M. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Natl. Acad. Sci. U. S. A. 104, 2761–2766 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny J. B. H., Eisen J. A., Penn K., Allison S. D. & Horner-Devine M. C. Drivers of bacterial β-diversity depend on spatial scale. Proc. Natl. Acad. Sci. U. S. A. 108, 7850–7854 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichstein J. Multiple regression on distance matrices: a multivariate spatial analysis tool. Plant Ecol. 188, 117–131 (2007). [Google Scholar]
- Van der Gucht K. et al. The power of species sorting: local factors drive bacterial community composition over a wide range of spatial scales. Proc. Natl. Acad. Sci. U. S. A. 104, 20404–20409 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duivenvoorden J. F., Svenning J.-C. & Wright S. J. Beta diversity in tropical forests. Science 295, 636–637 (2002). [DOI] [PubMed] [Google Scholar]
- Tuomisto H., Ruokolainen K. & Yli-Halla M. Dispersal, environment, and floristic variation of western Amazonian forests. Science 299, 241–244 (2003). [DOI] [PubMed] [Google Scholar]
- Jones M. M., Tuomisto H., Clark D. B. & Olivas P. Effects of mesoscale environmental heterogeneity and dispersal limitation on floristic variation in rain forest ferns. J. Ecol. 94, 181–195 (2006). [Google Scholar]
- Qian H. & Ricklefs R. E. Disentangling the effects of geographic distance and environmental dissimilarity on global patterns of species turnover. Global Ecol. Biogeogr. 21, 341–351 (2012). [Google Scholar]
- Astorga A. et al. Distance decay of similarity in freshwater communities: do macro- and microorganisms follow the same rules? Global Ecol. Biogeogr. 21, 365–375 (2012). [Google Scholar]
- Sogin M. L. et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U. S. A. 103, 12115–12120 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump B. C., Hopkinson C. S., Sogin M. L. & Hobbie J. E. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70, 1494–1505 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C. A. & Knight R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U. S. A. 104, 11436–11440 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logares R. et al. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 17, 414–422 (2009). [DOI] [PubMed] [Google Scholar]
- Wang X. K. et al. Effects of hypomagnetic field on magnetosome formation of Magnetospirillum magneticum AMB-1. Geomicrobiol. J. 25, 296–303 (2008). [Google Scholar]
- Pan Y. et al. Reduced efficiency of magnetotaxis in magnetotactic coccoid bacteria in higher than geomagnetic fields. Biophys. J. 97, 986–991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhilber F. et al. 9,400 years of cosmic radiation and solar activity from ice cores and tree rings. Proc. Natl. Acad. Sci. U. S. A. 109, 5967–5971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N. & Jackson R. B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103, 626–631 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker R. J., Grogan D. W. & Taylor J. W. Geographic barriers isolate endemic populations of hyperthermophilic Archaea. Science 301, 976–978 (2003). [DOI] [PubMed] [Google Scholar]
- Reche I., Pulido-Villena E., Morales-Baquero R. & Casamayor E. O. Does ecosystem size determine aquatic bacterial richness? Ecology 86, 1715–1722 (2005). [Google Scholar]
- Green J. L. et al. Spatial scaling of microbial eukaryote diversity. Nature 432, 747–750 (2004). [DOI] [PubMed] [Google Scholar]
- Borcard D., Legendre P. & Drapeau P. Partialling out the spatial component of ecological variation. Ecology 73, 1045–1055 (1992). [Google Scholar]
- Smith T. W. & Lundholm J. T. Variation partitioning as a tool to distinguish between niche and neutral processes. Ecography 33, 648–655 (2010). [Google Scholar]
- Flies C. B., Peplies J. & Schüler D. Combined approach for characterization of uncultivated magnetotactic bacteria from various aquatic environments. Appl. Environ. Microbiol. 71, 2723–2731 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M., Canter K., Mahler I. & Tornheim A. Observation of magnetoreceptive behavior in a multicellular magnetotactic prokaryote in higher than geomagnetic fields. Biophys. J. 88, 1496–1499 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Li J., Schüler D., Jogler C. & Pan Y. Diversity analysis of magnetotactic bacteria in Lake Miyun, northern China, by restriction fragment length polymorphism. Syst. Appl. Microbiol. 32, 342–350 (2009). [DOI] [PubMed] [Google Scholar]
- DeSantis T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker G. C., Smith J. J. & Cowan D. A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55, 541–555 (2003). [DOI] [PubMed] [Google Scholar]
- Cole J. R. et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott R. W. Virtues of the Haversine. Sky & Telescope 68, 159 (1984). [Google Scholar]
- Borgatti S. P., Everett M. G. & Freeman L. C. Ucinet 6 for Windows: software for social network analysis. (Analytic Technologies, Harvard, MA, 2002). [Google Scholar]
- Rosenberg M. S. & Anderson C. D. PASSaGE: pattern analysis, spatial statistics and geographic exegesis. Version 2. Methods Ecol. Evol. 2, 229–232 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1