Abstract
2D image space methods are processing methods applied after the volumetric data are projected and rendered into the 2D image space, such as 2D filtering, tone mapping and compositing. In the application domain of volume visualization, most 2D image space methods can be carried out more efficiently than their 3D counterparts. Most importantly, 2D image space methods can be used to enhance volume visualization quality when applied together with volume rendering methods. In this paper, we present and discuss the applications of a series of 2D image space methods as enhancements to confocal microscopy visualizations, including 2D tone mapping, 2D compositing, and 2D color mapping. These methods are easily integrated with our existing confocal visualization tool, FluoRender, and the outcome is a full-featured visualization system that meets neurobiologists’ demands for qualitative analysis of confocal microscopy data.
Index Terms: J.3 [Life and Medical Sciences], Biology and genetics—; I.3.8 [Computer Graphics], Application—
1 Introduction
In neurobiology research, laser scanning confocal microscopy, which is capable of capturing 3D volumes and 4D time sequences of biological samples, is an essential tool for neurobiologists to study the structures of and structural differences between samples. The data acquired from confocal microscopy are abundant with finely detailed biological structures resulting fromfluorescent staining. In order to faithfully reconstruct the 3D structural relationships and enhance the fine details from confocal volumes, specialized visualization tools are always demanded by neurobiologists, and biologists in general. FluoRender is such an interactive visualization tool that we developed along with our neurobiologist collaborators. It has a specially designed volume transfer function with suitable parameters for adjusting and fine-tuning visualization results of confocal volumes; it incorporates three render modes for combining and mixing of multiple confocal channels; and it supports rendering semi-transparent polygonal mesh together with volume data, for better definition of important biological boundaries. A preliminary comparison [23] of an early version of FluoRender with other visualization tools commonly used by neurobiologists has shown its advantage for visualizing finely detailed biological data.
Since its initial release, we continued the development of FluoRender with an emphasis on detail enhancement. The user group of FluoRender has expanded beyond our collaborating neurobiologists, and brought new challenges and problems that we have overlooked in our initial work. One problem that we started looking at were the features presented in 2D image processing packages, however commonly missing from volumetric visualization tools. We noticed that most biologists working with microscopy data are actually experts on image processing packages such as Photoshop, which are used for a variety of tasks including combining images, adjusting brightness and contrast, adding annotations, etc. They also have been using tools such as Photoshop with volumetric data visualization results including those from Maximum Intensity Projection (MIP) and Direct Volume Rendering (DVR). The familiarity with results from MIP rather than DVR usually makes biologists regard MIP advantageous at rendering sharp details, and this is more common with neurobiologists working with confocal microscopy data, which have an abundance of detail. We have convinced many that DVR can bring out details even better with properly adjusted volume transfer function settings, and will also correctly render the spatial relationship of confocal data. However, users of FluoRender still relied on image processing packages and attempted to enhance details from their retouching work. The retouching work with tools such as Photoshop is usually fraught with frustrations, because the commonly used image file formats for data exchange between the visualization tools and image processing packages lack the precision needed for further adjustment, and these packages are designed for photography rather than confocal data visualization.
The contributions of this paper are methods and techniques that can easily be used for detail enhancement and integrated into FluoRender. For easier brightness/contrast adjustments and detail enhancement, we use 2D tone-mapping operators, including gamma, luminance and scale-space equalization. We improve 2D composting for multiple channels by the introduction of groups. To enhance surface details and depth perception, we use 2D compositing to combine a shading and/or a shadow layer with MIP rendering. The rest of this paper is organized as follows: Section 2 discusses related work; Section 3 looks at the volume renderer within FluoRender and discusses how we ensure rendering precision; Section 4 presents the 2D image space methods that we choose to integrate into FluoRender for confocal data visualization enhancement; Section 5 discusses certain implementation details of the system; Section 6 demonstrates the improvements by case studies; we then conclude in Section 7.
2 Related Work
As stated in the introduction, the work presented in this paper is a continuation of our previous development of FluoRender. In [23], we focused on customizing a volume renderer for confocal microscopy data visualization in neurobiology research. Most of the techniques presented in [23] are applied in the 3D object space, such as transfer function design, rendering volumetric data combined with semi-transparent polygonal meshes with depth peeling, and mesh-based volume editing. However, two of the three render modes proposed (layered mode and composite mode) are 2D compositing methods applied after volumetric data are rendered, while only depth mode is the rendering method for multi-channel data commonly found in similar visualization tools. In this paper, we shift our focus to the applications of 2D image space methods, such as 2D compositing, tone mapping, and filtering.
In addition to volume transfer function design, many methods have been proposed to enhance features of volume data. Ebert and Rheingans [5] presented an object space volume illustration approach, which uses nonphotorealistic rendering to enhance important features. Kuhn et al. [13] designed an image-recoloring technique and applied it to volume rendering for highlighting important visual details. To improve visualization experiences for individuals with color vision deficiency, Machado et al. [15] proposed a physiologically-based model for re-coloring visualization results, including volume-rendered scientific data. For illustrative visualization, Wang et al. [24] presented a framework to aid users to select colors for volume rendering, in which case color mixing effects usually limit the choice of colors. While there are several commercial and academic visualization packages that neurobiologists have been using for confocal microscopy data, such as Amira [21], Imaris [2], and Volocity [17], 2D image space methods for detail enhancement are generally absent from these tools. In fact, choosing and designing proper 2D image space methods and parameters, integration of 2D and 3D methods, as well as their applications are interesting research topics for volume visualization in general. In [4], Bruckner et al. presented a framework for compositing of 3D renderings, and use the framework for interactively creating illustrative renderings of medical data. Tikhonova et al. [19] [20] proposed visualization by proxy, which is a framework for visualizing volume data that enables interactive exploration using proxy images. For fast prototyping and method/parameter searching, computer scientists often use comprehensive visualization and image processing libraries, such as VTK [11] and ITK [10]. Experimental applications with customized visualization pipelines are generated. However this is often regarded as impractical by neurobiologist users, since they usually demand a reliable tool with seamlessly integrated functions that are only relevant to their specific application scenario. Furthermore, the high throughput confocal microscopy data generated in biological experiments, including multi-channel data and time-sequence data, need customized data I/O and processing in order to be loaded promptly and visualized in real-time on typical personal computers.
3 High Precision Volume Renderer for Confocal Microscopy
Since the 2D image space methods discussed in this paper are applied to the rendering results of the volume renderer in FluoRender, it is worthwhile examining some of its features that ensure faithful rendering. The fine details of confocal microscopy data are prone to quality degeneration if the volume renderer’s precision is not adequate. We ensure the high precision of our volume renderer using three approaches.
Input precision
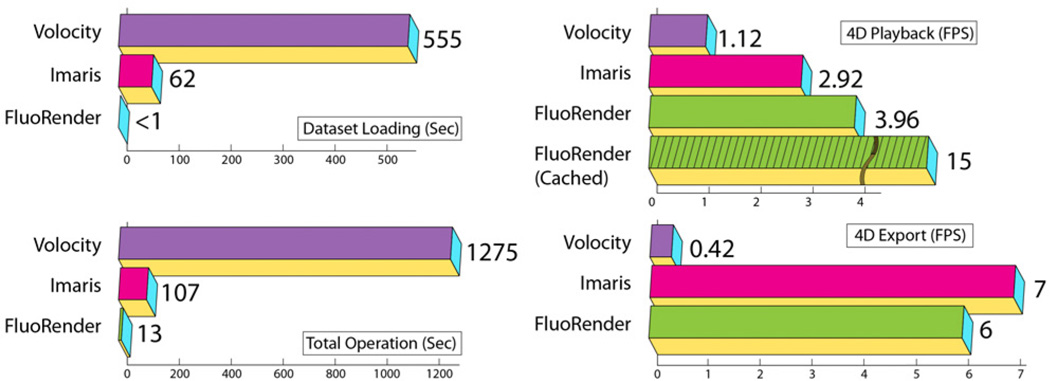
Confocal laser scanning allows simultaneous acquisition of multiple channels of differently stained biological structures. Depending on the model of the photomultiplier used with the microscope, the bit-depth of each channel varies from 8-bit to 16-bit. FluoRender can directly read most raw formats from confocal microscope manufacturers, such as Olympus and Zeiss. 16-bit 3D textures are used to store the confocal volume in graphics memory for rendering when the data file has greater than 8-bit depth. Many confocal visualization tools require data format conversion or perform down sampling and quantization, which not only compromise precision but also cause great latency for data loading. By minimizing data pre-processing and optimizing codes for data reading, FluoRender has negligible latency for common confocal datasets (cf. Figure 13), which is helpful especially for multiple samples and 4D sequences.
Figure 13.
Speed comparison. We test all speeds on the same PC with Intel Core i7 3.2GHz, 12GB memory, single 7200 RPM SATA disk, nVIDIA GTX280 and Microsoft Windows XP 64bit. The dataset is a two channel 4D confocal dataset with 210 frames, which occupies 3.43GB on disk. Volocity is 64bit at version 5.1.0. Imaris is 64bit at version 6.3.0. FluoRender is 64bit at version 2.9.0
Transfer function precision
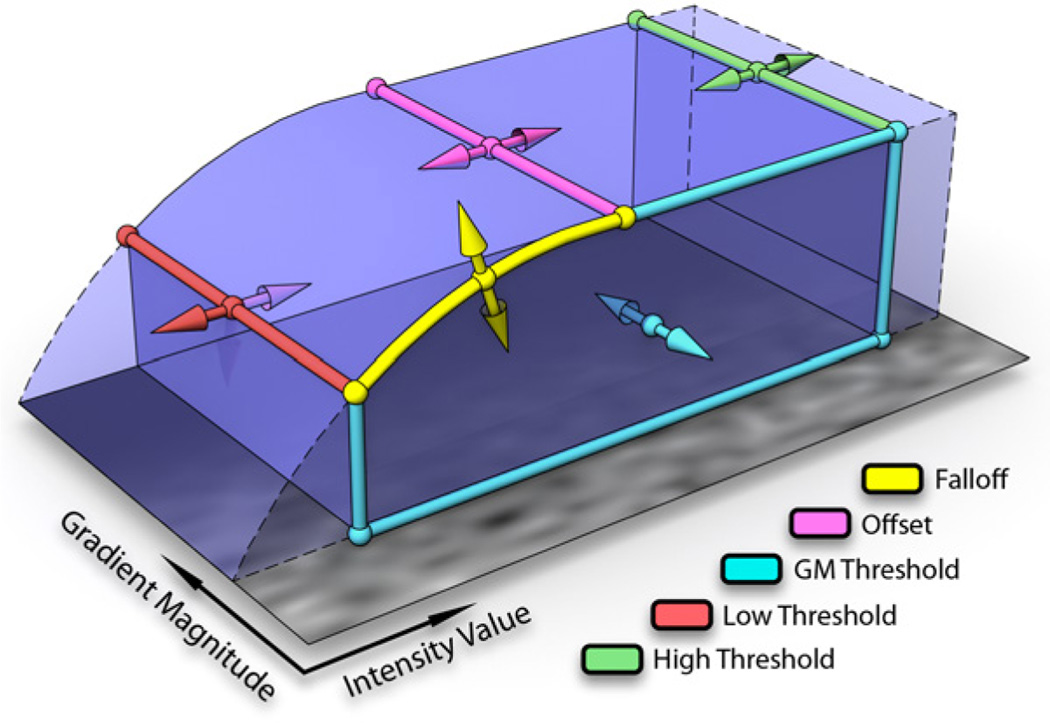
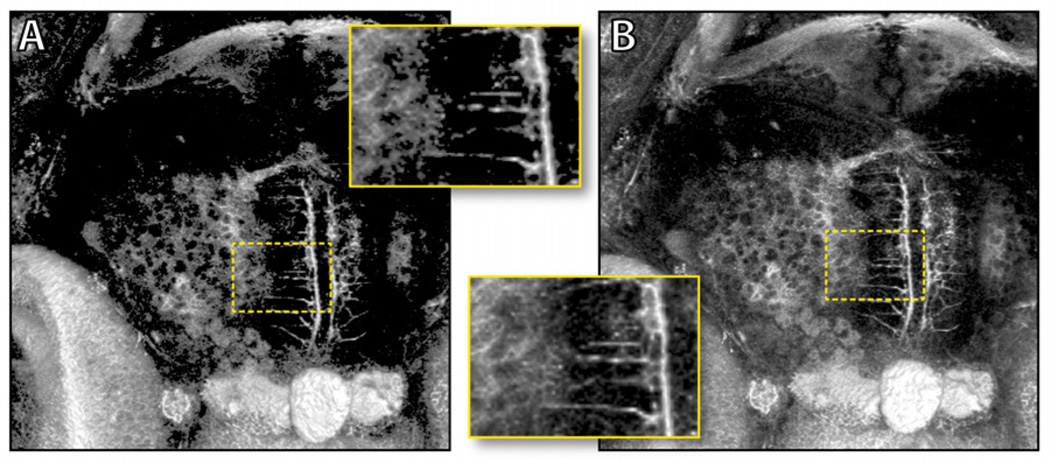
FluoRender uses a two-dimensional transfer function (Kniss et al. [12]) with five parameters to adjust the rendering result for each confocal channel (Figure 1). It is a common practice that the volume transfer function is rasterized as a texture, and updated every time the parameters change. Two problems may occur if the texture transfer function is used. First, there is quantization error, especially when the transfer function is nonlinear, since texture lookup only uses linear interpolation. Second, it is impractical to build a texture transfer function for 16-bit data due to texture size limitations. Since our 2D transfer function has only five customized parameters for confocal visualization, we pass the parameters into the shader computing the volume rendering result, and evaluate the volume transfer function on the fly. The real-time evaluation of the transfer function ensures the rendering quality of the low intensity signals in confocal data. Figure 2 compares the resulting difference between pre-quantized and real-time evaluation of the transfer function. While many other tools either do not have the flexibility of changing volume transfer function, or provide too many parameters and widgets that make evaluation on the fly impossible, FluoRender lets its users quickly adjust the volume transfer function for finely-detailed visualizations.
Figure 1.
An illustation of the 2D volume transfer function in Fluo-Render. The colored arrows indicate the possible adjustments for the parameters, which are: falloff, offset, gradient magnitude threshold, low scalar intensity threshold and high scalar intensity threshold. All parameters are adjusted with sliders as shown in Figure 10.
Figure 2.
Results from pre-quantized (A) and on the fly (B) evaluation of the transfer function. The two results are generated with the same transfer function settings. The pre-quantized transfer function clips many details in the low intensity regions, which are preserved with on the fly evaluation of the transfer function. The dataset shows tectal neurons of a 5-day-post-fertilization (5dpf) zebrafish.
Output precision
The volume rendering results of Fluo-Render always output to 32-bit floating-point framebuffers, and then all the 2D image space methods discussed subsequently are calculated with 32-bit precision. This is essential to our tool for high-precision adjustments, but unnecessary for other tools because of their lack of 2D image space methods.
4 2D Image Space Methods
2D image space methods are processing methods applied after the volumetric data are projected and rendered into the 2D image space, including straightforward techniques such as contrast and gamma corrections, and methods that require rendering and combining different customized layers. In this section, we chronicle the development of our work, and discuss each technique that we found useful and thus customized for confocal data visualization.
4.1 2D Tone Mapping
2D tone-mapping operators can be found in many image processing packages but are absent from confocal visualization tools and most volume visualization tools in general. When only the volume transfer function is adjusted, neurobiologist users sometimes found it difficult to achieve both satisfactory brightness and details for volume rendering outputs. They requested the use of tone mappings for the rendering results. This can be explained with the volume rendering integral. Consider the commonly used emission-absorption model, where the resulting intensity is calculated as [16]:
| (1) |
In Equation 1, q is the emission term, and κ is the absorption term. As shown in the two attached plots of Figure 3A and B, when we apply a monotonic adjustment (for example, gamma correction, which preserves the order of its input intensity values so that they are globally darkened or brightened) f(q,p) with a parameter p on q (or κ), the output intensity (I(D)) is generally not changing monotonically with p, due to the complexity of q (or κ) along the integration path (ray profile). The adjustment is usually embedded within volume transfer functions, and this non-monotonic relationship between parameter and result makes it difficult for users to adjust for desired brightness. Searching for adjustments and parameters that have monotonic influence on output intensity of volume rendering is out of the scope of this paper, and can be found in the work of Wan and Hansen [22]. Here we apply tone mapping with monotonic adjustments on volume rendering outputs, which is intuitive to use for adjusting brightness.
Figure 3.
The volume transfer function falloff and the 2D rendering gamma work well together. The confocal dataset shows three channels of a 5dpf zebrafish embryo: eye muscles (red), neurons (green), and nuclei (blue). A: The initial rendering without any adjustment. B: The result when the falloff is increased. The rendering does not become brighter as many users may expect. The attached plots show the change of one ray profile after the falloff is increased. Though the adjustment is monotonic and increases all values along the ray profile, the blended output becomes darker due to quick accumulation of low intensity values. C: The result when the gamma is increased. The rendering result is brightened, but noise becomes prominent in the regions indicated by yellow arrowheads. D: The satisfactory result achieved by decreasing falloff and increasing gamma. Neuron fibers are visualized clearly with less noise (in the same regions indicated in C).
The general definition of tone mapping is the mapping of one set of colors to another, but the term is mostly used with high dynamic range images (HDRI) (Reinhard et al. [18]), where the meaning narrows to compression of the high dynamic range of light information to a lower dynamic range. Since confocal data are acquired up to 16 bits per channel at present and strictly speaking are not HDR, we use the term tone mapping in this paper in respect of the general definition. However, the objectives of HDR tone mapping still apply to confocal data visualization, i.e. rendering all possible tone ranges at the same time and preserving the details with local contrast in order to obtain a natural look. We implemented the following three tone-mapping operators, and made certain customizations specifically for confocal data.
Gamma correction is the most-used non-linear operator in all image processing work. We follow convention and calculate the output color Cout with Equation 2:
| (2) |
The non-linear adjustment of the low intensity falloff in Figure 1 is essentially the same gamma correction embedded within the transfer function. However its actual influence on brightness is quite different from applying it in 2D: increasing the transfer function falloff enhances details for low intensity voxels (Figure 3B), which usually makes the rendering result less bright, and vice versa. The transfer function falloff is a parameter neurobiologist users frequently use to either enhance or suppress low intensity signals in confocal data. However the brightness of the results cannot be adjusted the same as one would expect from gamma correction (Figure 3C). By adding gamma correction in 2D as an independent parameter of the transfer function falloff, neurobiologists can adjust both details and brightness easily. For example, in Figure 3D, the volume transfer function falloff is decreased to suppress noise signals, and the gamma is increased to reveal the fine details of the neuron fibers.
Luminance is usually called exposure in photo-editing tools. It is a scalar multiplier on the input color, which is used to brighten/darken the overall rendering and expand/compress the contrast linearly. In order for the user to adjust the luminance intuitively, we customize its parameter L by mapping it to the actual factor with a piecewise function f(L).
| (3) |
Figure 4 shows f(L) in both linear and logarithmic scale plots. The function is pieced together from a linear function and a non-linear curve, and is C1 smooth. The user-adjustable parameter L has range [0; 2). It darkens the result by compressing the dynamic range within [0; 1), and brightens the result by expanding the dynamic range within (1; 2). In the logarithmic scale plot, the curve is anti-symmetric at the center point (1; 1), so in addition to a monotonic adjustment, our luminance operator gives an intuitive feel that the output is equally brightened or darkened when L is increase or decreased.
Figure 4.
The mapping of the user-adjustable parameter L and the scaling factor f(L), in linear scale (left) and logarithmic scale (right) plots.
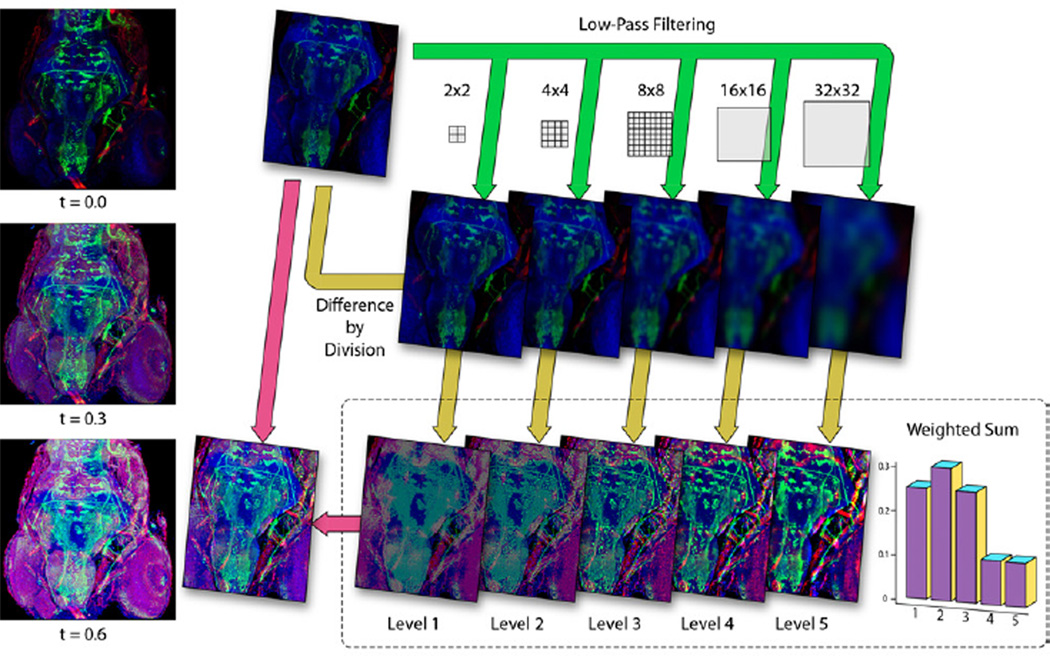
Scale-Space Equalization is a local tone-mapping operator that equalizes the uneven brightness and enhancing the fine details of confocal microscopy data. Using levels-of-detail with the scale space for tone mapping can be found in the work of Jobson et al. [9]. Instead of using logarithmic mappings for dynamic range compression, which is widely used in HDRI processing, we divide the input color (Cin) by the scale space color (), which is an average calculated by low-pass filtering. Thus the input color is equalized at a series of detail levels, hence the name scale-space equalization. The output color of this operator is calculated by a weighted sum of the equalized colors and then blended with the input color, as in Equation 4:
| (4) |
In Equation 4, vi is a set of weighting factors, which are empirically determined by experimenting with typical confocal datasets. A plot of the vi we use in FluoRender is shown in Figure 5. The only parameter exposed to the end-user is the blending factor t, which linearly blends the equalized color with the input color. This linear blending, which is missing even in most HDRI processing software, ensures a monotonic change to brightness as previously. Figure 5 illustrates the equalizing process. It also shows the results when the blending factor changes. The originally dark rendering of the confocal dataset is brightened, yet the fine details are still clearly visualized. For noisy confocal data, increasing the blending factor also enhances high frequency noise. Thus noise removal through pre- or post-processing is usually desired.
Figure 5.
The scale-space equalization process. The example dataset has three channels of stained muscles, neurons and nuclei of the zebrafish head.
The importance of scale-space equalization for confocal visualization is normalizing brightness – along the Z-axis for 3D channels and through time for 4D sequences, which are detailed in the case studies of Section 6.
4.2 2D Compositing with Groups
When different fluorescent dyes and proteins are used in biological samples for confocal laser scanning, the resulting datasets have multiple channels. In most confocal visualization tools having multi-channel support, the channels are combined in 3D object space, and this is usually done by considering inputs as one RGB volume. Though it maintains correct spatial relationship between the channels, neurobiologist users still seldom use it, because it becomes difficult to emphasize certain features from just one or two channels, which usually have more importance than others. In our previous work [23], we proposed two 2D compositing schemes, i.e. layered and composite render modes, in addition to 3D compositing, which is called depth mode in [23]. The application of 2D compositing is surprisingly successful and appreciated by neurobiologist users, since working with 2D compositing by regarding the confocal channels as 2D image layers is intuitive and channels of greater importance are easily emphasized.
However, compositing all channels with a single render mode may not be sufficient, especially when there are many confocal channels, including derived data from segmentation and analysis. Users want to group certain channels and combine specific channel groups differently with 2D or 3D compositing. Our collaborating neurobiologists resorted to 2D image processing packages for group compositing, since the early versions of FluoRender were not capable of rendering groups with different render modes. Figure 6 shows a simple example of three confocal channels, which are sufficient to demonstrate the problem. Figure 6A, B, and C are the three render modes in [23]. The dilemma for the user is which to choose in order to visualize the correct spatial relationship between the neuron and muscle channels, but leave the nuclei channel as the context. We extend the render modes by simply organizing data channels into groups. A group contains an arbitrary number of channels and has an independent render mode for combining its channels. Different groups are again combined with a render mode. Figure 6D shows the result that neurobiologists were pleased with: both the muscle and neuron channels are visualized, with the correct spatial relationship, and they have a clear context.
Figure 6.
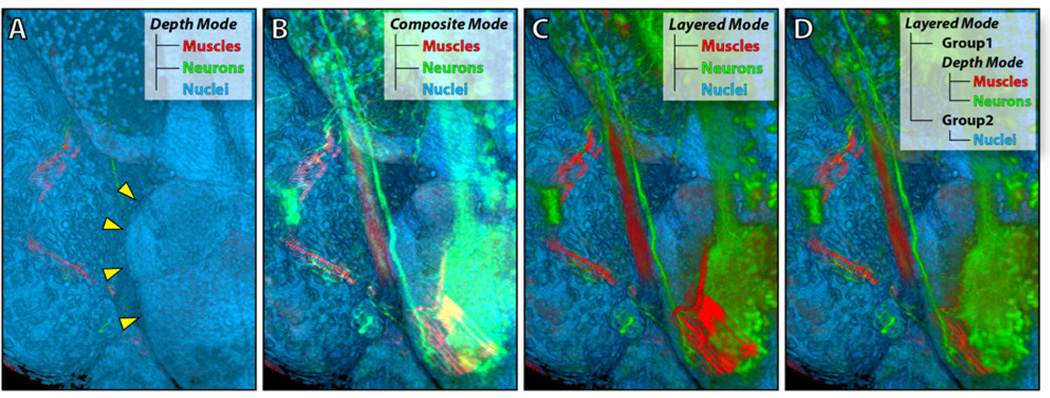
A confocal dataset of a 5dpf zebrafish embryo has three channels: eye muscles (red), neurons (green), and nuclei (blue). A: The channels are combined with 3D compositing. The muscle and neuron channels are barely seen. Yellow arrowheads indicate the boundary of the brain, which is on the right side of the eye when visualized as in the figure. B: The channels are composited with 2D addition. Highlight details are over-saturated, due to the additive compositing. C: The channels are composited with 2D layering. Details of the muscle and neuron channels are visualized, but the spatial order of the two is incorrect. D: The muscle channel and the neuron channel are grouped and combined with 3D compositing, which renders their spatial relationship correctly; the nuclei channel is in a separate group. The two groups are composited with 2D layering. The nuclei channel is a context layer, showing the boundary between the brain and the eye.
Groups also facilitate the parameter adjustment of the confocal channels. The user can group certain confocal channels, and set the parameters of the channels within the group to synchronize. Changes to the parameters of one channel are then automatically propagated to other channels of the group. Section 5 further discusses details on parameter linking and synchronizing.
4.3 Color Mapping and MIP
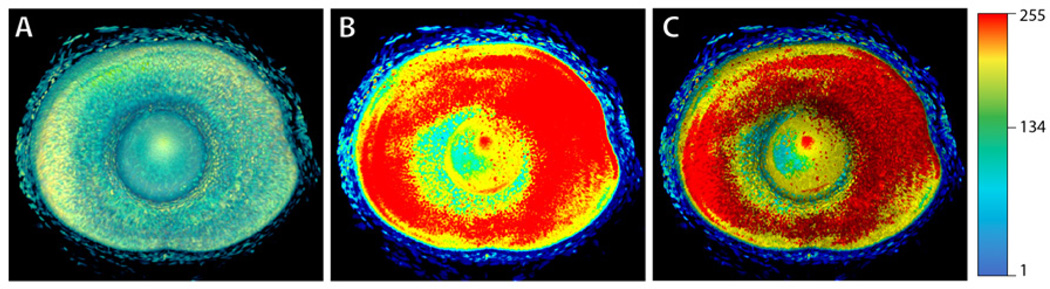
The confocal channels are scalar volumetric data, whose values represent the fluorescent intensities, which in turn measure amounts of biological expression. Biologists often want to assess the amount of gene/protein expression with better quantification than just rendering intensities. Furthermore, since high intensity values represent strong biological expression, it is important to visualize them over low intensity signals. Color mapping is an effective and intuitive method, but not without problems for normal volume renderings. Figure 7A shows a confocal channel rendered with a rainbow colormap as the transfer function. Neurobiologist users often feel that it does not fit into their research purposes well, because the colors in the result do not clearly correspond to those in the colormap due to compositing and voxels with high scalar intensities, which represent strong biological expression and are important to the research, are mostly occluded.
Figure 7.
Using a colormap as the volume transfer function and 2D color mapping of the MIP. All results have the same colormap, as shown on the right. A: The colormap is used as the volume transfer function. B: The colormap is applied to the MIP rendering output. C: The MIP rendering is overlaid with a shading layer (shading overlay is discussed in Section 4.4). The dataset shows a 5 dpf zebrafish eye.
The 2D color mapped maximum intensity projection solves both problems stated above. Figure 7B shows the result of a 2D color mapped MIP with the same colormap as in Figure 7A. Since MIP does not use normal volume compositing, the colors of its result represent the exact intensity values of the voxels, and high intensity voxels are always visualized. In fact, MIP is the only method recognized by neurobiologists for inspecting fluorescent staining intensities. However, there are also drawbacks to MIP, i.e. spatial relationships are unclear, and MIP renderings usually contain more high frequency noise than direct volume renderings. Noise signals can be reduced with common image processing methods, either on 3D volumes (pre-processing) or on 2D image space (post-processing). In the next sub-section, we focus on our method of enhancing spatial relationship for MIP renderings.
4.4 Shading and Shadow Overlays
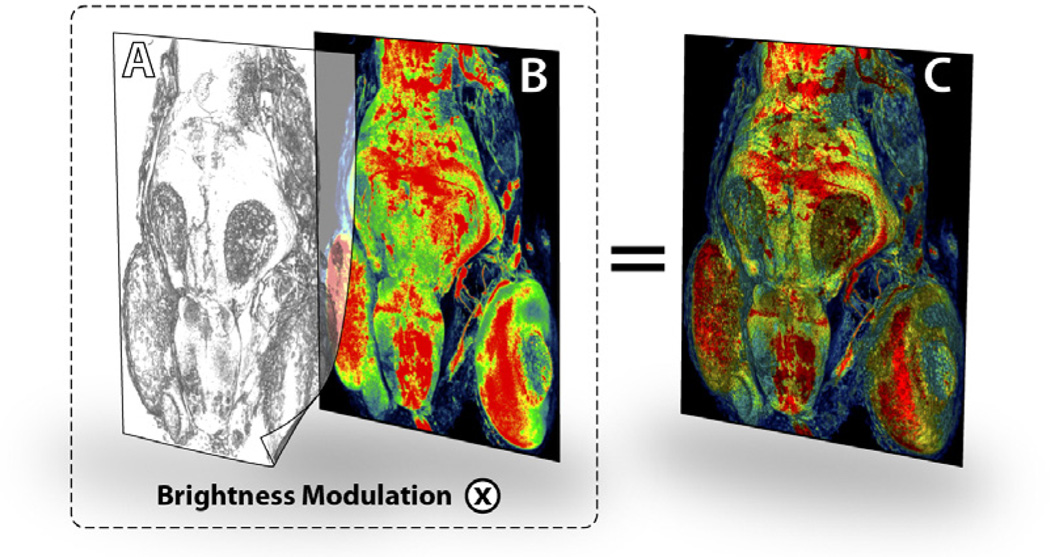
The way that MIP renders volume data can cause two problems for users. First, the orientation of the volume dataset under examination becomes obscure, which may confuse users especially when they rotate the data. Biologists usually prefer orthographic over perspective projection in order to better compare structure sizes. However it worsens the problem of orientation perception when MIP is used. Second, details of surface structures are lost, because unlike most other volumetric data, the structural details of confocal data are always comprised of less intensive signals surrounding high intensity ones, since the signals are generated by fluorescence emission. Adding global lighting effects, such as shadows, can help orient viewers to the renderings of volume data, thus solving the first problem. The second problem can be solved by incorporating local lighting effects, such as Phong shading. There are effective methods such as two-level volume rendering [7] and MIDA [3] that combine the advantages of MIP and shading effects from direct volume rendering. However, the results of above techniques are both somewhere between MIP and DVR, where one of the features of MIP that biologists appreciate, especially when a colormap is used, cannot be ensured, i.e. the colors of the result represent the exact intensity values of the voxels (Section 4.3). Furthermore, how global lighting effects, such as shadows, can be applied with above techniques is not clear. Fortunately, as mentioned above, one structure in confocal data is always comprised of low intensity details surrounding high intensity cores. This simplification of structures enables us to render MIP and lighting effects separately, and then combine them with 2D compositing. The MIP pass is color-mapped for examining the biological expression amount of structure cores; the shading and shadow passes render surface details and enhance orientation perception. The 2D compositing is completed by modulating the color brightness of the MIP rendering with the brightness of the effect passes.
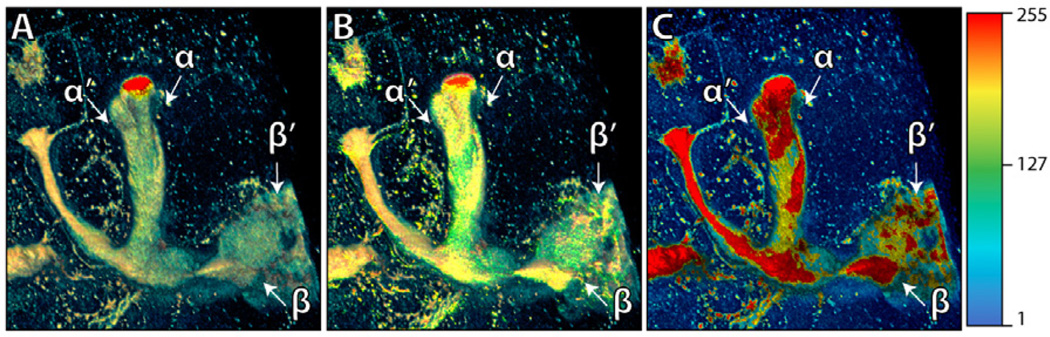
Figure 8 illustrates the 2D compositing and its result of a confocal dataset rendered with a shading layer. A shadow layer can be composited similarly. Neurobiologists can use the 2D color-mapped MIP with overlays for inspecting the amount of biological expression, because the result has a correct color correspondence with the colormap used (Figure 7C). The renderings of shading and shadow passes are grayscale images, and only the color brightness of the 2D color-mapped MIP is modulated, therefore the color hue stays the same, which is the actual variable used in the colormap. Figure 9 compares 2D compositing with DVR and MIDA [3], which uses a modified volume compositing scheme. Since other methods use compositions in 3D, the voxel colors are blended and cannot match the colors used in the colormap. However, for complex structures such as a network of blood vessels, this method has its limitation: shading/shadow and MIP cannot always be rendered consistently, since users have to adjust the volume transfer function for shading and shadow layers. In practice, this mode is used when important features are best represented by MIP and neurobiologist users want to add enhancements for surface details and orientation perception.
Figure 8.
A shading pass (A) is composited with the result of a 2D color-mapped MIP pass (B). The result (C) has the advantages of both MIP and DVR. The dataset has three confocal channels, including stained muscles, neurons, and nuclei.
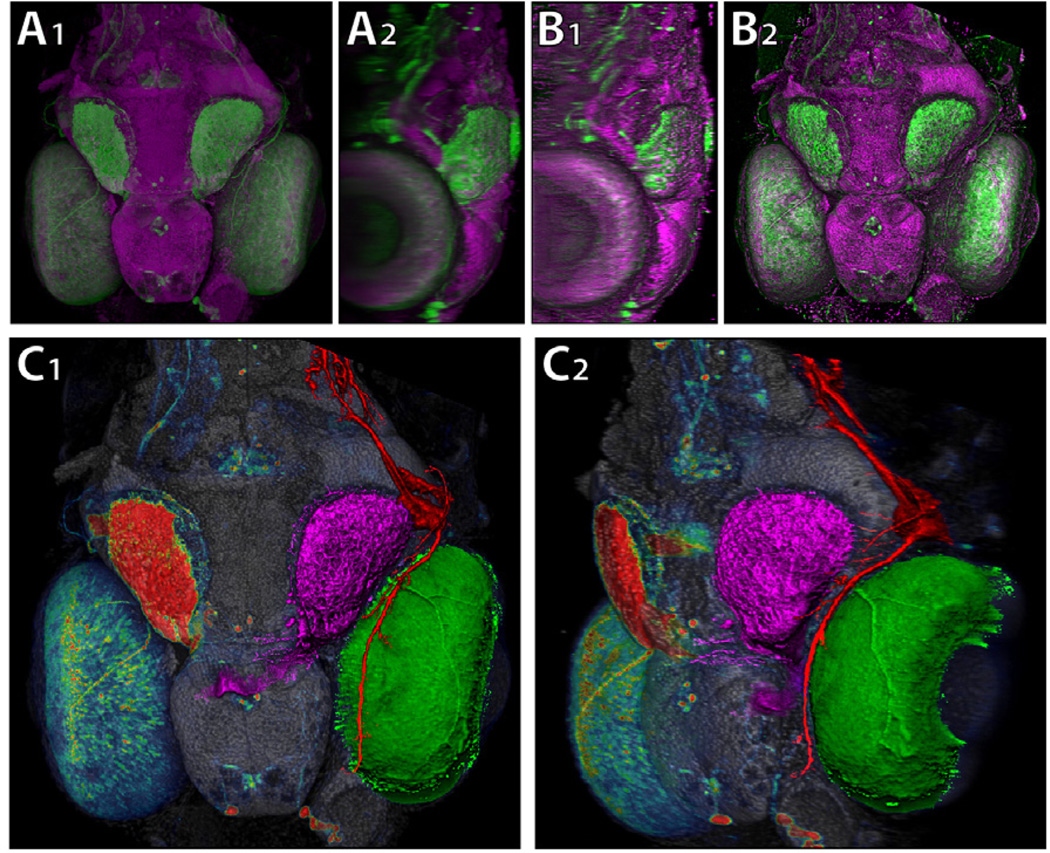
Figure 9.
A comparison of DVR (A), MIDA (B), and shading overlayon MIP (C). They all use the same colormap shown on the right. The dataset is the mushroom body (MB) of an adult Drosophila, stained with nsyb::GFP. This fluorescent protein specifically binds to presynaptic regions of neurons. Thus higher signal intensity indicates higher density of synapses of the mushroom body. By using MIP with 2D overlays, we can clearly see the head of α=α′ lobe has higher presynaptic density than its neck, which can be similarly observed for β=β′ lobe.
5 Implementation
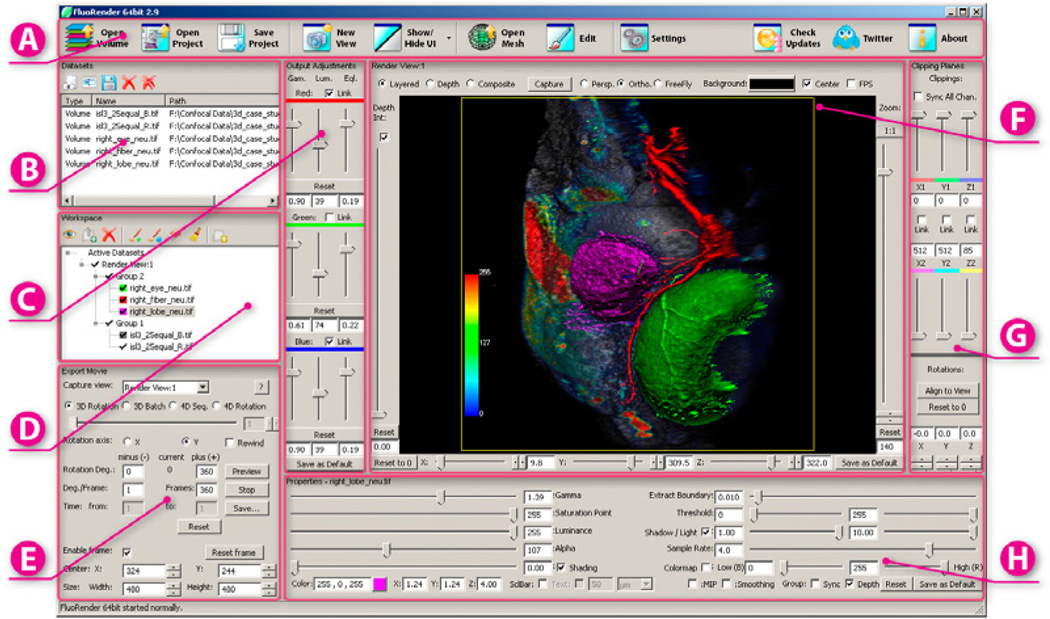
The work presented in this paper is an extension to FluoRender, an interactive visualization tool for confocal microscopy data. We use OpenGL and GLSL for the implementations of the techniques discussed in this paper, including on the fly evaluation of the volume transfer function, tone-mapping operator evaluations, shading and shadow calculations, compositing, and color mapping. While most of the implementations should be straightforward, there are some details worth mentioning. The three tone-mapping operators can concatenate and be evaluated at once – we first generate the scale space, apply gamma and luminance adjustment to all the levels, and then calculate equalization. For fast processing speed, we use the built-in mipmap generating function of OpenGL to approximate the scale space. For shadow overlay calculation, we use a 2D image space method similar to that of depth buffer unsharp masking [14]. Unlike other confocal visualization tools, such as Imaris and Volocity, which use ray tracing to pre-calculate shadows and are not real-time, we use 2D filtering on depth buffer. The rendering speed is real-time, which helps when multi-channel and time-sequence datasets are visualized. The 2D image space methods discussed in this paper are easily modularized, and each module can work independently of another. However, building an integrated visualization system that neurobiologists can easily use, especially when the amount of datasets visualized is large, still requires meticulous design of its user interactions. We develop the user interactions through close cooperation with frequent FluoRender users and experts in confocal microscopy. Figure 10 shows a screen capture of FluoRender’s main user interface. We feel the following design choices on user interactions make FluoRender capable of managing a large amount of datasets and adjustments with intuitiveness.
Figure 10.
FluoRender user interface. A: Toolbar; B: List of loaded datasets; C: Tone-mapping adjustment; D: Tree layout of current active datasets; E: Movie export settings; F: Render viewport; G: Clipping plane controls; H: Volume data property settings.
UI element layout. The most common operations, such as loading datasets, opening and saving projects, and UI management, are placed in the top toolbar as buttons with clear-meaning icons and text. Even first-time users can easily locate them and get started quickly. Sets of adjustments and settings are grouped, according to their category, into panels, which are usually distinguished by their layout. Settings and operations are placed where they can be easily accessed from pertinent selections.
Parameter linking and synchronizing simplifies adjustments when multi-channel datasets are presented. First, for data channels within a group, their properties can be linked, and then the settings of current selected channel are propagated to other channels of the same group. Later changes to any one channel of the group are also applied to the others. Second, parameters of the base and effect layers for shading and shadow are automatically linked. Third, for RGB color channels of the output from a group, the tone-mapping parameters are linked according to the color settings of the volume channels within the group. The tone-mapping parameter linking is decided automatically. The automatic linking simplifies user interactions yet users can still adjust per color channel, when primary colors are used for volume color settings.
Default settings coming with FluoRender installation packages are determined by our collaborating neurobiologists through their general workflows. All the volume properties and modes, 2D tone-mapping adjustments, and render view settings have default values, and they are automatically applied to loaded data.
6 Case Studies
FluoRender has been a free confocal visualization tool available for public download, and has several stable user groups of neurobiologists working with confocal microscopy data from different labs across the world. Its use in neurobiology research can be found in several publications in biology [1], [6], [8]. We present two case studies specifically on how neurobiologists use the newly added 2D image space methods for 3D and 4D confocal data visualizations.
Figure 11 shows the result of our first case study on using 2D image space methods for a two-channel confocal dataset. Neurobiologist users want to study the shape and spatial relationship of the neurons in the region between zebrafish eye and brain. The dataset is the nuclei (magenta channel in Figure 11A1) and neurons (green channel in Figure 11A1) of a 5dpf zebrafish head. Figure 11A1 (dorsal view) and A2 (lateral view) show the volume rendering result with no 2D image space methods applied and volume transfer function set to a linear ramp. They also represent the results from most other confocal visualization tools when the dataset is loaded. The results were still generated with FluoRender. We disabled default settings and set all parameters to neutral. Though the general shapes of its major structures can be visualized, such as the eyes, the brain and the tectum, many details, especially those in the neuron channel, are either occluded or not clearly seen. The lateral view of Figure 11A2 shows a common problem for confocal data: the brightness is decreasing along the Z-axis (from dorsal to ventral for this dataset). This is the direction in which the laser beam travels; due to scattering and tissue occlusion, signals become weaker as the scanning goes deeper along this direction. Since the brightness decrease is sample dependent, a simple calibration of the microscope cannot correct it. Figure 11B1 and B2 show the results from the same view directions, however rendered with FluoRender’s default transfer function settings and the enhancements discussed in this paper applied. With Fluo-Render’s default settings, this is also what a neurobiologist user gets from the start. Shading and shadow add depth and details to many surface structures; tone-mapping operators, especially scale-space equalization, brighten the signals deeper along the Z-axis. In Figure 11B1, the brightness is even, yet the details of the neural structures are enhanced as well. To emphasize and visualize important structures, the user has generated a derived volume which segments the tectum (magenta channel in Figure 11C1), the eye motor neuron (red channel in Figure 11C1), and the eye (green channel in Figure 11C1). The segmented data are in three separate channels, which are then grouped and set to depth mode [23]. With shading and shadow effects on, both spatial relationship and surface details are clearly visualized. The two original channels are grouped in composite mode. The nucleus channel is simply set to a dark gray color and used as background context; the neuron channel is set to MIP with overlaid shading, which shows the signal strength clearly. The two groups are further combined with layered render mode. Figure 11C2 shows the same rendering as in C1 from a different view. Since the 2D image space methods are calculated and applied after the volume datasets are rendered, real-time interactions are maintained.
Figure 11.
Case study 1. A1 and A2: dorsal and lateral views of a zebrafish head dataset, rendered without any enhancement; B1 and B2: the same dataset rendered from the same view directions, with enhancements applied; C1 and C2: groups and different render modes can create clear visualizations when derived channels are presented.
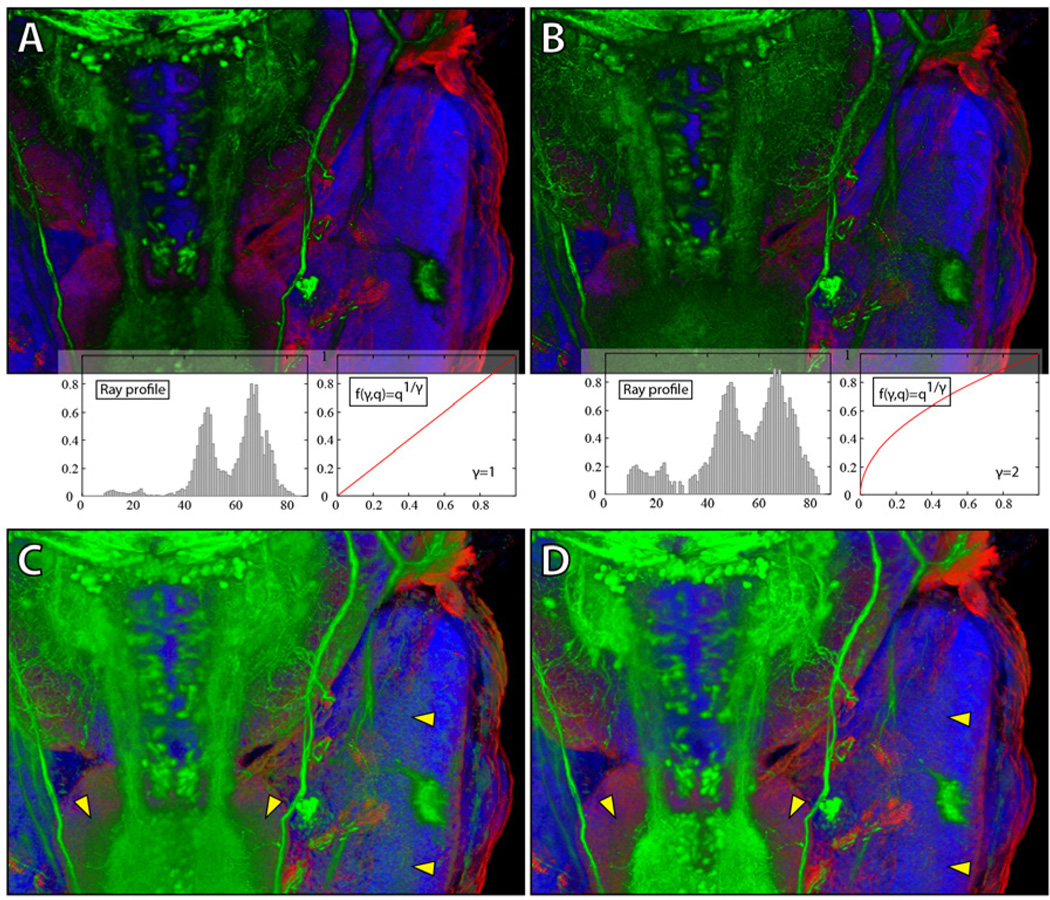
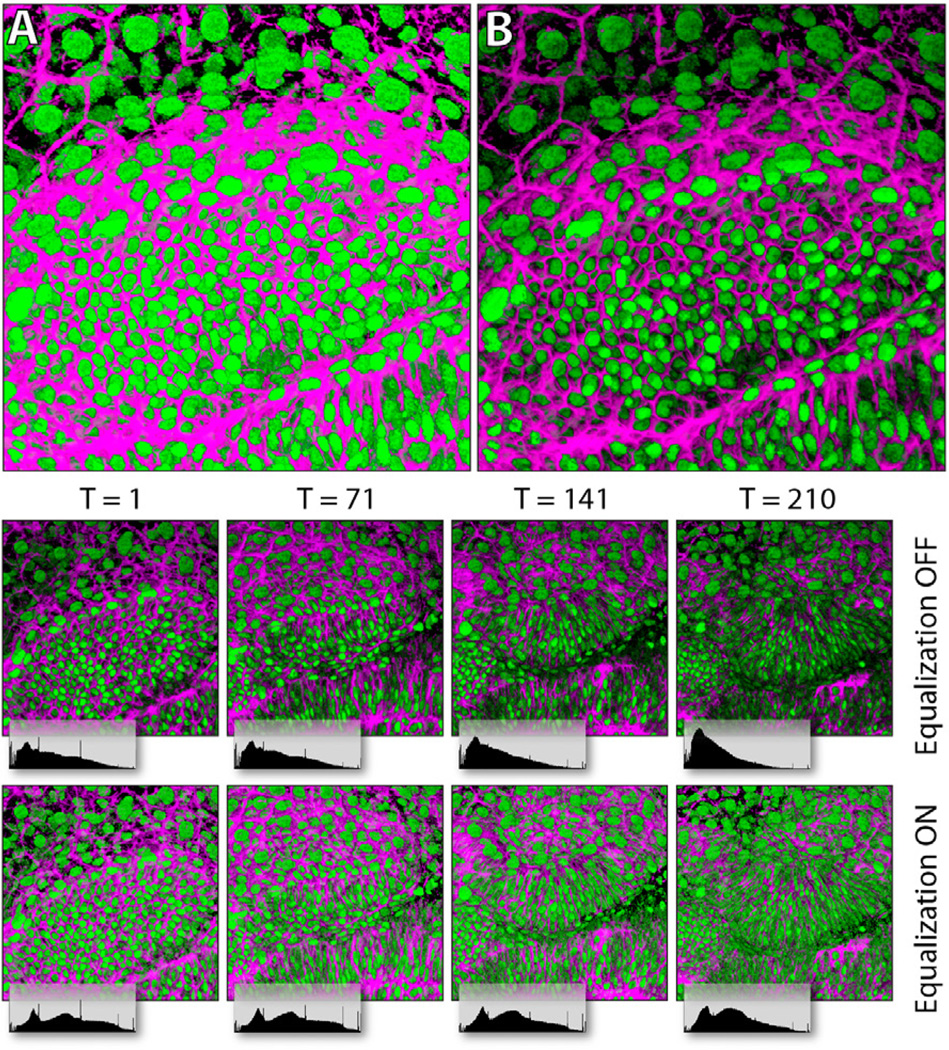
Figure 12 shows the second case study on a 4D confocal dataset, which is used to study zebrafish eye development. It has two channels of nuclei (green channel in Figure 12) and membranes (magenta channel in Figure 12), and 210 frames (12 hours of continuous imaging). Figure 12A and B compare the results before and after shadow effects are applied (the other parameters have been adjusted and are the same for A and B). Since the two channels are rendered in depth render mode, shadows created between them enhance details and the spatial relationship is more obvious. As mentioned in Section 4.1, scale-space equalization helps stabilizing brightness of 4D playbacks of confocal microscopy data. As can be seen in Figure 12, the signals of the dataset become continuously weaker during the 12-hour scanning process, due to bleaching of the fluorescent dyes. After the user turns on scale-space equalization, the brightness of the result is even through time. Biological phenomena, such as mitosis, can be visually detected from 4D playback of the dataset. Since 4D datasets are generally large in size, visualization speed becomes crucial in addition to rendering quality. We test and compare the visualization speeds of FluoRender and other two tools commonly used for 4D confocal visualization, Volocity and Imaris. The test results are shown in Figure 13, and comprised of four sub-tests. Dataset loading tests the time of loading the 3.43GB dataset. Since FluoRender only gathers file info at initial loading, the latency is negligible. Total operation time is from when application starts to when the first frame is visualized. It seems Volocity and Imaris do pre-processing, which causes considerable delay. The difference between 4D playback and 4D export is that 4D export saves the result, typically to an image sequence. However, Volocity uses lower resolution outputs for 4D playback. We calculate the speed by dividing the total number of frames (210) with playback time. FluoRender caches data using available system memory during the first playback, and the speed is considerably faster for later playback when data size is smaller than the system memory. Volocity and Imaris both require reading the whole dataset into memory for playback, which makes them impossible to use with datasets larger than the system memory. We learned from our neurobiologist users that Fluo-Render worked stably with a 4D dataset of 50GB on common PCs.
Figure 12.
Case study 2. A 4D dataset is visualized without (A) and with (B) shadow effect. The lower part of the figure compares the results with scale-space equalization off/on. The histogram of output pixel brightness is displayed under each image.
7 Conclusion
In this paper, we have discussed the application of a series of 2D image space methods as enhancements to confocal microscopy visualizations. By integrating the 2D image space methods with our existing confocal data visualization tool, FluoRender, we have implemented a full-featured visualization system for confocal microscopy data. Preliminary case studies have demonstrated the improvements after incorporating the methods presented in the paper.
Acknowledgements
This publication is based on work supported by Award No. KUS-C1-016-04, made by King Abdullah University of Science and Technology (KAUST), DOE SciDAC:VACET, NSF OCI-0906379, NIH-1R01GM098151-01. We also wish to thank the reviewers for their suggestions, and thank Chems Touati of SCI for making the demo video.
Contributor Information
Yong Wan, SCI Institute and the School of Computing, University of Utah.
Hideo Otsuna, Department of Neurobiology and Anatomy, University of Utah.
Chi-Bin Chien, Department of Neurobiology and Anatomy, University of Utah.
Charles Hansen, SCI Institute and the School of Computing, University of Utah.
References
- 1.Alexandre P, Reugels AM, Barker D, Blanc E, Clarke JDW. Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nature Neuroscience. 2010;13:673–679. doi: 10.1038/nn.2547. [DOI] [PubMed] [Google Scholar]
- 2.Bitplane AG. Imaris. 2011 http://www.bitplane.com/go/products/imaris.
- 3.Bruckner S, Gröller ME. Instant volume visualization using maximum intensity difference accumulation. Computer Graphics Forum. 2009;28(3):775–782. [Google Scholar]
- 4.Bruckner S, Rautek P, Viola I, Roberts M, Sousa MC, Gröller ME. Hybrid visibility compositing and masking for illustrative rendering. Computers and Graphics. 2010;34(4):361–369. doi: 10.1016/j.cag.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebert D, Rheingans P. Volume illustration: nonphotorealistic rendering of volume models. Proceedings of the conference on Visualization ’00; 2000. pp. 195–202. [Google Scholar]
- 6.Friedrich RW, Jacobson GA, Zhu P. Circuit neuroscience in zebrafish. Current Biology. 2010;20(8):R371–R381. doi: 10.1016/j.cub.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 7.Hauser H, Mroz L, Bischi GI, Gröller ME. Two-level volume rendering. IEEE Transactions on Visualization and Computer Graphics. 2001;7(3):242–252. [Google Scholar]
- 8.D HW, III, Westerfield M, Zon LI. The Zebrafish: Cellular and Developmental Biology, Part A. third edition. Elsevier Inc.; 2010. [DOI] [PubMed] [Google Scholar]
- 9.Jobson D, Rahman Z, Woodell G. A multiscale retinex for bridging the gap between color images and the human observation of scenes. IEEE Transactions on Image Processing. 1997;6(7):965–976. doi: 10.1109/83.597272. [DOI] [PubMed] [Google Scholar]
- 10.Kitware Inc. Insight Toolkit. 2011 http://www.itk.org/
- 11.Kitware Inc. Visualization Toolkit. 2011 http://www.vtk.org/
- 12.Kniss J, Kindlmann G, Hansen C. Multidimensional transfer functions for interactive volume rendering. IEEE Transactions on Visualization and Computer Graphics. 2002;8(3):270–285. [Google Scholar]
- 13.Kuhn G, Oliveira M, Fernandes L. An efficient naturalness-preserving image-recoloring method for dichromats. IEEE Transactions on Visualization and Computer Graphics. 2008;14(6):1747–1754. doi: 10.1109/TVCG.2008.112. [DOI] [PubMed] [Google Scholar]
- 14.Luft T, Colditz C, Deussen O. Image enhancement by un-sharp masking the depth buffer. ACM Transactions on Graphics. 2006;25(3):1206–1213. [Google Scholar]
- 15.Machado GM, Oliveira MM, Fernandes LAF. A physiologically-based model for simulation of color vision deficiency. IEEE Transactions on Visualization and Computer Graphics. 2009;15(6):1291–1298. doi: 10.1109/TVCG.2009.113. [DOI] [PubMed] [Google Scholar]
- 16.Max N. Optical models for direct volume rendering. IEEE Transactions on Visualization and Computer Graphics. 1995;1(2):99–108. doi: 10.1109/TVCG.2020.3030394. [DOI] [PubMed] [Google Scholar]
- 17.PerkinElmer Inc. Volocity 3D Image Analysis Software. 2011 http://www.perkinelmer.com/pages/020/cellularimaging/products/volocity.xhtml.
- 18.Reinhard E, Ward G, Pattanaik S, Debevec P. High Dynamic Range Imaging: Acuisition, Display and Image-Based Lighting. first edition. Elsevier Inc.; 2006. [Google Scholar]
- 19.Tikhonova A, Correa C, Ma K-L. Explorable images for visualizing volume data; IEEE Pacific Visualization Symposium (PacificVis); 2010. pp. 177–184. [Google Scholar]
- 20.Tikhonova A, Correa C, Ma K-L. Visualization by proxy: Anovel framework for deferred interaction with volume data. IEEE Transactions on Visualization and Computer Graphics. 2010;16(6):1551–1559. doi: 10.1109/TVCG.2010.215. [DOI] [PubMed] [Google Scholar]
- 21.Visage Imaging. Amira. 2011 http://www.amiravis.com.
- 22.Wan Y, Hansen C. Fast volumetric data exploration with importance-based accumulated transparency modulation; IEEE/EG International Symposium on Volume Graphics; 2010. pp. 61–68. [Google Scholar]
- 23.Wan Y, Otsuna H, Chien C-B, Hansen C. An interactive visualization tool for multi-channel confocal microscopy data in neurobiology research. IEEE Transactions on Visualization and Computer Graphics. 2009;15(6):1489–1496. doi: 10.1109/TVCG.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Giesen J, McDonnell K, Zolliker P, Mueller K. Color design for illustrative visualization. IEEE Transactions on Visualization and Computer Graphics. 2008;14(6):1739–1754. doi: 10.1109/TVCG.2008.118. [DOI] [PubMed] [Google Scholar]