Abstract
Murine bone marrow transplantation models provide an important tool in measuring hematopoietic stem cell (HSC) functions and determining genes/molecules that regulate HSCs. In these transplant model systems, the function of HSCs is determined by the ability of these cells to engraft and reconstitute lethally irradiated recipient mice. Commonly, the donor cell contribution/engraftment is measured by antibodies to donor- specific cell surface proteins using flow cytometry. However, this method heavily depends on the specificity and the ability of the cell surface marker to differentiate donor-derived cells from recipient-originated cells, which may not be available for all mouse strains. Considering the various backgrounds of genetically modified mouse strains in the market, this cell surface/ flow cytometry-based method has significant limitations especially in mouse strains that lack well-defined surface markers to separate donor cells from congenic recipient cells. Here, we reported a PCR-based technique to determine donor cell engraftment/contribution in transplant recipient mice. We transplanted male donor bone marrow HSCs to lethally irradiated congenic female mice. Peripheral blood samples were collected at different time points post transplantation. Bone marrow samples were obtained at the end of the experiments. Genomic DNA was isolated and the Y chromosome specific gene, Zfy1, was amplified using quantitative Real time PCR. The engraftment of male donor-derived cells in the female recipient mice was calculated against standard curve with known percentage of male vs. female DNAs. Bcl2 was used as a reference gene to normalize the total DNA amount. Our data suggested that this approach reliably determines donor cell engraftment and provides a useful, yet simple method in measuring hematopoietic cell reconstitution in murine bone marrow transplantation models. Our method can be routinely performed in most laboratories because no costly equipment such as flow cytometry is required.
Keywords: Medicine, Issue 73, Biomedical Engineering, Stem Cell Biology, Genetics, Immunology, Anatomy, Physiology, Cellular Biology, Surgery, Y Chromosome, Hematopoietic Stem Cells, HSC, stem cells, Bone Marrow Transplantation, Real-Time Polymerase Chain Reaction, rtPCR, PCR, Chimerism, Y chromosome specific gene, graft, engraftment, isolation, transplantation, cell culture, murine model, animal model
Introduction
Murine bone marrow (BM) transplantation model was first developed in 1960s1. This model has been extensively used for the study of donor hematopoietic stem cell (HSC) biology in a host recipient mouse. Murine bone marrow transplant model has provided us with valuable knowledge on HSC functions and their regulation and is indispensable in HSC research. Allogeneic bone marrow transplant model like C57Bl/6JH2b-Balb/CH2d or congenic transplant model like C56Bl/6JCD45.2-B6.SJLCD45.1 are used in many laboratories to study the gene function on HSC activity2, effect of drug treatment on HSC function3 or transplantation related diseases such as graft-versus-host disease (GvHD)4.
Cell surface markers such as MHC haplotype or CD45.1 are commonly used for distinguishing donor-derived cells from recipient-originated cells. C57Bl/6JH2b, CD45.2, Balb/CH2d and B6.SJLCD45.1 are the most commonly used mouse strains in bone marrow transplantation because the donor cell contribution can be easily assessed by flow cytometry measuring CD45.1 vs. CD45.2 or H2b vs. H2d. However, many other strains such as FVB/NJ5 and C3H are also often used to generate genetically engineered transgenic or knockout mice. These mice may be backcrossed to an inbred line and maintained in a mixed genetic/MHC background. In these cases, determining donor cell engraftment and HSC function could be difficult as donor specific- cell surface markers may not be available.
Using Y-chromosome-specific DNA probe to detect the donor male cells by southern blot in sex-mismatched bone marrow transplantation was first developed by Dr. Miwa's group 6. Then, a real-time PCR for the sex-determining region Y was found to be an accurate and highly specific method to quantitate male fetal cells in the maternal blood system7. This concept was adapted by Dr. Schwarzenberger's group for the development of a real-time PCR technique in a murine bone marrow transplantation model to determine donor cell engraftment 8. We further modified this method for the measurement of donor cell engraftment in FVB/NJ mouse bone marrow transplant model. This method is currently extensively utilized in our group for studying the role of Pim1 kinase in HSC biology.
Protocol
1. Bone Marrow Cell Isolation
Euthanize male donor FVB/NJ mice and female FVB/NJ mice using CO2 method followed by cervical dislocation. The female FVB/NJ bone marrow cells will be used as competitive cells.
Use small scissors and forceps, dissect out femurs and tibiaes from mice and place them in a 60 mm tissue culture dish containing 6 ml ice-cold RPMI1640 with 5% heat inactivated FBS. Use kimwipe tissue to remove muscle and other tissues. Cut off both ends of each bone shaft in the dish.
Connect the end of the bone with 23G needle on 3 cc syringe, flush out bone marrow with RPMI1640 with 5% heat inactivated FBS into the dish. Disaggregate bone marrow tissues by repeated aspirations using the same needle. Transfer the cell suspension to 15 ml centrifuge tube.
Spin down the cells for 5 min at 400 x g, remove the supernatant, resuspend the cells in 1 ml of room temperature red blood cell lysis buffer (155 mM potassium bicarbonate, 10 mM Ammonium chloride, 0.1 mM of EDTA, PH=7.4) and incubate at room temperature for 5 min then add 5-10 ml of RPMI 1640 with 5% heat inactivated FBS.
Pass the cells through a cell strainer. Collect the flow through to a new tube. Spin down for 5 min at 400 x g. Remove the supernatant; the cell pellet should not contain any red color. The absence of red color indicates a complete removal of red blood cells. Resuspend the cell pellet in 10 ml of RPMI1640 with 5% heat inactivated FBS. Gently vortex to make sure the cell suspension is completely uniform. Take an aliquot and count the cells in a hemacytometer. Calculate how much volume of cells needed for bone marrow transplantation and aliquot enough cells and mix male donor cells with competitor female cells at a ratio of 5:2. Spin down for 5 min at 400 x g, wash with PBS, resuspend in PBS with the final concentration of donor cells at 5×106/ml and competitor cells at 2×106/ml.
2. Competitive Bone Marrow Transplantation
Irradiate female recipient mice (8-12 wks of age) with 137Cs gamma rays radiator at a single dose of 11Gy 4-6 hr before bone marrow transplantation.
Place the irradiated female mice in a mouse restrainer. Inject the mixed donor and competitor cells via tail vein in 0.1 ml of total volume such that each mouse receives 5×105 donor cells and 2×105 competitor bone marrow cells.
3. Sample Collection
Couples weeks after bone marrow transplantation, collect peripheral blood samples (~50 μl) from female recipient mouse by retro-orbital bleeding under anesthesia condition. Samples are collected into EDTA coated tubes. BM samples can also be collected at the end of experiment, usually at least 4 months post transplantation, with same procedures as previous described (Step 1); usually 20-40% of 1 tibia BM cells are enough to make enough amount of DNA. Blood or BM samples from age matched normal female and male mice are also collected to prepare standard curve.
4. Genomic DNA Isolation
Add 4× volume of room temperature RBC lysis buffer (~ 200 μl) to each blood sample. Mix well and incubate at room temperature for 5 min. Add 1 ml of PBS, then spin down to remove most of the lysed RBC.
Isolate DNA using a blood DNA extraction kit (QIAmp DNA Blood extraction kit). Pre-warm the elution buffer (AE) at 37 °C to enhance the yield of eluted DNA. Male and female DNAs for standard curve are isolated similarly.
(Optional) Genomic DNA can be further purified or concentrated using EtOH precipitation method in the presence of 3 M Sodium Acetate (pH=5.5). The DNA pellets are then resuspended in 40-50 μl distilled water for analysis immediately or stored at -20 °C for future use.
Measure the DNA concentration with Nanodrop ND-1000 spectra photometer. Samples with OD 260/280 between 1.8-2.0 are used for further analysis.
Dilute DNA with DNAase free H2O to 4ng/μl in a total volume of 50 μl.
5. Standard Curve Preparation
Dilute male DNA and female DNA to a concentration of 4 ng/μl, and make the DNA mixture according the Table1
| % of male DNA in Female Background | Volume (μl) of 4ng/μl male DNA | Volume (μl) of 4ng/μl female DNA | Total volume(μl) |
| 0.2 | 1 | 499 | 500 |
| 0.5 | 1 | 199 | 200 |
| 2.5 | 5 | 195 | 200 |
| 12.5 | 25 | 175 | 200 |
| 50 | 100 | 100 | 200 |
| 87.5 | 175 | 25 | 200 |
| 100 | 200 | 0 | 200 |
Table 1. Sample preparation for standard curve. Normal male and female FVB/NJ mice at the age of 8-12wks were sacrificed. Blood and BM cells were collected. DNAs from male cells and female cells were isolated and re-suspended at a concentration of 4ng/μl. The male and female DNAs were mixed at various ratios to generate the DNA standard sample mixtures.
6. Real-time PCR
Set up the PCR reaction plate by mixing SYBR Green supermix reagent with primers and genomic DNA. The reaction volume (20 μl) contains 400 nM of each primer and 5 μl of blood cell genomic DNA (4ng/μl x5 μl=20 ng) in each reaction. DNA samples and standards were set up in triplicate. The sequences of primers are shown in Table 2.
| Gene name | Forward | Reverse |
| Bcl2 | 5′-AAGCTGTCACAGAGGGGCTA | 5′- CAGGCTGGAAGGAGAAGATG |
| Zfy1 | 5-TGGAGAGCCACAAGCTAACCA | 5'- CCCAGCATGAGAAAGATTCTTC |
Table 2. RT-PCR primer sequence for murine Bcl2 and Zfy1.
Perform PCR reaction using Biorad iQ5 PCR machine with the following conditions: 95 °C 3 min, 42 amplification cycles of 95 °C for 10 sec, 58 °C for 25 sec and 72 °C for 15 sec, followed by a melting-curve step.
Obtain Cycle threshold (Ct) values by Bio-Rad iQ5 2.1 Standard Edition Optical System. Calculate δCt (CtZfy1-CtBcl2) value, and Zfy1 expression level is calculated as the value of 2-δ Ct.
Establish standard curves for each reaction series using 2-δCt reading from known male/female standard mixtures. The standard curves are generated by plotting the mean of 2-δCt value of triplicates to the known % male DNA in the mixture with linear regression fitting.
Representative Results
Figure 1 and 2 showed examples of standard curves plotted with mean values of 2-δCt against percentages of male DNA. Figure 1A showed specific melting temperature for Bcl2 and Zfy1 amplicons localized at 78.5 °C and 88.5 °C, respectively. Bcl2 is used as a reference gene to normalize the total amount of loaded DNA in each PCR reaction. Bcl2 amplification curves (Log view) for each standard sample merge with each other independent of male DNA concentration, indicating equal amount of loaded DNA (Figure 1B). However, Zfy1 amplification curve migrated to the left with increasing amount of male DNA in the sample (Figure 1C). To validate our method, we tested male donor cell engraftment in transplanted mice receiving BM cells from WT vs. Pim triple knockout (TKO) mice at different time points post transplantation. Both peripheral blood (6wks) and BM samples (16wks) were analyzed. HSCs from Pim TKO mice have defects in reconstituting lethally irradiated mice (An, N et al., manuscript under revision). As expected, recipient mice transplanted with Pim TKO cells had a much lower percentage of male cells compared to mice transplanted with WT cells (Figure 3).
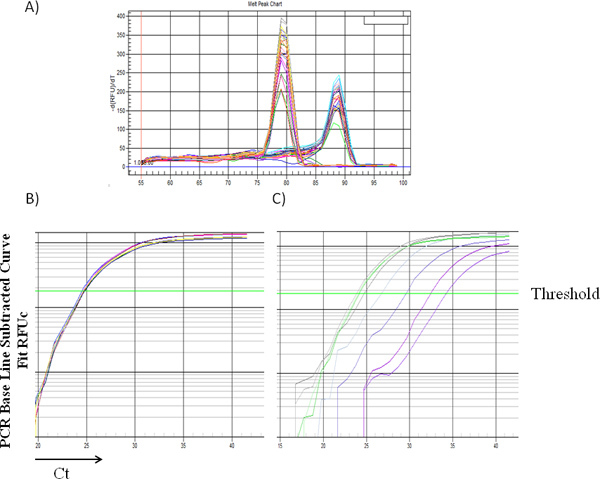 Figure 1.(A) Representative melting curve from multiple samples with different male DNA percentages. The peak at 78.5 indicates amplification of Bcl2, while the peak at 88.5 °C indicates amplification of Zfy-1. Representative amplification curve from multiple samples with different male DNA percentage for Bcl2 (B) and Zfy1 (C), Ct: cycle threshold. Click here to view larger figure.
Figure 1.(A) Representative melting curve from multiple samples with different male DNA percentages. The peak at 78.5 indicates amplification of Bcl2, while the peak at 88.5 °C indicates amplification of Zfy-1. Representative amplification curve from multiple samples with different male DNA percentage for Bcl2 (B) and Zfy1 (C), Ct: cycle threshold. Click here to view larger figure.
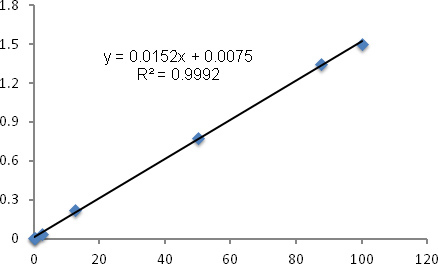 Figure 2. RT-PCR standard curve for male DNA percentage. Samples (20 ng DNA in 20 μl total volume) of male and female DNA mixtures obtained from bone marrow cells were processed for RT-PCR as described in protocol. The mean values of δCt (CtZfy1-CtBcl2) for individual samples were calculated and 2-δ Ct (Y-axis) were plotted against male DNA percentage (X-axis) (range from 0.2% to 100%) with linear regression fitting. R2 represents the coefficient of determination.
Figure 2. RT-PCR standard curve for male DNA percentage. Samples (20 ng DNA in 20 μl total volume) of male and female DNA mixtures obtained from bone marrow cells were processed for RT-PCR as described in protocol. The mean values of δCt (CtZfy1-CtBcl2) for individual samples were calculated and 2-δ Ct (Y-axis) were plotted against male DNA percentage (X-axis) (range from 0.2% to 100%) with linear regression fitting. R2 represents the coefficient of determination.
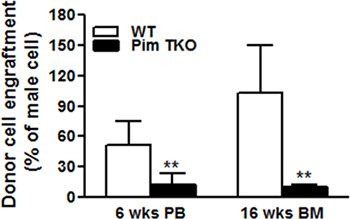 Figure 3. Competitive bone marrow transplant experiments. 5 x 105 total BM cells from male WT or age matched male Pim triple KO (TKO) mice along with 2 x 105 female competitor bone marrow cells were transplanted into irradiated female hosts. Peripheral blood (PB) chimerism at 6 weeks and BM chimerism at 16 wks post transplantation were estimated by percentage of male DNA by RT-PCR as decried in the protocol (*p<0.05, **P<0.01).
Figure 3. Competitive bone marrow transplant experiments. 5 x 105 total BM cells from male WT or age matched male Pim triple KO (TKO) mice along with 2 x 105 female competitor bone marrow cells were transplanted into irradiated female hosts. Peripheral blood (PB) chimerism at 6 weeks and BM chimerism at 16 wks post transplantation were estimated by percentage of male DNA by RT-PCR as decried in the protocol (*p<0.05, **P<0.01).
Discussion
The objective of our current study is to provide the audiences a PCR based technique to quantify donor cell engraftment in a competitive murine bone marrow transplantation model. Several studies have been reported using RT-PCR to detect donor cells in transplantation models 9-10. Dr. Schwarzenberger's group first developed a murine bone marrow transplantation model using real-time PCR to amplify y-chromosome-specific 8. This method was used to study Gli-1 function in HSC activity 11. We then further modified the protocol and provided a detailed, step-by-step experimental protocol in visualized format. The visualized presentation of our protocol allows audiences to follow our protocol easily. We also compared our PCR results using genomic DNA from BM samples vs. DNA from peripheral blood. The donor engraftment data from BM and blood samples are basically consistent, however, usually less variation was found in BM samples. This is likely due to residual hemoglobin after RBC lysis in the peripheral blood that may compromise the purity/quality of genomic DNA in the blood samples.
There are several advantages with our PCR-based technique to quantify donor cell engraftment. First; our technique can be performed in most of laboratories. There is no need for specialized and expensive equipment other than real time-PCR machine. We further modified PCR reaction component and used SYBR Green supermix instead of Taqman. This change further reduces the costs and expenses as we eliminate the fluorescent-labeled probe. Second, our technique is reliable and highly reproducible. We used the Bcl2 reference gene to normalize the total DNA amount as opposed to DNA quantitation kit. Bcl2 gene was used because: 1) this gene is not located on Y-chromosome. Therefore, its expression is not affected by male donor cell engraftment. 2). Bcl2 is not regulated under the current transplantation conditions. As shown in Figure 1, Bcl2 gene produces a reliable and highly consistent control for total DNA loading independent of loaded male DNA amount. Zfy1 is a zinc finger gene within testis determining region 12 and is located on the Y chromosome. It has been reported that the gene expression level of Zfy1 corresponds well to the amount of male cell DNAs in a mouse transplantation model8 . Third, our PCR based technique is highly sensitive, and can detect donor cell engraftment at <1%. Furthermore, samples can be stored for long time for later analysis. In contrast, the conventional flow cytometry-based method requires immediate sample processing. Finally, our technique is mouse strain independent and can be used for genetic background that lacks well-defined surface markers to separate donor cells from recipient cells. This is the most important advantage of this method.
Under our current experimental condition (donor/competitor ratio is 5/2), we observed around 80-90% donor engraftment at 16 weeks using the PCR-based method. Our data are quite comparable with conventional, flow cytometry based- method that showed ~95% donor cell engraftment at 16 weeks post transplantation with donor (CD45.2)/competitor (CD45.1) ratio at 4:12. Compared to the conventional method that uses cell surface marker to detect donor cells in recipient mice, the major disadvantage of our method is the inability to analyze the engraftment in various subpopulations of blood cells such as in B cells, T cells or Granulocytes. However, if needed, it is possible to collect each population of cells using cell sorting technique and then subject the samples for RT-PCR to determine donor cell contribution in each cell population. Since the donor cell engraftment was calculated based on standard curve, the sample readout is totally dependent on the slope and y intercept of standard curve. It is possible that we may get off-range readout (i.e. over 100% of engraftment). However, these "false positive or false negative" results can be avoided if: 1) the DNA isolation procedures for tested samples and standard curve samples are identical, and 2) the chosen range of standard curve is close to the expected sample readout. In addition, since there may be differences in each PCR reaction and in the calibration status of PCR machine, standard curve samples should be included in each PCR plate to avoid variations between each PCR reaction.
Finally, we validated our technique using our Pim triple KO mice. Consistent with previous report 13, we found that donor HSCs from Pim triple KO mice are defective in reconstituting lethally irradiated recipient mice.
Disclosures
We have no competing financial interests to disclose.
Acknowledgments
We thank Dr. Charles Greenberg for the use of his Real time PCR machine. This work is supported by MUSC Hollings Cancer Center Startup Fund, Hollings Cancer Center ACS IRG, ASCO Conquer Cancer Foundation Career Development Award, NIH 1K08HL 103780-01A1, and NIH 3P30CA138313-01S3. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding agents.
References
- McCulloch EA, Till JE. The radiation sensitivity of normal mouse bone marrow cells, determined by quantitative marrow transplantation into irradiated mice. Radiat. Res. 1960;13:115–125. [PubMed] [Google Scholar]
- Xiao N, et al. Hematopoietic stem cells lacking Ott1 display aspects associated with aging and are unable to maintain quiescence during proliferative stress. Blood. 2012;119:4898–4907. doi: 10.1182/blood-2012-01-403089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Chen BJ, Deoliveira D, Mito J, Chao NJ. Selective enhancement of donor hematopoietic cell engraftment by the CXCR4 antagonist AMD3100 in a mouse transplantation model. PLoS One. 2010;5:e11316. doi: 10.1371/journal.pone.0011316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi B, et al. GVHD after chemotherapy conditioning in allogeneic transplanted mice. Bone Marrow Transplant. 2008;42:807–818. doi: 10.1038/bmt.2008.261. [DOI] [PubMed] [Google Scholar]
- Taketo M, et al. FVB/N: an inbred mouse strain preferable for transgenic analyses. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2065–2069. doi: 10.1073/pnas.88.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki H, et al. Genotypic analysis using a Y-chromosome-specific probe following bone marrow transplantation. Am. J. Hematol. 1988;27:30–33. doi: 10.1002/ajh.2830270108. [DOI] [PubMed] [Google Scholar]
- Lo YM, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am. J. Hum. Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne P, et al. Chimerism analysis in sex-mismatched murine transplantation using quantitative real-time PCR. Biotechniques. 2002;32:279–280. [PubMed] [Google Scholar]
- Ma X, et al. Contribution of recipient-derived cells in allograft neointima formation and the response to stent implantation. PLoS One. 2008;3:e1894. doi: 10.1371/journal.pone.0001894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio E, et al. A comparison between real-time quantitative PCR and DNA hybridization for quantitation of male DNA following myoblast transplantation. Cell Transplant. 2004;13:817–821. doi: 10.3727/000000004783983369. [DOI] [PubMed] [Google Scholar]
- Merchant A, Joseph G, Wang Q, Brennan S, Matsui W. Gli1 regulates the proliferation and differentiation of HSCs and myeloid progenitors. Blood. 2010;115:2391–2396. doi: 10.1182/blood-2009-09-241703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine CM, Chan K, Hake LE, Lau YF. The two candidate testis-determining Y genes (Zfy-1 and Zfy-2) are differentially expressed in fetal and adult mouse tissues. Genes & development. 1990;4:63–74. doi: 10.1101/gad.4.1.63. [DOI] [PubMed] [Google Scholar]
- Grundler R, et al. Dissection of PIM serine/threonine kinases in FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4-mediated homing and migration. J. Exp. Med. 2009;206:1957–1970. doi: 10.1084/jem.20082074. [DOI] [PMC free article] [PubMed] [Google Scholar]


