Abstract
Intracellular organelles are highly dynamic structures with varying shape and composition, which are subjected to cell-specific intrinsic and extrinsic cues. Their membranes are often juxtaposed at defined contact sites, which become hubs for the exchange of signaling molecules and membrane components1,2,3,4. The inter-organellar membrane microdomains that are formed between the endoplasmic reticulum (ER) and the mitochondria at the opening of the IP3-sensitive Ca2+ channel are known as the mitochondria associated-ER membranes or MAMs4,5,6. The protein/lipid composition and biochemical properties of these membrane contact sites have been extensively studied particularly in relation to their role in regulating intracellular Ca2+ 4,5,6. The ER serves as the primary store of intracellular Ca2+, and in this capacity regulates a myriad of cellular processes downstream of Ca2+ signaling, including post-translational protein folding and protein maturation7. Mitochondria, on the other hand, maintain Ca2+ homeostasis, by buffering cytosolic Ca2+ concentration thereby preventing the initiation of apoptotic pathways downstream of Ca2+ unbalance4,8. The dynamic nature of the MAMs makes them ideal sites to dissect basic cellular mechanisms, including Ca2+ signaling and regulation of mitochondrial Ca2+ concentration, lipid biosynthesis and transport, energy metabolism and cell survival 4,9,10,11,12. Several protocols have been described for the purification of these microdomains from liver tissue and cultured cells13,14.
Taking previously published methods into account, we have adapted a protocol for the isolation of mitochondria and MAMs from the adult mouse brain. To this procedure we have added an extra purification step, namely a Triton X100 extraction, which enables the isolation of the glycosphingolipid enriched microdomain (GEM) fraction of the MAMs. These GEM preparations share several protein components with caveolae and lipid rafts, derived from the plasma membrane or other intracellular membranes, and are proposed to function as gathering points for the clustering of receptor proteins and for protein–protein interactions4,15.
Keywords: Neuroscience, Issue 73, Genetics, Cellular Biology, Molecular Biology, Biochemistry, Membrane Microdomains, Endoplasmic Reticulum, Mitochondria, Intracellular Membranes, Glycosphingolipids, Gangliosides, Endoplasmic Reticulum Stress, Cell Biology, Neurosciences, MAMs, GEMs, Mitochondria, ER, membrane microdomains, subcellular fractionation, lipids, brain, mouse, isolation, animal model
Protocol
The following protocol is intended for the isolation and purification of MAMs and GEMs from mouse brain.
Solutions required for Isolation of mitochondria, MAMs and GEMs.
Fractionation to obtain crude mitochondrial preparation:
Solution A: 0.32 M Sucrose, 1 mM NaHCO3, 1 mM MgCl2, 0.5 mM CaCl2 + Protease Inhibitors (add fresh, as needed).
Solution B: 0.32 M Sucrose, 1 mM NaHCO3 + Protease Inhibitors (add fresh, as needed).
MAMs Isolation:
Isolation Medium: 250 mM Mannitol, 5 mM HEPES pH 7.4, 0.5 mM EGTA, 0.1% BSA.
Gradient Buffer: 225 mM Mannitol, 25 mM HEPES pH 7.5, 1 mM EGTA, 0.1% BSA (final concentration).
GEMs Isolation:
GEM Extraction Buffer: 25 mM HEPES pH 7.5, 0.15 M NaCl, 1% Triton X-100 + Protease Inhibitors (add fresh, as needed).
GEM Solubilising Buffer: 50 mM Tris-HCl, pH 8.8, 5 mM EDTA , 1% SDS + Protease Inhibitors (add fresh, as needed).
1. First Step Subcellular Fractionation: Removal of Nuclei, Unlysed Cells and Cellular Debris
Euthanize mouse in CO2 chamber.
Immediately remove brain, halve, place in 2 ml tubes on ice and weigh.
Note: Keep brain halves separate throughout the entire procedure. The following applies to the treatment of a single brain half.
Place half brain in a pre-chilled 2 ml glass Dounce tissue grinder containing 1 ml cold Solution A. Homogenize with 15 total strokes of a large clearance pestle.
Transfer to a 15 ml falcon tube, on ice, and dilute homogenates up to 10 volumes w/v, e.g. 0.225 g to 2.25 ml.
Centrifuge sample at 1,400 x g for 10 min at 4 °C.
Carefully remove the supernatant and transfer to a 30 ml round-bottom glass centrifuge tube, on ice. Make sure not to disturb the pellet.
Resuspend the pellet in the same 10 volumes of Solution A. Homogenize in the same grinder, with 3-6 strokes of a small clearance pestle, 1 ml at a time.
Transfer to a fresh 15 ml falcon tube and centrifuge at 710 x g for 10 min at 4 °C. This results in a pellet of nuclei and cell debris.
Carefully remove the supernatant and pool with the supernatant saved in step 1.6.
2. Second Step: Crude Mitochondria Isolation
Centrifuge supernatants at 13,800 x g for 10 min at 4 °C.
Transfer supernatant to an Ultra-Clear Beckman Centrifuge Tube, on ice
Resuspend pellet in 10 volumes Solution A. Homogenize in the same grinder, with 3-6 strokes of a small clearance pestle, 1 ml at a time.
Centrifuge as step 2.1.
Repeat steps 2.2-2.4.
The resulting pellet is an enriched mitochondrial fraction. Pool the supernatants (cytosol and ER), cover with parafilm and keep on ice.
Resuspend the pellet with 6 strokes of a small clearance pestle, in 4.8 ml/g (g is referred to the original brain weight) of Solution B, in a fresh homogeniser.
Using glass Pasteur pipettes, prepare a Discontinuous Sucrose Gradient in an Ultra-Clear Beckman Centrifuge Tube, adding subsequently to the bottom of the tube as follows, carefully, ensuring no bubbles are formed: Resupended Pellet (0.32 M Sucrose) 3 ml 850 mM Sucrose (in 1 mM NaHCO3) 3 ml 1 M Sucrose (in 1 mM NaHCO3) 3 ml 1.2 M Sucrose (in 1 mM NaHCO3)
Centrifuge at 82,500 x g for 2 hr at 4 °C. The resulting separation at the end of the gradient will produce three bands and a pellet: 1) myelin and other membrane contaminants (0.32 M - 0.85 M interface); 2) ER, Golgi, plasma membranes (0.85 M - 1 M interface); 3) synaptosomes (1 M - 1.2 M interface); 4) crude mitochondria (pellet) that is used for the subsequent purification steps
3. Isolation of Mitochondria-associated ER Membrane, MAMs
Resuspend the freshly isolated crude mitochondria from one brain half, in 2 ml Isolation Medium containing freshly added protease inhibitors.
Prepare a 30% Percoll gradient with Gradient Buffer 8 ml per half brain, and place in an Ultra-Clear Beckman Centrifuge Tube.
* Note: Make gradient buffer more concentrated (1.43 times) to achieve correct final concentration after diluting Percoll to 30%.
Layer mitochondrial suspension on top of prepared gradient SLOWLY to avoid any bubbles.
Centrifuge at 95,000 x g for 30 min, 4 °C.
- Remove the Heavy Fraction (lower band) FIRST with a glass Pasteur pipette and transfer to a fresh round-bottom glass tube, on ice.
- Remove the Light Fraction (upper band) SECOND with another glass Pasteur pipette and transfer to a separate fresh round-bottom glass tube, on ice.
Dilute both fractions collected in step 3.5 with 10 ml Isolation Medium and centrifuge at 6,300 x g for 10 min at 4 °C.
Discard the supernatant from the Heavy Fraction obtained in step 3.6, resuspend the pellet with another 10 ml Isolation Medium and centrifuge again, as step 3.6. The resulting pellet will be the pure mitochondrial fraction.
Transfer the supernatant from the Light Fraction obtained in step 3.6 to an Ultra-Clear Beckman Centrifuge Tube, on ice. Discard the pellet.
Add sufficient Isolation Medium to the supernatant obtained in step 3.8 to fill the tube and centrifuge at 100,000 x g for 1 hr, 4 °C. The resulting pellet will be the MAMs fraction. Carefully remove and discard the supernatant.
Note: At this step, you may also centrifuge at 100,000 x g for 1 hr, 4 °C the supernatant saved at step 2.6. The pellet will be the ER fraction and supernatant the Cytosolic fraction.
4. Extraction of Glycosphingolipid-enriched Microdomains, GEMs
Lyse pure mitochondria and/or MAM fractions in 500 μl-1 ml of Extraction Buffer for 20 min on ice.
Centrifuge lysates at 15,300 x g for 2 min at 4 °C.
Collect supernatants (Triton X-100 soluble material) and re-centrifuge pellets for 2 min to remove remaining soluble material. (Triton X-100 extracted Mitochondria and/or Triton X-100 extracted MAMs).
Solubilize pellets in Solubilising Buffer. This solubilized material represents the GEM fractions and can be used for further analysis.
Representative Results
Based on our experience with using this protocol we can safely recommend it for the isolation and purification of MAMs, GEMs and mitochondrial fractions from mouse brain. The procedure as outlined is highly reproducible and consistent. In Figure 1 we show a representative image of the way pure mitochondria and MAMs layer on a Percoll gradient (step 3.4). A defined, milky band containing purified mitochondria (Mito-P) segregates at the bottom of the ultracentrifuge tube, while the MAM fraction makes up a diffuse and broad band above the mitochondria. After careful recovery of the individual gradient bands, the purified fractions are resuspended in a lysis buffer, separated on SDS polyacrylamide gels and blotted onto PVDF membranes. Blots are then probed with a battery of antibodies for specific protein markers that will ascertain the purity of the fractions and their protein composition. Figure 2A shows the distribution of cytosolic, ER and mitochondrial markers in the isolated MAMs and mitochondrial fractions. Pure mitochondria preparations should be devoid of both ER and cytosolic markers. The close apposition between ER and mitochondrial membranes at the MAM contact sites explains the presence of calreticulin (ER marker) and Tom-20 (mitochondria marker) in these purified preparations. It is also possible to use MAMs specific markers FACL4 and PACS2, as well as other markers enriched within these domains, such as IP3R-1 and GRP754. The MAMs can be further extracted with Triton X-100 to obtain the GEMs. These microdomains, which contain components of lipid rafts and/or caveolae are caveolin-1 positive as shown in Figure 2B.
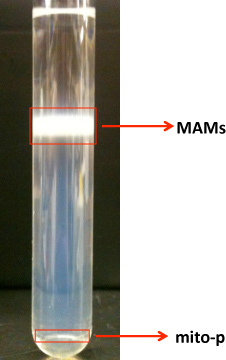 Figure 1. Shows a picture of the layering of the mitochondrial-associated ER membrane (MAMs) and the pure mitochondrial fractions (mito-p).
Figure 1. Shows a picture of the layering of the mitochondrial-associated ER membrane (MAMs) and the pure mitochondrial fractions (mito-p).
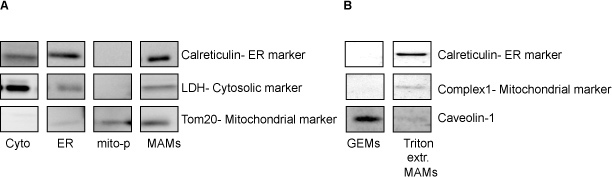 Figure 2. A) Shows western blots run to check the purity and the distribution of cytosolic, ER and mitochondrial markers in Cytosolic (cyto)-, ER-, pure mitochondrial fractions (mito-p)- and MAM-fractions. B) Western blots of the purified glycosphingolipid enriched microdomains (GEMs) and Triton X-100 extracted (Triton extr. MAMs) fractions isolated from MAMs.
Figure 2. A) Shows western blots run to check the purity and the distribution of cytosolic, ER and mitochondrial markers in Cytosolic (cyto)-, ER-, pure mitochondrial fractions (mito-p)- and MAM-fractions. B) Western blots of the purified glycosphingolipid enriched microdomains (GEMs) and Triton X-100 extracted (Triton extr. MAMs) fractions isolated from MAMs.
Discussion
The sites of contact between intracellular membranes or between organelles and the plasma membrane of cells represent dynamic signaling platforms for basic cellular processes. The accurate characterization of their function and composition under both physiological and pathological conditions requires reliable and reproducible purification protocols. The methods detailed here have been specifically optimized by our lab for the isolation and purification of the MAMs and their corresponding GEMs from the adult mouse brain. This protocol has been successfully implemented for the identification of the molecular effectors that specify these microdomains, and underlie the apoptotic pathway downstream of Ca2+ imbalance leading to neuronal cell death in a neurodegenerative metabolic disease in children4.
Disclosures
None of the authors have any conflicts of interest to declare.
Acknowledgments
We acknowledge the contribution of Renata Sano in conceiving the initial protocol. A.d’A. holds the Jewelers For Children (JFC) Endowed Chair in Genetics and Gene Therapy. This work was funded in part by NIH grants GM60905, DK52025 and CA021764, and the American Lebanese Syrian Associated Charities (ALSAC).
References
- Poburko D, Kuo KH, Dai J, Lee CH, van Breemen C. Organellar junctions promote targeted Ca2+ signaling in smooth muscle: why two membranes are better than one. Trends Pharmacol. Sci. 2004;25(1):8–15. doi: 10.1016/j.tips.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Pani B, Ong HL, Liu X, Rauser K, Ambudkar IS, Singh BB. Lipid rafts determine clustering of STIM1 in endoplasmic reticulum-plasma membrane junctions and regulation of store-operated Ca2+ entry (SOCE. J. Biol. Chem. 2008;283(25):17333–17340. doi: 10.1074/jbc.M800107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T, Rabouille C. Endoplasmic reticulum: one continuous network compartmentalized by extrinsic cues. Curr. Opin. Cell Biol. 2005;17(4):362–368. doi: 10.1016/j.ceb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Sano R, Annunziata I, Patterson A, Moshiach S, Gomero E, Opferman J, Forte M, d'Azzo A. GM1-ganglioside accumulation at the mitochondria-associated ER membranes links ER stress to Ca(2+)-dependent mitochondrial apoptosis. Mol. Cell. 2009;36(3):500–511. doi: 10.1016/j.molcel.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raturi A, Simmen T. Where the endoplasmic reticulum and the mitochondrion tie the knot: The mitochondria-associated membrane (MAM) Biochim. Biophys. Acta. 2012. [DOI] [PubMed]
- Giorgi C, De Stefani D, Bononi A, Rizzuto R, Pinton P. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. Int. J. Biochem. Cell Biol. 2009;41(10):1817–1827. doi: 10.1016/j.biocel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Azzo A, Tessitore R. Sano Gangliosides as apoptotic signals in ER stress response. Cell Death Differ. 2006;13:404–414. doi: 10.1038/sj.cdd.4401834. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Murgia M, Pozzan T. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 1993;262:744–747. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- Lynes EM, Bui M, Yap MC, Benson MD, Schneider B, Ellgaard L, Berthiaume LG, Simmen T. Palmitoylated TMX and calnexin target to the mitochondria-associated membrane. EMBO J. 2011;31(2):457–470. doi: 10.1038/emboj.2011.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Hayashi T, Su TP. The role of cholesterol in the association of endoplasmic reticulum membranes with mitochondria. Biochem. Biophys. Res. Commun. 2012;417(1):635–639. doi: 10.1016/j.bbrc.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S. The ER-mitochondria interface: the social network of cell death. Biochim. Biophys. Acta. 2012;1823(2):327–334. doi: 10.1016/j.bbamcr.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 2009;4(11):1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- Pizzo P, Viola A. Lymphocyte lipid rafts: structure and function. Curr. Opin. Immunol. 2003;15(3):255–260. doi: 10.1016/s0952-7915(03)00038-4. [DOI] [PubMed] [Google Scholar]


